Insights into the Genital Microbiota of Women Who Experienced Fetal Death in Utero
Abstract
1. Introduction
2. Methods
2.1. Participants
2.2. Ethics Approval
2.3. Sample Collection
2.4. DNA Extraction, Amplification and Sequencing
2.5. Data Analyses
3. Results
3.1. Characteristics of Participants
3.2. Composition of the Microbiomes
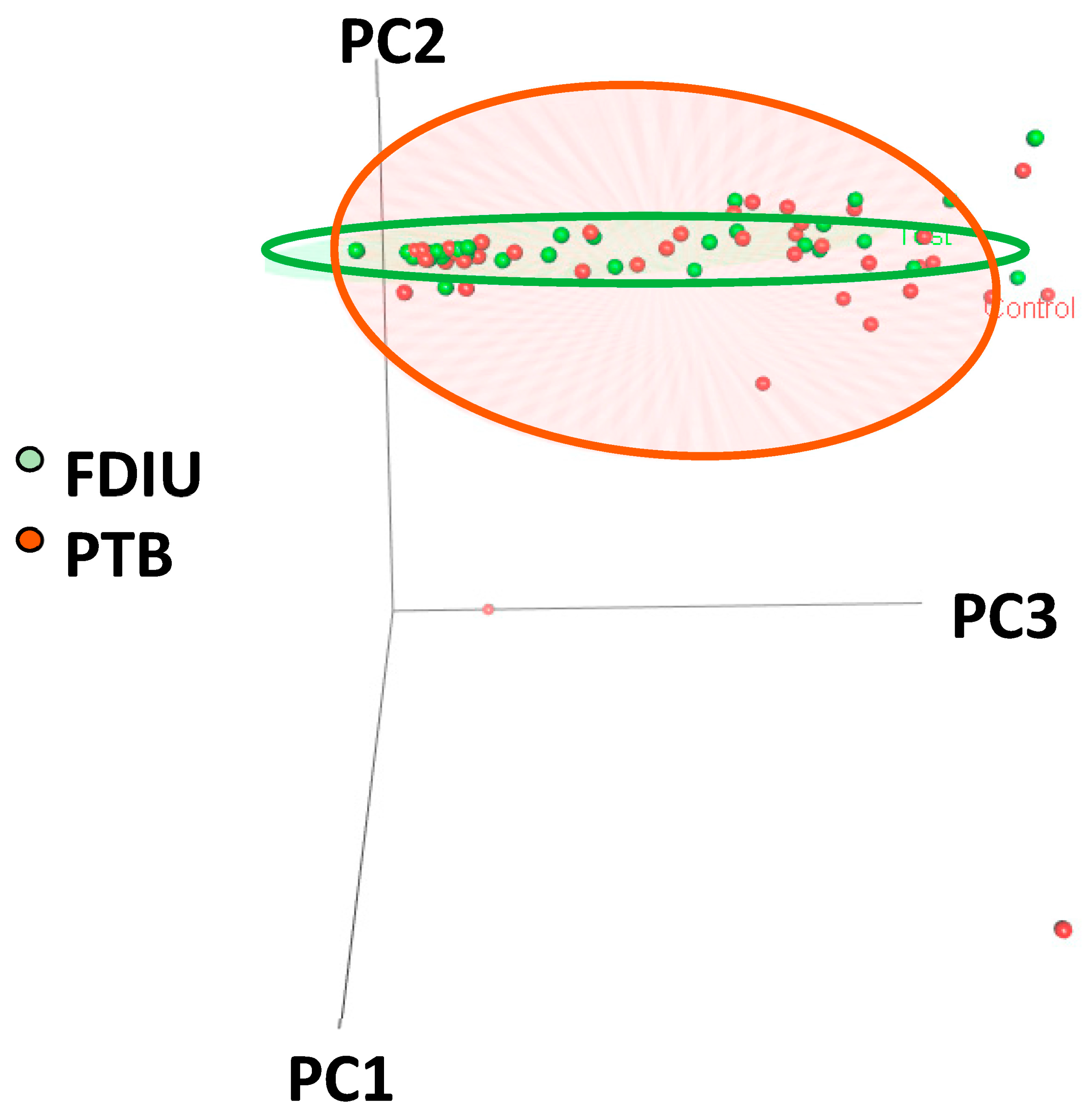
3.3. Composition and Diversity of the PTB and FDIU Genital Microbiomes
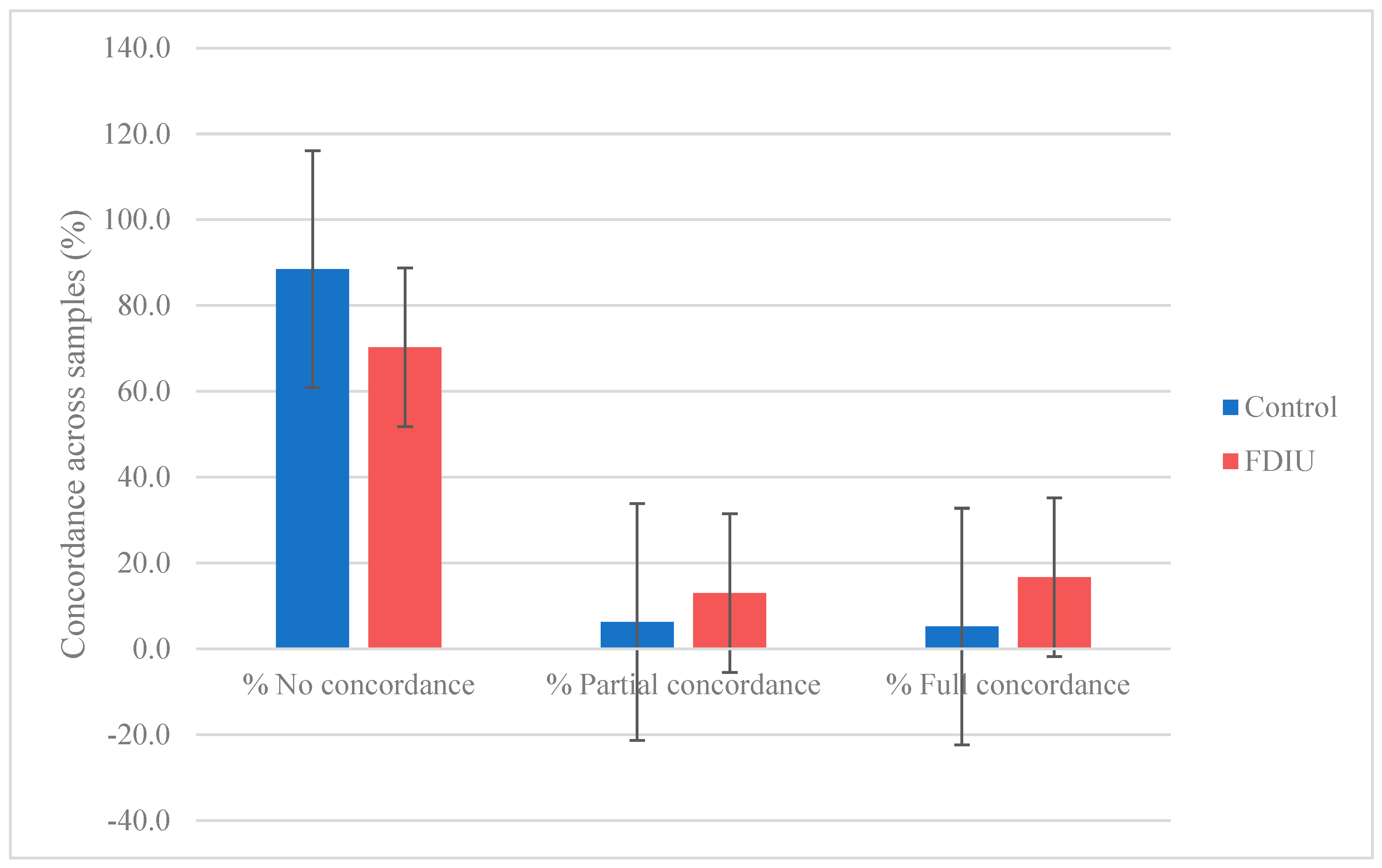
3.4. Relative Abundance and Frequency of Taxa
3.5. Lactobacillus Relative Abundances
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flenady, V.J.; Middleton, P.; Wallace, E.M.; Morris, J.; Gordon, A.; Boyle, F.M.; Homer, C.S.E.; Henry, S.; Brezler, L.; Wojcieszek, A.M.; et al. Stillbirth in Australia 1: The road to now: Two decades of stillbirth research and advocacy in Australia. Women Birth 2020, 33, 506–513. [Google Scholar] [CrossRef]
- Lawn, J.J.E.; Blencowe, H.; Waiswa, P.; Amouzou, A.; Mathers, C.; Hogan, D.; Flenady, V.; Frøen, J.F.; Qureshi, Z.U.; Calderwood, C.; et al. Stillbirths: Rates, risk factors, and acceleration towards 2030. Lancet 2016, 387, 587–603. [Google Scholar] [CrossRef] [PubMed]
- UN Inter-Agency Group for Child Mortality Estimation. A Neglected Tragedy: The Global Burden of Stillbirth Report 2020; UNICEF: New York, NY, USA, 2020; Available online: https://www.unicef.org/media/84851/file/UN-IGME-the-global-burden-of-stillbirths-2020.pdf (accessed on 30 July 2022).
