Abstract
A new member of the DegP-type periplasmic serine endoproteases of the S1C family from the marine bacterium Cobetia amphilecti KMM 296 (CamSP) was expressed in Escherichia coli cells. The calculated molecular weight, number of amino acids, and isoelectric point (pI) of the mature protein CamSP are 69.957 kDa, 666, and 4.84, respectively. The proteolytic activity of the purified recombinant protease CamSP was 2369.4 and 1550.9 U/mg with the use of 1% bovine serum albumin (BSA) and casein as the substrates, respectively. The enzyme CamSP exhibited maximum activity at pH 6.0–6.2, while it was stable over a wide pH range from 5.8 to 8.5. The optimal temperature for the CamSP protease activity was 50 °C. The enzyme required NaCl or KCl at concentrations of 0.3 and 0.5 M, respectively, for its maximum activity. The Michaelis constant (Km) and Vmax for BSA were determined to be 41.7 µg/mL and 0.036 µg/mL min−1, respectively. The metal ions Zn2+, Cu2+, Mn2+, Li2+, Mg2+, and Ca2+ slightly activated CamSP, while the addition of CoCl2 to the incubation mixture resulted in a twofold increase in its protease activity. Ethanol, isopropanol, glycerol, and Triton-X-100 increased the activity of CamSP from two- to four-times. The protease CamSP effectively degraded the wheat flour proteins but had no proteolytic activity towards soybean, corn, and the synthetic substrates, α-benzoyl-Arg-p-nitroanilide (BAPNA) and N-Succinyl-L-alanyl-L-alanyl-L-prolyl-L-phenylalanine 4-nitroanilide (SAPNA).
1. Introduction
Proteases (peptidases, proteolytic enzymes) belong to a large group of hydrolases with different structures and biological functions that catalyze the cleavage of peptide bonds in proteins. They are widely distributed in nature, both in microorganisms and in animal and plant cells [1]. Proteases control the activation, synthesis, and turnover of proteins for various physiological processes in cells [2,3,4]. Depending on their localization in the cell, and the arrangement of amino acid (aa) residues in the active center and mechanism of action, proteases are classified into exo- or endoproteases belonging to different families, such as serine, aspartic, cysteine, and many others [5,6,7].
The representatives of each structural family are described in detail in the peptidase database MEROPS (http://merops.sanger.ac.uk/, accessed on 20 April 2023). Each peptidase is assigned to a family based on statistically significant similarities in the amino acid sequence, and families that are thought to be homologous grouped together in a Clan by indexes [5]. The S1C subfamily of DegP-type periplasmic serine endoproteases is a group of proteins that have a chymotrypsin-like fold, with a single domain, and play a dual role in bacterial cells [5,8]. They act as chaperones at low temperatures, helping to fold and stabilize other proteins, but switch to the peptidase function at high temperatures, degrading unfolded or misfolded proteins that accumulate in the periplasm following heat shock or other stress conditions [9,10,11,12]. These enzymes are essential for bacterial survival and virulence at high temperatures (above 42 °C), which generate misfolded proteins and participate in the biogenesis of partially folded outer-membrane proteins (OMPs). Therefore, the bacterial DegP may be used as the target in both drug discovery and infectious disease treatment [13,14]. In mammals, the dysfunction of protease HtrA2, which is structurally similar to the bacterial DegP-type proteases, is associated with cancer and Alzheimer’s disease [8,15]. HtrA/DegP/Q-type enzymes are known to be allosterically regulated and have the catalytic triad of His-Asp-Ser and two PDZ domains that mediate substrate recognition and oligomerization, with a preference for cleaving peptide bonds between hydrophobic residues, such as Val-Val or Ile-Xaa [9,11,12,13,14,16]. Allosteric effects may be observed between the individual domains of monomeric proteins DegP or between the protomers of the oligomeric protein complexes. Moreover, allosteric switching between their active and inactive conformations is frequently accompanied by changes in their oligomeric states (up to 24-mers) [10].
Proteases have been isolated and characterized from various sources, but microorganisms are the most preferred source for industrial purposes due to their biochemical diversity, rapid growth in a limited space, and a simple nutrition medium, as well as established genetic methods [17,18]. Bacterial proteases are easily produced in large quantities and are most often thermostable and active over a wide pH range, making them available for industrial use. The enzymes isolated from the microorganisms Bacillus sp. [19,20,21,22], Aspergillus sp. [23], and some other bacteria [24] are among the proteases used in various industries, such as leather, textile, pharmaceutical, detergent, and food, including baking. Acidic proteases are obtained from fungi, while neutral proteinases are mainly of plant origin. The isolation of both acidic and neutral proteases from fungi and plants is relatively laborious and uneconomical, while alkaline proteases obtained from different types of bacteria are in great demand in industry [17].
Efforts are currently underway to find and isolate new bacteria from poorly understood habitats that can produce enzymes with unique properties suitable for industrial applications [25]. The World Ocean can become such a unique source of producers of new proteases. Much of the deep-sea environment is subjected to high pressure, high salt concentration, and low temperature, and the organisms living in these conditions have adapted to them. Great interest in the enzymes isolated from marine microorganisms is associated with their activity and stability under extreme conditions due to the habitat of these bacteria [26].
The present study presents data on the recombinant production and purification of the new neutral DegP-type protease CamSP from the marine bacterium C. amphilecti KMM 296 (Collection of Marine Microorganisms, G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch, the Russian Academy of Sciences (PIBOC FEB RAS)), isolated from the coelomic fluid of the mussel Crenomytilus grayanus, and the physical–chemical and catalytic properties of the enzyme are described.
2. Materials and Methods
2.1. Reagents
High-purity-grade reagents from Merck (Munich, Germany), Sigma (OOO Sigma-Aldrich Rus, Moscow, Russia), and Helicon (Moscow, Russia) were used. Kits for DNA extraction, restriction, and ligation, oligonucleotides, and Taq polymerase were purchased from Evrogen (Moscow, Russia) and Thermo Fisher Scientific RU (Moscow, Khimki, Russia); kanamycin was produced by Sintez (Moscow, Russia). Yeast extract, bactoagar, tryptone, and peptone were purchased from “Helicon” and “Dia-M” (Moscow, Russia). DNA and protein molecular weight markers were purchased from BioRad (Hercules, CA, USA).
2.2. Construction of the Plasmid pET40 CamSP
The recombinant plasmid pET40 CamSP was constructed by inserting into the NcoI/XhoI region of the plasmid pET-40b(+) (Thermo Fisher Scientific-RU, Moscow, Khimki, Russia) the full-length protease-encoding gene, synthesized by polymerase chain reaction (PCR) without signal peptide, using the genomic DNA of C. amphilecti KMM 296 (Collection of Marine Microorganisms, PIBOC FEB RAS). The reaction mixture contained per 10 µL: 1 µL–10× Encyclo buffer, 0.2 µL–50× Encyclo polymerase mix (Encyclo PCR kit; Evrogen, Moscow, Russia), 0.2 µL–50× dNTP mix (10 mM each), forward and reverse primers (1 µL of 5 µM each), and 1 µL–20 ng DNA. The amplification process consisted of 38 PCR cycles (15 s–95 °C, 1.4 min–72 °C). After amplification, the PCR product was purified via electrophoresis in 1% agarose gel.
The PCR product (1 µg) was treated with the restriction enzymes NcoI and XhoI in an optimal buffer (Thermo Fisher Scientific RU) for 3 h at 37 °C, which were removed from the reaction mixture using a phenol treatment (1:1). To the aqueous fraction containing the PCR product, 1/10 volume of 0.3 M sodium acetate (pH 5.2) and 1/2 volume of isopropyl alcohol were added, then incubated at −20 °C for 30 min. After centrifugation at 14,000× g rpm for 20 min, the precipitate was washed with 75% ethanol and then dried at a room temperature. The precipitate was dissolved in 20 µL of deionized water. A total of 2 µg of the pET-40b(+) DNA (Thermo Fisher Scientific-RU) was treated with NcoI and XhoI, as described above. The resulting fragment of the CamSP gene and the NcoI/XhoI part of pET-40b(+) were ligated in 50 μL of ligation buffer according to the instructions (Novagen). Then, 10 μL of the reaction mixture was used to transform competent E. coli Rosetta (DE3) cells. The transformants were grown on Luria-Bertani (LB) agar containing 50 μg/mL kanamycin. After incubation for 16 h at 37 °C, the clones were screened, and the target plasmid DNA was isolated and sequenced.
2.3. Production of the Recombinant Protease CamSP
The recombinant strain of E. coli Rosetta (DE3) was grown in 25 mL of the liquid LB medium containing 25 mg/mL of kanamycin at 200 rpm for 16 h at 37 °C. Then, the cells were placed in a fresh LB medium (1 L) containing kanamycin at the concentration of 25 mg/mL and incubated at 37 °C on a shaker at 200 rpm until the optical density at 600 nm was 0.6–0.8. After that, 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce the CamSP gene expression, and incubation was continued at 37 °C for 6 h at 200 rpm. Cells were pelleted via centrifugation at 4000× g rpm for 15 min at 8 °C, suspended in 35 mL of 25 mM Tris-HCl buffer (pH 7.5), and subjected to an ultrasonic treatment at a frequency of 22 kHz and 0–4 °C at intervals 30 s to clarify the suspension. The suspension was centrifuged at 11,000× g rpm for 30 min at 8 °C, the precipitate was discarded, and the proteolytic activity of CamSP was determined in the resulting extract (described below). Protein concentration was measured according to the Bradford method [27] using bovine serum albumin (BSA) as a reference.
2.4. Isolation and Purification of the Recombinant Protease CamSP
For the CamSP isolation, the resulting supernatant after ultrasonic treatment was applied to a 25 × 3.2 cm Ni-IMAC-Sepharose column (Cytiva (GE Healthcare) Life Sciences, Buckinghamshire, UK), equilibrated with 50 mM Tris-HCl, pH 7.5 (buffer A), and washed with five volumes of the same buffer. The recombinant protein was eluted with a linear gradient of 0–0.5 M imidazole in 50 mM Tris-HCl buffer, pH 7.5, and 0.5 M NaCl (6 volume of the column) at a rate of 1.3 mL min−1. The fraction containing CamSP was purified on a 10 × 1.4 cm Source 15 Q column (Cytiva (GE Healthcare) Life Sciences) and equilibrated by buffer A with 1 mM MgCl2 (buffer B). The protein was eluted with a linear gradient of 0–0.5 M NaCl in buffer B. Ion exchange chromatography was performed at 1 mL min−1; the volume of fractions was 1 mL. The fractions containing CamSP were collected and treated with enterokinase at a final concentration of 1 unit per 1 mg of protein for 22 h at 25 °C. After the enterokinase treatment, the protein solution was applied to a HisTrap™ High-Performance column (Cytiva (GE Healthcare) Life Sciences) and pre-equilibrated with buffer B. The recombinant protein was eluted with 10 volumes of buffer B by a linear gradient of 0–0.5 M imidazole and 0.5 M NaCl at a rate of 0.5 mL min−1. The fractions containing CamSP were collected and chromatographed using a mono-Q HR (4 × 0.8 cm) column (Cytiva (GE Healthcare) Life Sciences, UK), equilibrated with buffer B, and then washed with 10 column volumes of buffer B. The target protein was eluted by linear gradient of 0–0.5 M NaCl in buffer B at a rate of 0.5 mL min−1 as 1 mL fractions. The purified CamSP preparation was used to study the physical–chemical properties and substrate specificity.
2.5. Determination of the CamSP Proteolytic Activity
The protease activity was determined using the method described earlier [28], using 1% casein or bovine serum albumin (BSA) as the substrates (Sigma-Aldrich, St. Louis, MO, USA). The enzyme solution, diluted to the required concentration, was mixed with 0.5 mL of 50 mM Tris-HCl buffer, pH 7.5, containing 1% casein (or BSA) and incubated for 20 min at 50 °C. The reaction was stopped by adding cold 10% trichloroacetic acid (TCA). The acid-soluble product was determined spectrophotometrically at a wavelength of 280 nm. The standard curve was built using the solutions of tyrosine at a concentration of 0–100 µg L−1. One unit (U) of protease activity was defined as the amount of the enzyme that releases 1 µg of tyrosine in 1 min under the experimental conditions used. Protease activity is the average of two measurements taken in triplicate. The difference between the values did not exceed 5%.
2.6. Influence of pH on the CamSP Proteolytic Activity
The optimal pH was determined in a range of pH 3.2–10.0, using 1% casein as a substrate. The following buffer solutions were used: 100 mM sodium acetate (pH 3.2–6.2), 100 mM Tris-HCl (pH 6.0–10.0), glycine-NaOH (pH 8.0–10.0), and phosphate buffer (5.2–8.2).
2.7. Determination of the CamSP Thermal Stability and Optimum Temperature
The effect of temperature on the CamSP stability and the optimum temperature for manifestation of its proteolytic activity were determined by incubating the standard incubation mixture for the proteolytic activity measurement at 15 to 65 °C at intervals of 5 to 10 °C. The proteolytic activity was determined using the standard method described above.
2.8. Effect of Divalent Metal Ions and Inhibitors on the CamSP Proteolytic Activity
The effect of divalent metal ions on the CamSP proteolytic activity was evaluated using the standard method for determining the proteolytic activity by the addition of Mg2+, Ca2+, Mn2+, Zn2+, Co2+, Ni2+, Cu2+, and Li2+ to the incubation mixture at a final concentration of 2–10 mM. The effects of chelating agents (EGTA, EDTA), solvents, and detergents on the CamSP proteolytic activity were evaluated by the addition of PMSF, triton-X-100, SDS, ethanol, isopropanol, and glycerol to the incubation mixture in various concentrations. The incubation mixture without cations, chelating agents, solvents, and detergents was used as the control.
2.9. Effect of NaCl and KCl on the CamSP Proteolytic Activity
The salt tolerance was studied by adding NaCl and KCl to the standard incubation mixture at a concentration of 0–1.5 M. The CamSP proteolytic activity was determined as described above.
2.10. Determination of the CamSP Molecular Weight
The molecular weight of CamSP was determined using polyacrylamide gel electrophoresis under denaturing conditions (SDS-PAGE, polyacrylamide gel electrophoresis with sodium dodecyl sulfate) according to the Laemmli method [29] and gel filtration on a calibrated Superdex 200 PG column (105 × 2 cm) (GE Healthcare) Life Sciences, Buckinghamshire, UK).
2.11. Determination of the CamSP Substrate Specificity
The substrate proteolytic specificity of CamSP was determined using the synthetic chromogenic substrates α-benzoyl-Arg-p-nitroanilide (BAPNA) and N-succinyl-L-alanyl-L-alanyl-L-prolyl-L-phenylalanine 4-nitroanilide (SAPNA) (Sigma-Aldrich, St. Louis, MO, USA) and natural substrates, such as casein, bovine serum albumin (BSA), as well as corn, wheat, and soybeans, purchased at a domestic market.
The standard 25 mM solutions of BAPNA and SAPNA were prepared in 5% dimethyl sulfoxide (DMSO) immediately before the experiment. Protease activity was analyzed by mixing 0.5 mg of the purified enzyme CamSP and 1 mL of assay buffer containing one of the substrates (1 mM BAPNA or 20 mM SAPNA with 50 mM Tris-HCl, pH 7.5). After the 30 min incubation at 50 °C, the reaction was stopped by adding 1 mL of cold 10% trichloroacetic acid (TCA) and kept at a room temperature for 15 min. The increase in absorbance due to protein hydrolysis and release of p-nitroanilide were measured at the corresponding wavelength of the original substrate. One unit of activity was taken as the amount of the enzyme that released 1.0 mM p-nitroanilide per min, the concentration of which was determined using a molar absorption coefficient of 10,500 M–1 cm–1 at a wavelength of 410 nm [30].
The flour was ground from each type of grain (soybean, wheat, or corn) in a household blender; then, a 1% solution of the resulting flour was prepared in buffer A (50 mM tris-HCl, pH 7.0, 0.3 M NaCl, 10% ethanol) and incubated at 37 °C for 60 min, with constant stirring. After incubation, the solutions were centrifuged for 5 min at 10,000× g rpm, and the optical density in the supernatant was measured. After that, each sample was diluted with buffer A to an optical density of approximately 0.5 at 280 nm, and the protease CamSP was added. The reaction was carried out for 60 min at 50 °C and then stopped with one volume of the cold 10% TCA. The optical density was measured at 280 nm. Buffer A without the flour, but with the addition of the CamSP enzyme, was used as the control.
2.12. Determination of Kinetic Parameters for the Protease CamSP
The kinetic parameters of CamSP were calculated from the initial reaction rates using bovine serum albumin (BSA) as a substrate at concentrations from 10 to 100 μg mL−1 in 50 mM Tris-HCl, pH 7.5, at 50 °C. Each reaction was carried out for 5 min in triplicate. The Michaelis constant (Km), the maximum reaction rate (Vmax), and the turnover number (kcat) were determined by plotting the Lineover–Burk plot using the OriginPro 8.5 program.