- Hug, L.; You, D.; Blencowe, H.; Mishra, A.; Wang, Z.; Fix, M.J.; Wakefield, J.; Moran, A.C.; Gaigbe-Togbe, V.; Suzuki, E.; et al. Global, regional, and national estimates and trends in stillbirths from 2000 to 2019: A systematic assessment. Lancet 2021, 398, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G. Stillbirth statistics in Australia. Research Paper Series. Parliament of Australia. 2022. Available online: https://www.aph.gov.au/About_Parliament/Parliamentary_Departments/Parliamentary_Library/pubs/rp/rp2122/StillbirthStatisticsAustralia (accessed on 30 September 2022).
- Eunice Kennedy Shriver National Institute of Child Health and Human Development, What Are Possible Causes of Stillbirth? Available online: https://www.nichd.nih.gov/health/topics/stillbirth/topicinfo/causes# (accessed on 22 December 2022).
- Megli, C.J.; Coyne, C.B. Infections at the maternal–fetal interface: An overview of pathogenesis and defence. Nat. Rev. Microbiol. 2022, 20, 67–82. [Google Scholar] [CrossRef]
- Stillbirth, Cleveland Clinic. 2023. Available online: https://my.clevelandclinic.org/health/diseases/9685-stillbirth (accessed on 15 February 2023).
- Fettweis, J.M.; Brooks, J.P.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss, J.F. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272–2282. [Google Scholar] [CrossRef]
- Dinsdale, N.K.; Castaño-Rodriguez, N.; Quinlivan, J.A.; Mendz, G.L. Comparison of the genital microbiomes of pregnant aboriginal and non-aboriginal women. Front. Cell. Infect. Microbiol. 2020, 10, 523764. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Y.; Chen, T.; Li, R. The female vaginal microbiome in health and bacterial vaginosis. Front. Cell. Infect. Microbiol. 2021, 11, 631972. [Google Scholar] [CrossRef]
- Saraf, V.S.; Sheikh, S.A.; Ahmad, A.; Gillevet, P.M.; Bokhari, H.; Javed, S. Vaginal microbiome: Normalcy vs. dysbiosis. Arch. Microbiol. 2021, 203, 3793–3802. [Google Scholar] [CrossRef]