2.13. Analysis of Nucleotide and Amino Acid Sequences
Analysis of the CamSP homology was carried out using the BLAST program available on the portal of the US National Center for Biotechnology Information (NCBI), with Nucleotide blast algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 20 April 2023), MEROPS (http://merops.sanger.ac.uk/, accessed on 20 April 2023), and UNIPROT (http://uniprot.org, accessed on April, 2023). The amino acid sequence of signal peptide was determined using the SignalP-5.0 server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0, accessed on 20 April 2023). The protein CamSP characteristics (molecular weight, amino acid composition, aliphatic index, GRAVY index) were obtained using the ProtParam program on the ExPASy portal (http://web.expasy.org/protparam/, accessed on 20 April 2023). Secondary structure of the protein CamSP was calculated using the SOPMA program (https://npsa-prabi.ibcp.fr/cgi-in/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 20 April 2023). Tertiary structure of the protein CamSP was predicted via AlphaFold v2 with the use of 3D model of the predicted DegP-type protease from Cobetia sp. ICG0124 (85.654% sequence identity) as a template found by AFDB search (A0A3T0K7V4|SWISS-MODEL Repository (expasy.org), accessed on 31 May 2023). Phylogenetic analysis was carried out using MEGA 11 (Molecular Evolutionary Genetics Analysis) software package (http://www.megasoftware.net/, accessed on 20 April 2023). Ancestral states were inferred using the Maximum Likelihood method and JTT matrix-based model. The tree used a set of possible amino acids (states) at each ancestral node based on their inferred likelihood at site 1. For each node, only the most probable state was shown. Initial trees for the heuristic search were obtained by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with superior log likelihood value. The rates among sites were treated as being uniform among sites (uniform rates option). The analysis involved 16 amino acid sequences, which were taken from the NCBI and MEROPS databases according to the BLAST search results against the protease CamSP as the query sequence. There were 482 positions in the final dataset in total.
3. Results and Discussion
3.1. Structural Classification and 3D Modelling of CamSP
Based on the structural classification and sequence analysis in the MEROPS database (http://merops.sanger.ac.uk/, accessed on 20 April 2023), the protein CamSP from C. amphilecti KMM 296 was concluded to belong to the S1C subfamily of DegP-type periplasmic serine endoproteases of clan PA(S) [5]. Among available structures in the SWISS-MODEL repository of the MEROPS database [5,31], the AlphaFold model of Cobetia sp. ICG0124 (gene: A0A3T0K7V4_9GAMM, seq identity 85.65%) built itself on the base of the crystal structure of Helicobacter pylori HtrA (7xs0.1.A), with an average model confidence (pLDDT) equal to 76.62. The crystal structures of H. pylori HtrA (monomer and homo-trimer PDB ID: 7xs0.1.A, seq identity 35.80%); E. coli DegP (homo-12-mer PDB ID: 6jjk.1.G, seq identity 35.87%); Legionella pneumophila proteinase Do (homo-12-mer PDB ID: 4ynn.1.A, seq identity 35.41%), Campylobacter jejuni proteinase DegQ (homo-12-mer PDB ID: 6z05.1.A, seq identity 36.17%); chloroplastic protease Do-like 2 and 9 (homo-hexamer PDB ID: 5ilb.1.A, seq identity 27.22%) were chosen by AlphaFold for the construction of eight 3D models for the C. amphilecti KMM 296 protease CamSP (Figure S1a–c).
Due to a common HtrA/DegP/Q catalytic triad of His-Asp-Ser and two PDZ domains, which mediate substrate recognition and oligomerization [9,13,14,16], the C. amphilecti KMM 296 protease CamSP has 36.15% identity with the high-temperature requirement human mitochondrial trimeric protease HtrA2 (Protein Data Bank (PDB) ID: 5m3n.1, chain A). The human HtrA2 is in a list of the top templates for 3D modelling CamSP, according to the Swiss-Prot analysis [5] (Table S1). The oligomerization up to 24-mers for the HtrA/DegP/Q-type enzymes is thought to be an allosteric regulation to switch the active and inactive conformations. Allosteric effects may be observed between the individual domains of monomeric proteins DegP or between the protomers of the oligomeric protein complexes [10].
3.2. Calculated Structural Properties and Phylogenetic Relatedness of CamSP
The C. amphilecti KMM 296 mature protease CamSP consists of 666 aa residues and has a putative signal peptide of 25 aa residues, identified using the SignalP-5.0 server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0, accessed on 20 April 2023) [32]. Analysis of the aa sequence of the C. amphilecti protease CamSP by the SOPMA server (NPS@: SOPMA secondary structure prediction (ibcp.fr), accessed on 20 April 2023) revealed four classes of secondary structures, namely: random turns, α-helices, extended strands, and β layers in the percentages of 36.49, 27.33, 24.92, and 11.26, respectively [33]. The GRAVY index of CamSP, analyzed by the ProtParam server and equal to −0.097, indicates that the protease has hydrophilic properties and is highly soluble in aqueous buffer solutions [33].
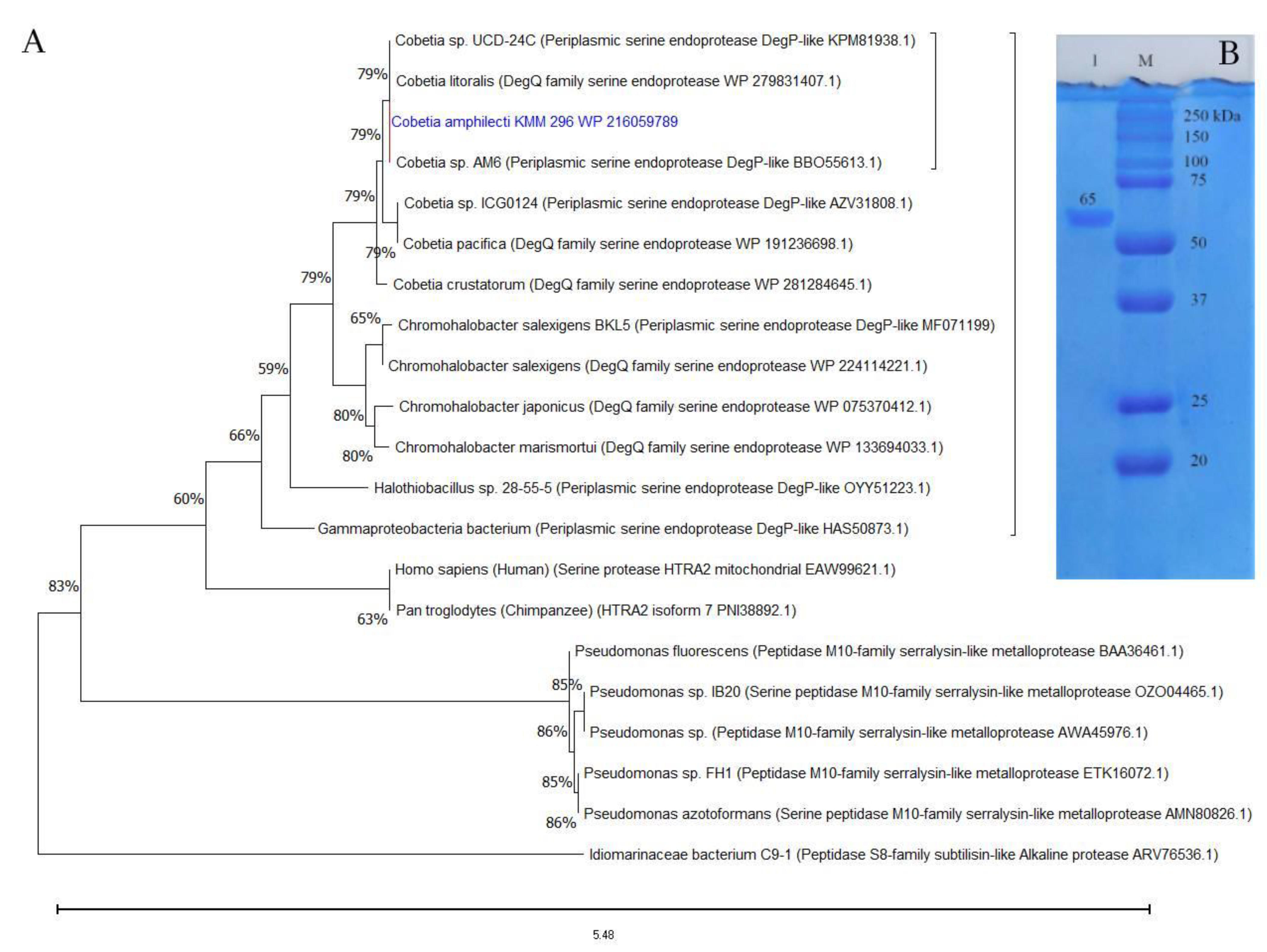
According to the BLAST-based search results in MEROPS and NCBI databases, the aa sequences of CamSP-like proteins from the species of Cobetia are much closer to the biochemically studied highly halothermotolerant proteases from Chromohalobacter sp. TVSP101 [34] and Chromohalobacter salexigens BKL5 [35], with an identity of approximately 62%. The phylogenetic tree reconstruction allowed for clustering the CamSP-like proteins of Cobetia spp. separately from other S1C subfamily proteins, indicating a new structural member of the DegP/Q-type serine protease family (Figure 1A).
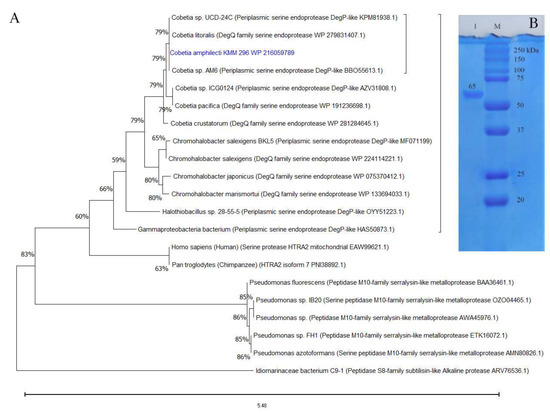
Figure 1.
The protein structure analysis and molecular weight of the C. amphilecti KMM 296 protease CamSP: (A) Phylogeny reconstruction for protease CamSP and closest relatives using the Maximum Likelihood method and JTT matrix-based model (MEGA11, http://www.megasoftware.net/, accessed on 20 April 2023); (B) SDS-PAGE of the purified recombinant protein CamSP (line 1); M—molecular weight marker (BioRad).
3.3. Heterologous Expression and Isolation of CamSP
Heterologous expression of the C. amphilecti KMM 296 gene (GenBank ID: WP_216059789.1) corresponding to the mature protein CamSP in the E. coli Rosetta DE(+) cells resulted in the production of a soluble recombinant protein CamSP, with a proteolytic activity of 1550.9 U/mg towards the casein substrate (1%) after its purification, according to the scheme described in Table 1.

Table 1.
Purification scheme for the recombinant C. amphilecti KMM 296 protease CamSP.
The isolation of enzymatically active recombinant protein CamSP confirmed the C. amphilecti ability to produce the functionally active DegP-type protease, with a calculated molecular weight of 69,957 kDa for the mature protein, which is consistent with the estimation of its molecular weight using polyacrylamide gel electrophoresis (PAGE) and gel filtration on Superdex-200 (Figure 1B). According to the results of SDS-PAGE, the molecular weight of CamSP is 65 ± 5 kDa (Figure 1), which coincides with the Chromohalobacter sp. TVSP101 protease of 66 kDa [34], but it is higher than the weight of DegP protease of 45 kDa isolated from C. salexigens BKL5 [35] that has been found to form the active multimers of 297.9 and 579.12 kDa [35].
3.4. Effect of pH on CamSP Activity
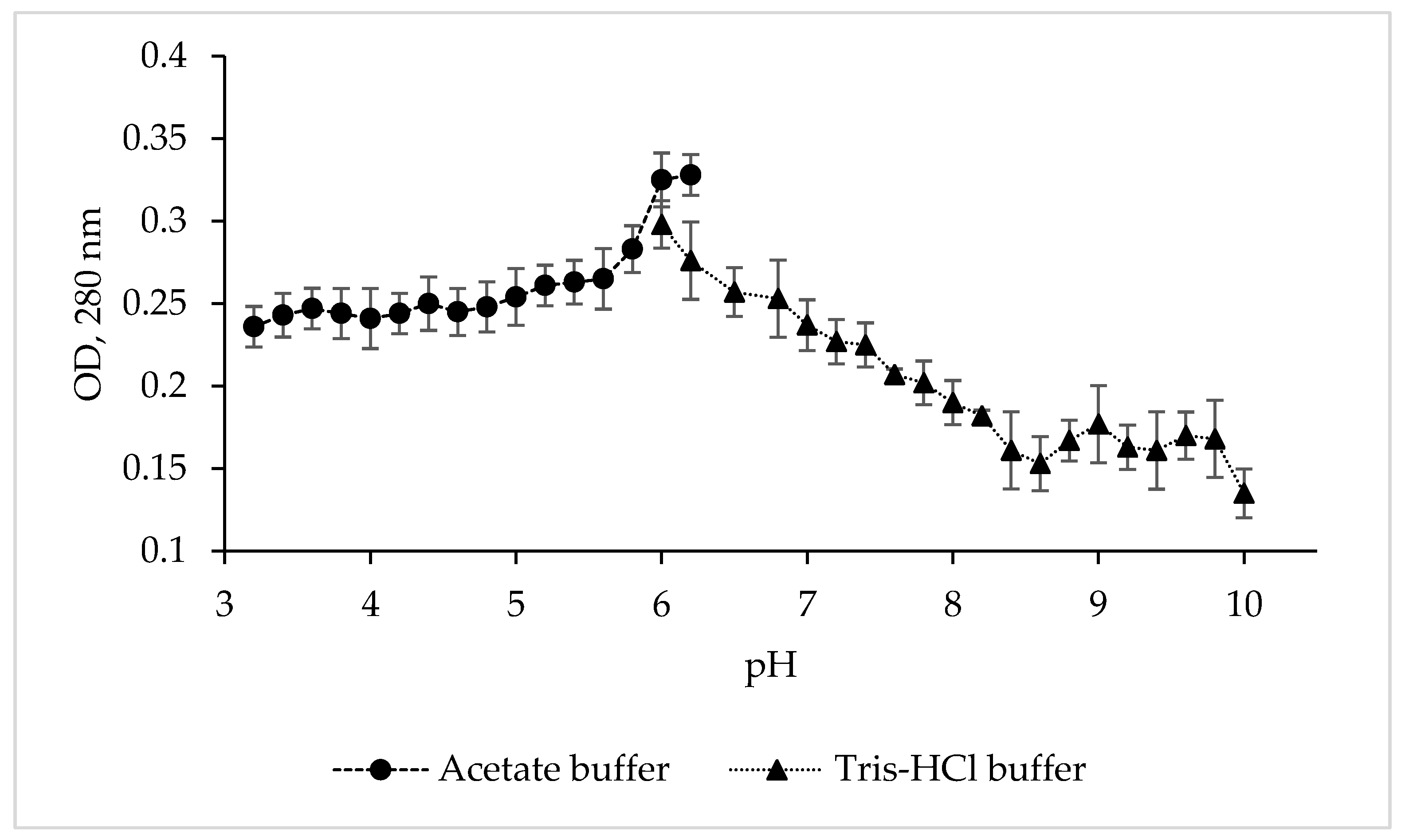
To determine the pH optimum and stability of the protease CamSP, the enzyme activity was studied in various buffer solutions over a wide pH range (Figure 2).
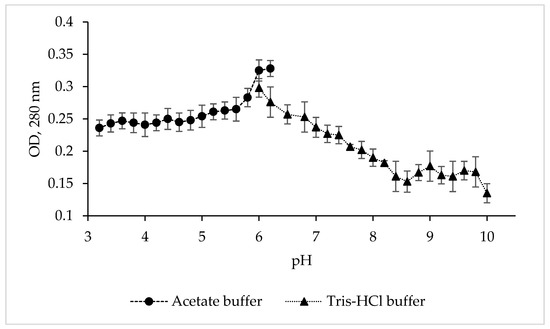
Figure 2.
Effect of pH and buffers on the proteolytic activity of C. amphilecti KMM 296 protease CamSP with the use of 1% casein as substrate.
Although CamSP is stable from pH 5.8 to 8.5, its activity was at the highest level at pH 6.0–6.2 in 0.1 M acetate buffer (Figure 2), similar to the serine proteases from Thermoactinomyces sp., Alkalihalobacillus lehensis JO-26, and Pseudoaltermonas sp. SM9913 (Table 2). The glycine-NaOH (pH 8.0–10.0) and phosphate (5.2–8.2) buffers strongly decreased the activity of CamSP. The other most characterized bacterial proteases have much higher levels of pH optimum at 8.0 to 11.0, especially thermo- and salt-tolerant enzymes (Table 2). However, DegP-like proteases have been reported to be a virulence factor in pathogenic bacteria, which are often endocytosed into the lysosomes, an acidic subcellular compartment, of the host cells [11]. Moreover, enteric bacteria like pathogenic E. coli have evolved an acid stress resistance mechanism involving the DegP protease function for surviving in the host stomach conditions. Although the E. coli DegP protease activity is completely lost under acidic conditions, it is substantially restored after neutralization at around pH 5.5 and participates in the degradation of acid-denatured periplasmic proteins toxic to the cells [11,12].

Table 2.
Comparative physical–chemical and catalytic characteristics of microbial proteases.