- Pekmezovic, M.; Mogavero, S.; Naglik, J.R.; Hube, B. Host-pathogen interactions during female genital tract infections. Trends Microbiol. 2019, 27, 982–996. [Google Scholar] [CrossRef]
- Shahid, M.; Quinlivan, J.A.; Peek, M.; Castaño-Rodriguez, N.; Mendz, G.L. Is there an association between the vaginal microbiome and first trimester miscarriage? A prospective observational study. J. Obstet. Gynaecol. Res. 2022, 48, 119–128. [Google Scholar] [CrossRef]
- Jiao, X.; Zhang, L.; Du, D.; Wang, L.; Song, Q.; Liu, S. Alteration of vaginal microbiota in patients with recurrent miscarriage. J. Obstet. Gynaecol. 2022, 42, 248–255. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.; Gao, J.; Wu, M.; Li, C.; Wang, Z.; Huang, N.; Cui, L.; Du, M.; Ying, C. Characterization of vaginal microbiota in women with recurrent spontaneous abortion that can be modified by drug treatment. Front. Cell. Infect. Microbiol. 2021, 11, 680643. [Google Scholar] [CrossRef] [PubMed]
- Gudnadottir, U.; Debelius, J.W.; Du, J.; Hugerth, L.W.; Danielsson, H.; Schuppe-Koistinen, I.; Fransson, E.; Brusselaers, N. The vaginal microbiome and the risk of preterm birth: A systematic review and network meta-analysis. Sci. Rep. 2022, 12, 7926. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The vaginal microbiome and preterm birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Seale, A.C.; Bianchi-Jassir, F.; Russell, N.J.; Kohli-Lynch, M.; Tann, C.J.; Hall, J.; Madrid, L.; Blencowe, H.; Cousens, S.; Baker, C.J.; et al. Estimates of the burden of Group B streptococcal disease worldwide for pregnant women, stillbirths, and children. Clin. Infect. Dis. 2017, 65 (Suppl. S2), S200–S219. [Google Scholar] [CrossRef]