3.5. Effect of Ionic Strength on CamSP Activity
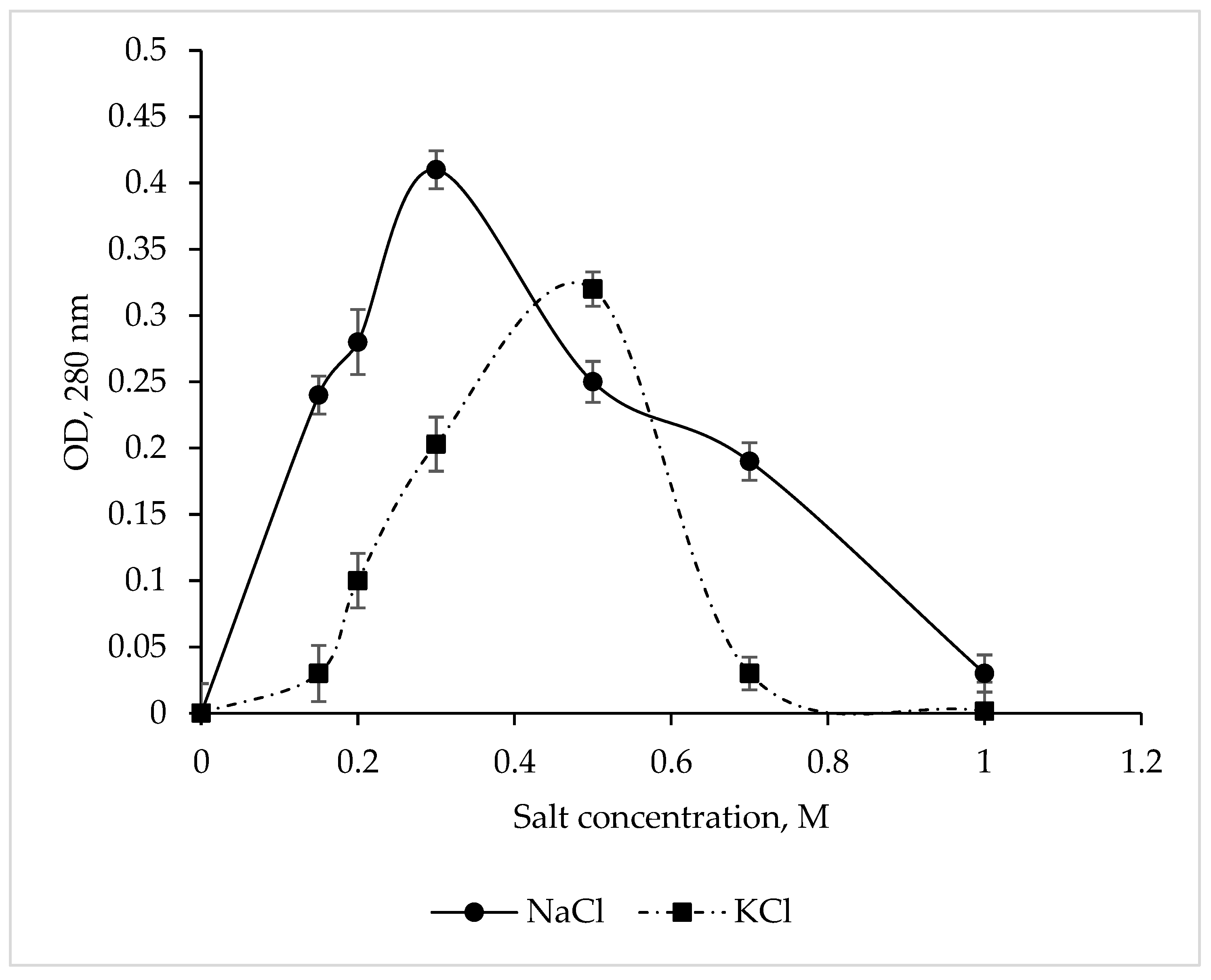
The CamSP protease exhibited maximum activity at 0.3 M NaCl and 0.5 M KCl (Figure 3), while Na+ and K+ salts at a concentration of 1 M completely inhibited the enzyme. These results are in complete agreement with the data obtained for CamSP in silico using the ProtParam program (http://web.expasy.org/protparam/, accessed on 20 April 2023) [44]. The C. amphilecti KMM 296 alkaline phosphatases CmAP and CmPhoD have the same moderate salt dependence, indicating their extracellular functions and the marine environment habitat of their producing microorganism [45,46].
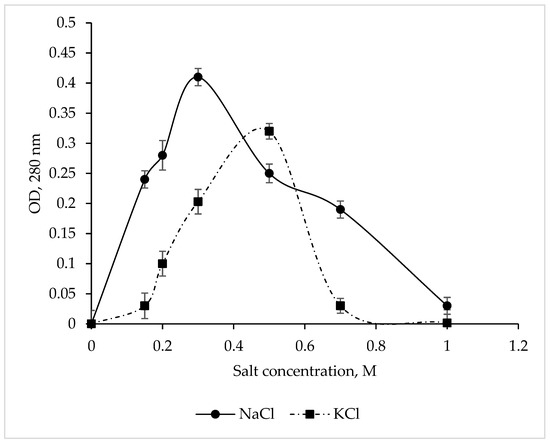
Figure 3.
Effect of NaCl and KCl on the C. amphilecti KMM 296 protease CamSP activity.
The in silico analysis of the aa residues in CamSP showed that the protein composition is dominated by acidic amino acids, while the ratio of alkaline and acidic amino acids is 0.9. Comparing the obtained ratio with nonhalophilic and extremely halophilic archeons [35], the experimental data on the CamSP protease confirmed that it is a nonhalophilic protein.
3.6. Effect of Divalent Metals on CamSP Activity
The various salts of Zn2+, Cu2+, Mn2+, Li2+, Mg2+, and Ca2+ slightly activated CamSP, while adding CoCl2 to the incubation mixture resulted in a twofold increase in the protease activity (Table 3), similar to the Co2+ dependence of the C. amphilecti KMM 296 alkaline phosphatase CmPhoD [46]. In addition, the marine microorganism C. amphilecti KMM 296 was found to possess a lot of magnesium and cobalt transporters, efflux proteins (CorC, A), and cobalt–zinc–cadmium resistance proteins (CzcD), suggesting significant environmental exposure to these metals and an involvement of the mollusk-associated strain KMM 296 in bioremediation and redox cycling of the transition metals [47,48,49].

Table 3.
Influence of metal salts on the CamSP proteolytic activity.
3.7. Effect of Detergents, Chelators, and Organic Solvents on CamSP Activity
The effects of anionic (SDS) and nonionic (Triton X-100) surfactants, the inhibitors for serine proteases (PMSFs) and metalloproteases (EDTA and EGTA), organic solvents (ethanol and isopropanol), and glycerol on the C. amphilecti KMM 296 protease CamSP are presented in Table 4. The protease CamSP was slightly inhibited by the chelating agents and 50% by SDS (Table 4). With increasing the EDTA concentration, the activity of CamSP decreased, confirming the metal ions’ influence on the level of protease activity. The enzyme expectedly exhibited reduced activity in the presence of the serine protease inhibitor PMSF, suggesting that the CamSP protease belongs to the class of serine proteases (Table 4). Ethanol and isopropanol increased the activity of CamSP by an average of 4-times and glycerol and Triton-X-100 by 2-times, which are consistent with the data on some proteases, for example, a Serratia marcescens metalloprotease [25], making the C. amphilecti protease useful for industrial applications such as peptide synthesis [39,50].

Table 4.
Influence of different detergents, chelators, and organic solvents on the CamSP proteolytic activity.
3.8. Effect of Temperature on CamSP Activity
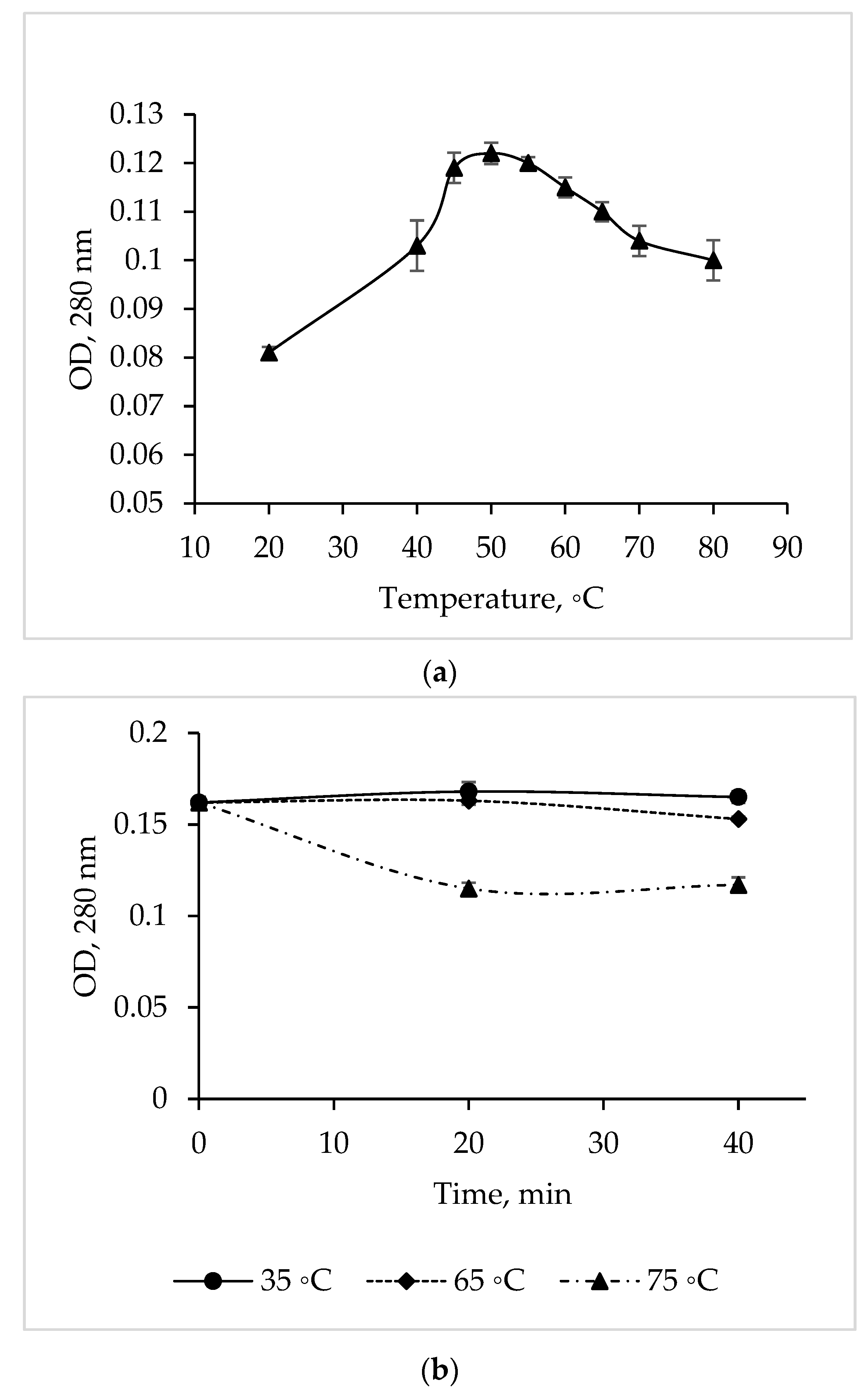
The temperature dependence study showed that the maximum activity of the C. amphilecti KMM 296 protease is at 50 °C, but the enzyme activity remained at a high level in a temperature range of 45–55 °C (Figure 4a).
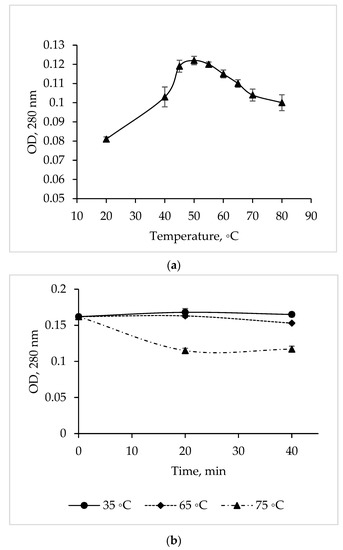
Figure 4.
Temperature optimum (a) and thermal stability (b) of the C. amphilecti KMM 296 protease CamSP.
The CamSP proteolytic activity was 100% from 35 to 65 °C for 40 min (Figure 4b). In addition, the activity of CamSP decreased by 27% after 20 min of its pre-incubation at 75 °C (before the proteolysis reaction); then, the enzyme retained 73% of the initial activity for another 20 min (Figure 4b). The long-time thermostability of CamSP is in accordance with the known biological functions of the DegP-like proteases, being essential for bacterial growth under high-temperature conditions [11]. According to Figure 4 and the aliphatic index of 96.25 obtained by analyzing the CamSP aa sequence with the ProtParam program (http://web.expasy.org/protparam/, accessed on 20 April 2023), the C. amphilecti KMM 296 protease is a moderately thermostable enzyme, similar to the serine metalloproteases from the marine host- and sediment-associated Bacillus subtilis GA CAS8 and Pseudoaltermonas sp. SM9913 MPC-02, respectively [22,26].
3.9. Substrate Specificity of CamSP
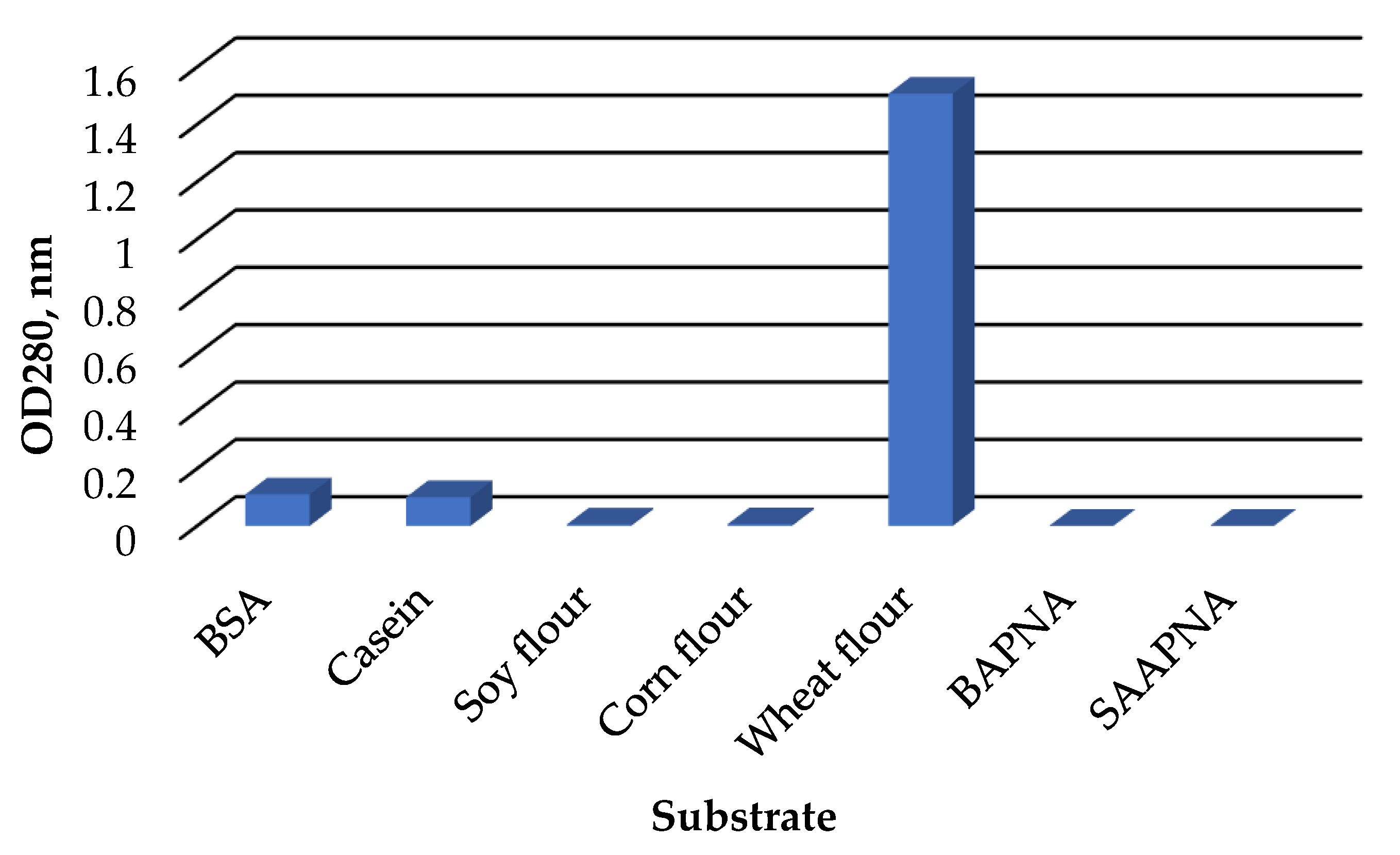
The C. amphilecti KMM 296 protease CamSP exhibited the proteolytic activity towards natural substrates, such as casein, bovine serum albumin, and wheat flour proteins (Figure 5). However, CamSP did not degrade soybean, corn, and synthetic substrates α-benzoyl-Arg-p-nitroanilide (BAPNA), N-Succinyl-L-alanyl-L-alanyl-L-prolyl-L-phenylalanine 4-nitroanilide (SAPNA) (Figure 5).
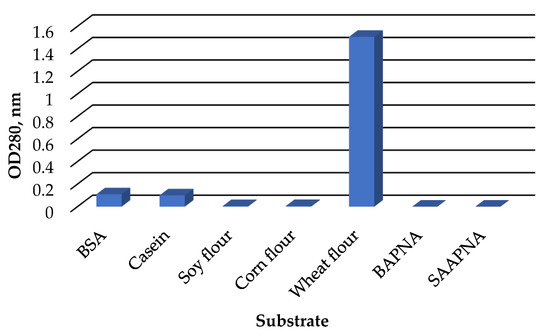
Figure 5.
Relative proteolytic activity of C. amphilecti KMM 296 protease CamSP towards proteins of different origin.
The widely used substrate for the different studies on bacterial serine proteases with various proteolytic activities was casein (Table 2), although the E. coli DegP-type protease activity towards this substrate and several other nonnative substrates was weak [12]. The natural DegP targets were identified as the colicin A lysis protein, bacterial pilins, nonpilus adhesins, and partially unfolded proteins to gain access to their catalytic sites for cleaving by DegP protease between paired hydrophobic residues [12]. Generally, HtrA/DegP/Q-type proteases cleave peptide bonds preferentially between such hydrophobic residues as Val-Val or Ile-Xaa [9,13,14,16].