- De Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Stinson, L.; Boyce, M.C.; Payne, M.S.; Keelan, J.A. The not-so-sterile womb: Evidence that the human fetus is exposed to bacteria prior to birth. Front. Microbiol. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Klopp, J.; Ferretti, P.; Meyer, C.U.; Hilbert, K.; Haiß, A.; Marißen, J.; Henneke, P.; Hudalla, H.; Pirr, S.; Viemann, D.; et al. Meconium microbiome of very preterm infants across Germany. mSphere 2022, 7, e00808-21. [Google Scholar] [CrossRef]
- Urushiyama, D.; Suda, W.; Ohnishi, E.; Araki, R.; Kiyoshima, C.; Kurakazu, M.; Sanui, A.; Yotsumoto, F.; Murata, M.; Nabeshima, K.; et al. Microbiome profile of the amniotic fluid as a predictive biomarker of perinatal outcome. Sci. Rep. 2017, 7, 12171. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Huang, Y.; Li, D.; Hu, D.; Jin, C.; Sadykova, A.; Cai, W.; Liao, C.; Pan, S. Amniotic fluid and vaginal microbiota in pregnant women with gestational diabetes mellitus by metagenomics. Med. Microecol. 2023, 15, 100074. [Google Scholar] [CrossRef]
- Brosius Lutz, A.; Al-Nasiry, S.; Kramer, B.W.; Mueller, M. Understanding host-pathogen interactions in acute chorioamnionitis through the use of animal models. Front. Cell. Infect. Microbiol. 2021, 11, 709309. [Google Scholar] [CrossRef]
- Dos Anjos Borges, L.G.; Pastuschek, J.; Heimann, Y.; Dawczynski, K.; PEONS Study Group; Schleußner, E.; Pieper, D.H.; Zöllkau, J. Vaginal and neonatal microbiota in pregnant women with preterm premature rupture of membranes and consecutive early onset neonatal sepsis. BMC Med. 2023, 21, 92. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org/ (accessed on 10 July 2022).