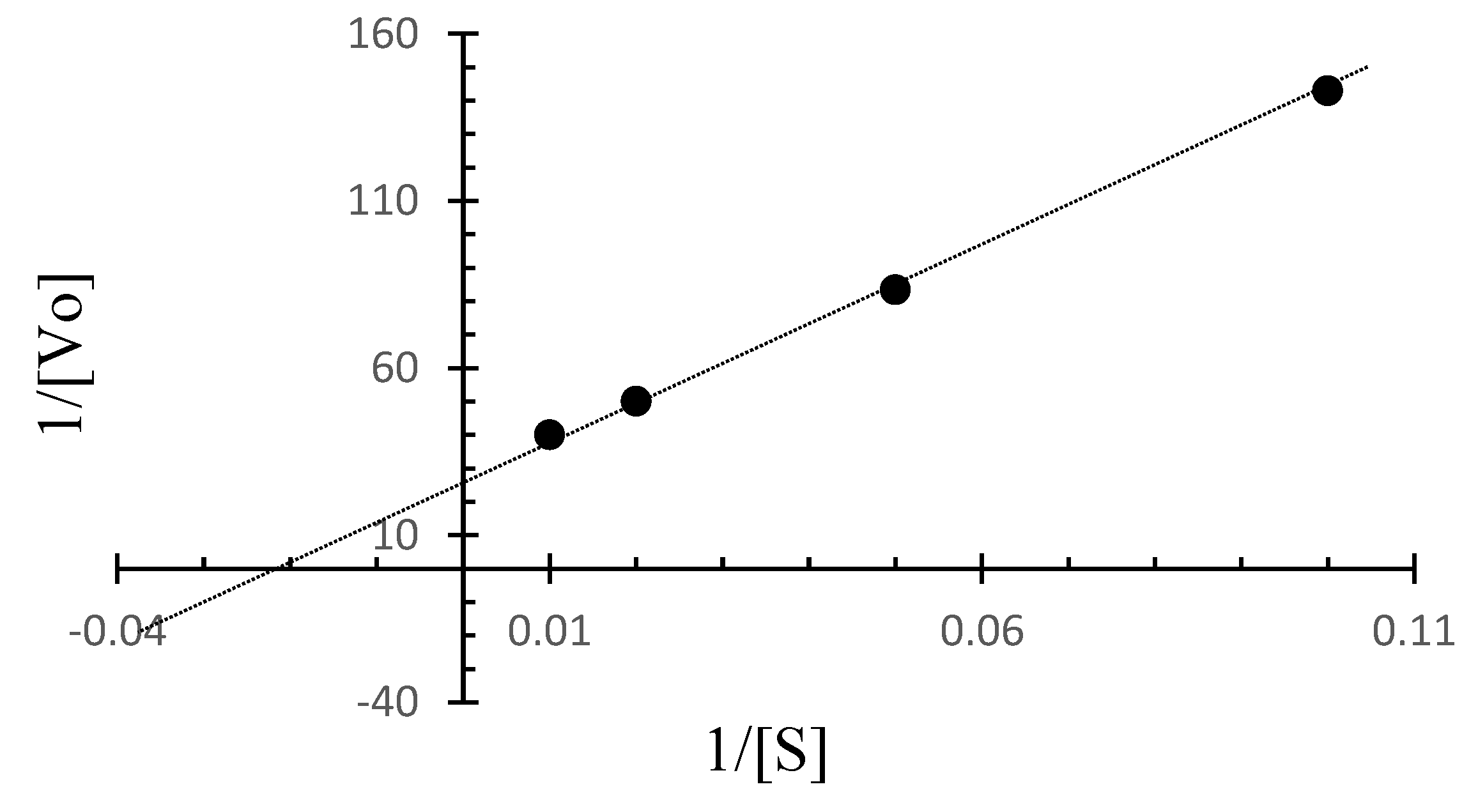
The Lineweaver–Burk plot for C. amphilecti KMM 296 protease CamSP was built based on the initial rates for BSA as the substrate at different concentrations and at a temperature of 50 °C (Figure 6). Based on the Lineweaver–Burk plot, the kinetic parameters for CamSP are Vmax = 0.036 µg/mL min−1 and Km = 41.7 µg/mL, 1/2 Vmax = 0.018 µg/mL min−1, kcat = 529.42 min−1, and kcat/Km = 14.706 min−1.
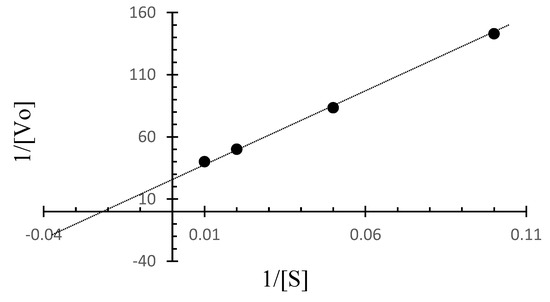
Figure 6.
The Lineweaver-Burk plot for CamSP based on the initial rates obtained for BSA at different concentrations at 50 °C.
The substrate specificity and catalytic properties of the enzyme CamSP may be applicable in the pharmaceutical, food, and bakery industries for the hydrolysis of wheat gluten and the production of gluten-free products for medical nutrition [17].
4. Conclusions
The gene of the marine bacterium C. amphilecti KMM 296 under the GenBank ID: WP_216059789.1, expressed in E. coli cells, was confirmed to encode for the metabolically active protease belonging to the structurally new periplasmic serine endoprotease/chaperon of the DegP type of clan PA(S). The C. amphilecti KMM 296 protease CamSP is a moderately thermostable, metal-dependent, and solvent-tolerant proteolytic enzyme, which is in a monomeric state under the used conditions (50 mM Tris-HCl buffer, pH 7.5) and preferentially cleaves the proteins of milk, wheat, and blood serum at neutral pH, suggesting its food and pharmaceutical use potential.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11071852/s1, Table S1: Template search results against the query amino acid sequence of C. amphilecti KMM 296 protease CamSP obtained by the automated protein structure homology-modelling server SWISS-MODEL; Figure S1. (a): The homology modelling of the C. amphilecti KMM 296 protease CamSP monomer build with the use of AlphaFold DB model of A0A3T0K7V4_9GAMM (gene: A0A3T0K7V4_9GAMM, organism: Cobetia sp ICG0124); (b): The homology modelling of the C. amphilecti KMM 296 protease CamSP build with the use of the crystal structure of H. pylori proteinase HtrA homo-trimer (PDB ID: 7xs0.1.A); (c): The homology modelling of the C. amphilecti KMM 296 protease CamSP build with the use of the crystal structure of L. pneumophila proteinase Do homo-12-mer (PDB ID: 4ynn.1.A).