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Serrano, M.G.; Parikh, H.I.; Brooks, J.P.; Edwards, D.J.; Arodz, T.J.; Edupuganti, L.; Huang, B.; Girerd, P.H.; Bokhari, Y.A.; Bradley, S.P.; et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019, 5, 1001–1011. [Google Scholar] [CrossRef]
- Ncib, K.; Bahia, W.; Leban, N.; Mahdhi, A.; Trifa, F.; Mzoughi, R.; Haddad, A.; Jabeur, C.; Donders, G. Microbial diversity and pathogenic properties of microbiota associated with aerobic vaginitis in women with recurrent pregnancy loss. Diagnostics 2022, 12, 2444. [Google Scholar] [CrossRef]
- Oerlemans, E.F.M.; Wuyts, S.; Bellen, G.; Wittouck, S.; De Boeck, I.; Ruban, K.; Allonsius, C.N.; van den Broek, M.F.L.; Donders, G.G.G.; Lebeer, S. The dwindling microbiota of aerobic vaginitis, an inflammatory state enriched in pathobionts with limited TLR stimulation. Diagnostics 2020, 10, 879. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Shim, S.S.; Park, H.J.; Cha, D.H. Influence of Maternal Microbiome and Inflammatory Response in Preterm Birth: Recent Aspects of the Prevention of Preterm Birth. Microbiol. Res. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Doster, R.S.; Kirk, L.A.; Tetz, L.M.; Rogers, L.M.; Aronoff, D.M.; Gaddy, J.A. Staphylococcus aureus infection of human gestational membranes induces bacterial Bbofilm formation and host production of cytokines. J. Infect. Dis. 2017, 215, 653–657. [Google Scholar]
- Shadbolt, R.; We, M.L.S.; Kohan, R.; Porter, M.; Athalye-Jape, G.; Nathan, E.; Shrestha, D.; Strunk, T. Neonatal Staphylococcus aureus sepsis: A 20-year Western Australian experience. J. Perinatol. 2022, 42, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.P.; Cheah, F.C.; Wong, K.K.; Shah, S.A.; Phon, S.E.; Ng, B.K.; Lim, S.P.; Khong, T.Y.; Tan, G.C. Gardnerella vaginalis infection in pregnancy: Effects on placental development and neonatal outcomes. Placenta 2022, 120, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Serrano, M.G.; Fettweis, J.M.; Basta, P.; Rosen, E.; Ludwig, K.; Sorgen, A.A.; Blakley, I.C.; Wu, M.C.; Nancy Dole, N. Race, the vaginal microbiome, and spontaneous preterm birth. mSystems 2022, 7, 3. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Krypotou, E.; Scortti, M. Listeria placental infection. mBio 2017, 8, 3. [Google Scholar] [CrossRef]
- Borges, S.F.; Silva, J.G.L.; Teixeira, P.C.M. Survival and biofilm formation of Listeria monocytogenes in simulated vaginal fluid: Influence of pH and strain origin. FEMS Immunol. Med. Microbiol. 2011, 62, 315–320. [Google Scholar]
- Majewska, A.; Kierzkowska, M.; Kawecki, D. What we actually know about the pathogenicity of Bacteroides pyogenes. Med. Microbiol. Immunol. 2021, 210, 157–163. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Quinlivan, J.A.; Mendz, G.L. Bacteroides and Hafnia infections associated with chorioamnionitis and preterm birth. J. Clin. Gynaecol. Obstet. 2014, 3, 76–79. [Google Scholar] [CrossRef][Green Version]
- Souto Rodrigues, P.; de Lima, L.P.; de Mesquita Lorete-Terra, A.; Seabra, S.H.; Bittencourt Dias Vieira, J.M. Bacteroides fragilis induces conformational alterations in Trichomonas vaginalis ultrastructure in an in vitro interaction. J. Trop. Pathol. 2020, 49, 11–20. [Google Scholar] [CrossRef]
- Aguinaga, M.; Valdespino, Y.; Medina, D.; Espino y Sosa, S.; Sevilla, R.; Miranda, O.; Acevedo, S.; Monroy, I.E.; Helguera, A.C.; Pérez, J.; et al. Causal analysis of fetal death in high-risk pregnancies. J. Perinat. Med. 2021, 49, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Vander Haar, E.L.; Wu, G.; Gyamfi-Bannerman, C.; Thomas, C.; Wapner, R.J.; Reddy, U.M.; Zhao, L.; Silver, R.M.; Goldenberg, R.L.; Han, Y.W. Microbial analysis of umbilical cord blood reveals novel pathogens associated with stillbirth and early preterm birth. mBio 2022, 13, e0203622. [Google Scholar] [CrossRef]
| PTB Group | FDIU Group | |
|---|---|---|
| Number of participants | 11 | 11 |
| Mean maternal age (years and range) | 29.1 (18–38) | 30.2 (15–42) |
| Mean gestational age at delivery (weeks and range) | 33.5 (30–36) | 24.6 (23–31) |
| Parity | ||
| 1 | 5 (46%) | 6 (55%) |
| 2 | 4 (36%) | 2 (18%) |
| ≥3 | 2 (18%) | 3 (27%) |
| Infection during pregnancy * | 5 (46%) | 7 (64%) |
| Complications | ||
| None | 5 (46%) | 4 (36%) |
| Pre-eclampsia | 1 (1%) | 1 (1%) |
| Preterm premature rupture of membranes | 1 (1%) | 1 (1%) |
| Other ** | 4 (36%) | 5 (46%) |
| Index | PTB Controls | FDIU Cases | ||
|---|---|---|---|---|
| Vagina | Amniotic | Vagina | Amniotic | |
| S | 210 | 379 | 194 | 353 |
| T | 38 | 147 | 44 | 111 |
| d | 3.637 | 4.990 | 3.784 | 4.709 |
| J | 0.322 | 0.599 | 0.613 | 0.643 |
| H | 1.172 | 2.991 | 2.319 | 3.028 |
| Taxon | PTB | FDIU | ||
|---|---|---|---|---|
| Vagina | Amniotic | Vagina | Amniotic | |
| E. coli/Shigella | --- | 1/(++); 7/(+) | 6/(+) | 1/(+++); 5/(++); 2/(+) |
| Staphylococcus | 2/(+++), 2/(+) | 1/(++): 7/(+) | 1/(+++), 1/(++); 4/(+) | 2/(+++); 8/(+) |
| Gardnerella | 1/(+++); 1/(+) | 2/(+) | 4/(++) | 2/(+++) |
| Cloacibacterium | 2/(+) | 3/(++); 5/(+) | 1/(+) | 8/(+) |
| Enterococcus | 1/(+) | 3/(+) | 2/(++) | 1/(+++); 3(+) |
| Anoxybacillus | 1/(+) | 10/(+) | 2/(+) | 10/(+) |
| Streptococcus | 3/(+) | 10/(+) | 3/(+) | 9/(+) |
| Tepidomonas | 2/(+) | 10/(+) | 1/(+) | 9/(+) |
| Paracoccus | 1/(+) | 6/(++); 3/(+) | 1/(+) | 8/(+) |
| Listeria | --- | --- | 2/(+++) | 2/(+++) |
| Acinetobacter | 2/(+) | 9/(+) | 3/(+) | 7/(+) |
| Kocuria | 1/(+) | 9/(+) | 1/(+) | 1/(++); 6(+) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holliday, M.; Uddipto, K.; Castillo, G.; Vera, L.E.; Quinlivan, J.A.; Mendz, G.L. Insights into the Genital Microbiota of Women Who Experienced Fetal Death in Utero. Microorganisms 2023, 11, 1877. https://doi.org/10.3390/microorganisms11081877
Holliday M, Uddipto K, Castillo G, Vera LE, Quinlivan JA, Mendz GL. Insights into the Genital Microbiota of Women Who Experienced Fetal Death in Utero. Microorganisms. 2023; 11(8):1877. https://doi.org/10.3390/microorganisms11081877
Chicago/Turabian StyleHolliday, Mira, Kumar Uddipto, Gerardo Castillo, Luz Estela Vera, Julie A. Quinlivan, and George L. Mendz. 2023. "Insights into the Genital Microbiota of Women Who Experienced Fetal Death in Utero" Microorganisms 11, no. 8: 1877. https://doi.org/10.3390/microorganisms11081877
APA StyleHolliday, M., Uddipto, K., Castillo, G., Vera, L. E., Quinlivan, J. A., & Mendz, G. L. (2023). Insights into the Genital Microbiota of Women Who Experienced Fetal Death in Utero. Microorganisms, 11(8), 1877. https://doi.org/10.3390/microorganisms11081877