Author Contributions
Conceptualization, methodology, and investigation, L.B. and Y.N.; resources, L.T.; project administration, O.S.; writing—original draft preparation, Y.N.; writing—review and editing, L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of Russian Federation, grant number 15.BRK.21.0004 (Contract No. 075-15-2021-1052).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Niyonzima, F.N.; More, S. Detergent-compatible proteases: Microbial production, properties, and stain removal analysis. Prep. Biochem. Biotechnol. 2015, 45, 233–258. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Structural bioinformatics and its impact to biomedical science. Curr. Med. Chem. 2004, 11, 2105–2134. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Structural bioinformatics and its impact to biomedical science and drug discovery. Front. Med. Chem. 2006, 3, 455–502. [Google Scholar] [CrossRef]
- Chou, K.-C.; Howe, W.J. Prediction of the tertiary structure of the β-secretase zymogen. Biochem. Biophys. Res. Commun. 2002, 292, 702–708. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic. Acids. Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar] [CrossRef]
- Gupta, R.; Beg, Q.K.; Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002, 59, 15–32. [Google Scholar] [CrossRef]
- Mönttinen, H.A.M.; Ravantti, J.J.; Poranen, M.M. Structural comparison strengthens the higher-order classification of proteases related to chymotrypsin. PLoS ONE 2019, 14, e0216659. [Google Scholar] [CrossRef]
- Huesgen, P.F.; Miranda, H.; Lam, X.; Perthold, M.; Schuhmann, H.; Adamska, I.; Funk, C. Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics and mechanism. Biochem. J. 2011, 435, 733–742. [Google Scholar] [CrossRef]
- Merdanovic, M.; Burston, S.G.; Schmitz, A.L.; Köcher, S.; Knapp, S.; Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. Activation by substoichiometric inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 1414–1418. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Y.; Shao, H.; Ma, J.; Song, X.; Zhang, M.; Chang, Z. DegP functions as a critical protease for bacterial acid resistance. FEBS J. 2018, 285, 3525–3538. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Dexter, P.; Evans, A.K.; Liu, C.; Hultgren, S.J.; Hruby, D.E. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: The PapA pilin. J. Bacteriol. 2002, 184, 5762–5771. [Google Scholar] [CrossRef] [PubMed]
- Shehzad, S.; Pandey, R.; Malhotra, P.; Gupta, D. Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP. Molecules 2021, 26, 2742. [Google Scholar] [CrossRef]
- Cho, H.; Choi, Y.; Min, K.; Son, J.B.; Park, H.; Lee, H.H.; Kim, S. Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy. Commun. Biol. 2020, 3, 547. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Bose, R.; Bose, K. Unraveling the Dichotomy of Enigmatic Serine Protease HtrA2. Front. Mol. Biosci. 2022, 9, 824846. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, M.; Li, X.; Zhao, S.; Pu, H.; Shen, J.; Adam, Z.; Clausen, T.; Zhang, L. The crystal structure of Deg9 reveals a novel octameric-type HtrA protease. Nat. Plants 2017, 3, 973–982. [Google Scholar] [CrossRef]
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. in Bioeng. Biotechol. 2019, 7, 110. [Google Scholar] [CrossRef]
- Kocher, G.S.; Mishra, S. Immobilization of Bacillus circulans MTCC 7906 for enhanced production of alkaline protease under batch and packed bed fermentation conditions. Int. J. Microbiol. 2009, 7, 359–378. [Google Scholar] [CrossRef]
- Singhal, P.; Nigam, V.; Vidyarthi, A. Studies on production, characterization and applications of microbial alkaline proteases. Int. J. Adv. Biotechnol. Res. 2012, 3, 653–669. [Google Scholar]
- Singh, P.; Rani, A.; Chaudhary, N. Isolation and characterization of protease producing Bacillus sp. from soil. Int. J. Pharm. Sci. Res. 2015, 6, 633–639. [Google Scholar]
- Sellami-Kamoun, A.; Haddar, A.; Ali, N.E.-H.; Ghorbel-Frikha, B.; Kanoun, S.; Nasri, M. Stability of thermostable alkaline protease from Bacillus licheniformis RP1 in commercial solid laundry detergent formulations. Microbiol. Res. 2008, 163, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Sathishkumar, R.; Ananthan, G.; Arun, J. Production, purification and characterization of alkaline protease by ascidian associated Bacillus subtilis GA CAS8 using agricultural wastes. Biocatal. Agric. Biotechnol. 2015, 4, 214–220. [Google Scholar] [CrossRef]
- Singh, R.; Mittal, A.; Kumar, M.; Mehta, P.K. Microbial protease in commercial applications. J. Pharm. Chem. Biol. Sci. 2016, 4, 365–374. [Google Scholar]
- Satyanarayana, T.; Sharma, A.; Mehta, D.; Puri, A.K.; Kumar, V.; Mohanan, N.; Joshi, S. Biotechnological applications of biocatalysts from the Firmicutes bacillus and Geobacillus species. In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B., Prakash, A., Eds.; Springer: Dordrecht, Germany, 2012; pp. 343–379. [Google Scholar] [CrossRef]
- Thakur, S.; Sharma, N.K.; Thakur, N.; Bhalla, T.C.S. Organic solvent tolerant metallo protease of novel isolate Serratia marcescens PPB-26: Production and characterization. 3 Biotech 2016, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Zhang, Y.Z.; Gao, P.J.; Luan, X.-W. Two different proteases produced by a deep-sea psychrotrophic bacterial strain, Pseudoaltermonas sp. SM9913. Mar. Biol. 2003, 143, 989–993. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Oda, K.; Murao, S. Purification and some properties of acid proteinase A and B of Scytalidium lignicolum ATCC 24568. Agric. Biol. Chem. 1974, 38, 2435–2444. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural protein during the assembly of the head 385 of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Erlanger, B.F.; Kokowsky, N.; Cohen, W. The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 1961, 95, 271–278. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic. Acids. Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. Physico-chemical characterization and topological analysis of pathogenesis-related proteins from Arabidopsis thaliana and Oryza sativa using in-silico approaches. PLoS ONE 2020, 15, e0239836. [Google Scholar] [CrossRef]
- Vidyasagar, M.; Prakash, S.; Mahajan, V.; Shouche, Y.S.; Sreeramulu, K. Purification and characterization of an extreme halothermophilic protease from a halophilic bacterium Chromohalobacter sp. TVSP101. Braz. J. Microbiol. 2009, 40, 12–19. [Google Scholar] [CrossRef]
- Fitriani, D.; Saifur, R.M.; da Irfan, D.P. Proteolytic activity of recombinant DegP from Chromohalobacter salexigens BKL5. Electron. J. Biotechnol. 2017, 29, 7–12. [Google Scholar] [CrossRef]
- Fullana, N.; Braña, V.; Marizcurrena, J.J.; Morales, D.; Betton, J.-M.; Marín, M.; Castro-Sowinski, S. Identification, recombinant production and partial biochemical characterization of an extracellular cold-active serine-metalloprotease from an Antarctic Pseudomonas isolate. AIMS Bioeng. 2017, 4, 386–401. [Google Scholar] [CrossRef]
- Sánchez-Porro, C.; Mellado, E.; Bertoldo, C.; Antranikian, G.; Ventosa, A. Screening and characterization of the protease CP1 produced by the moderately halophilic bacterium Pseudoalteromonas sp. strain CP76. Extremophiles 2003, 7, 221–228. [Google Scholar] [CrossRef]
- Kalwasińska, A.; Jankiewicz, U.; Felföldi, T.; Burkowska-But, A.; Brzezinska, M.S. Alkaline and Halophilic Protease Production by Bacillus luteus H11 and Its Potential Industrial Applications. Food Technol. Biotechnol. 2018, 56, 553–561. [Google Scholar] [CrossRef]
- Bhatt, H.B.; Singh, S.P. Cloning, Expression, and Structural Elucidation of a Biotechnologically Potential Alkaline Serine Protease from a Newly Isolated Haloalkaliphilic Bacillus lehensis JO-26. Front. Microbiol. 2020, 11, 941. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00941 (accessed on 20 April 2023). [CrossRef] [PubMed]
- Zabolotskaya, M.V.; Demidyuk, I.V.; Akimkina, T.V.; Kostrov, S.V. A novel neutral protease from Thermoactinomyces species 27a: Sequencing of the gene, purification, and characterization of the enzyme. Protein. J. 2004, 23, 483–492. [Google Scholar] [CrossRef]
- Damare, S.; Mishra, A.; D’Souza-Ticlo-Diniz; Krishnaswamy, D.A.; Raghukumar, C. A deep-sea hydrogen peroxide-stable alkaline serine protease from Aspergillus flavus. 3 Biotech 2020, 10, 528. [Google Scholar] [CrossRef] [PubMed]
- Bansal, P.; Kumar, R.; Singh, J.; Suman, D. Production of Extracellular Alkaline Serine Protease from Pediococcus acidilactici NCDC 252: Isolation, Purification, Physicochemical and Catalytic Characterization. Catal. Lett. 2021, 151, 324–337. [Google Scholar] [CrossRef]
- Zhou, C.; Qin, H.; Chen, X.; Zhang, Y.; Xue, Y.; Ma, Y. A novel alkaline protease from alkaliphilic Idiomarina sp. C9-1 with potential application for eco-friendly enzymatic dehairing in the leather industry. Sci. Rep. 2018, 8, 16467. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–608. [Google Scholar]
- Golotin, V.A.; Balabanova, L.A.; Likhatskaya, G.N.; Rasskazov, V.A. Recombinant production and characterization of a highly active alkaline phosphatase from marine bacterium Cobetia marina. Mar. Biotechnol. 2015, 17, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Noskova, Y.; Likhatskaya, G.; Terentieva, N.; Son, O.; Tekutyeva, L.; Balabanova, L. A Novel Alkaline Phosphatase/Phosphodiesterase, CamPhoD, from Marine Bacterium Cobetia amphilecti KMM 296. Mar. Drugs 2019, 17, 657. [Google Scholar] [CrossRef]
- Nordin, N.; Guskov, A.; Phua, T.; Sahaf, N.; Xia, Y.; Lu, S.; Eshaghi, H.; Eshaghi, S. Exploring the structure and function of Thermotoga maritima CorA reveals the mechanism of gating and ion selectivity in Co2+/Mg2+ transport. Biochem. J. 2013, 451, 365–374. [Google Scholar] [CrossRef]
- Dulay, H.; Tabares, M.; Kashefi, K.; Reguera, G. Cobalt Resistance via Detoxification and Mineralization in the Iron-Reducing Bacterium Geobacter sulfurreducens. Front. Microbiol. 2020, 11, 600463. [Google Scholar] [CrossRef]
- Arif, S.; Nacke, H.; Schliekmann, E.; Reimer, A.; Arp, G.; Hoppert, M. Composition and niche-specific characteristics of microbial consortia colonizing Marsberg copper mine in the Rhenish Massif. Biogeosciences 2022, 19, 4883–4902. [Google Scholar] [CrossRef]
- Kumar, D.; Bhalla, T.C. Microbial proteases in peptide synthesis: Approaches and applications. Appl. Microbiol. Biotechnol. 2005, 68, 726–736. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).