Altitudinal Gradient and Soil Depth as Sources of Variations in Fungal Communities Revealed by Culture-Dependent and Culture-Independent Methods in the Negev Desert, Israel
Abstract
1. Introduction
2. Materials and Methods
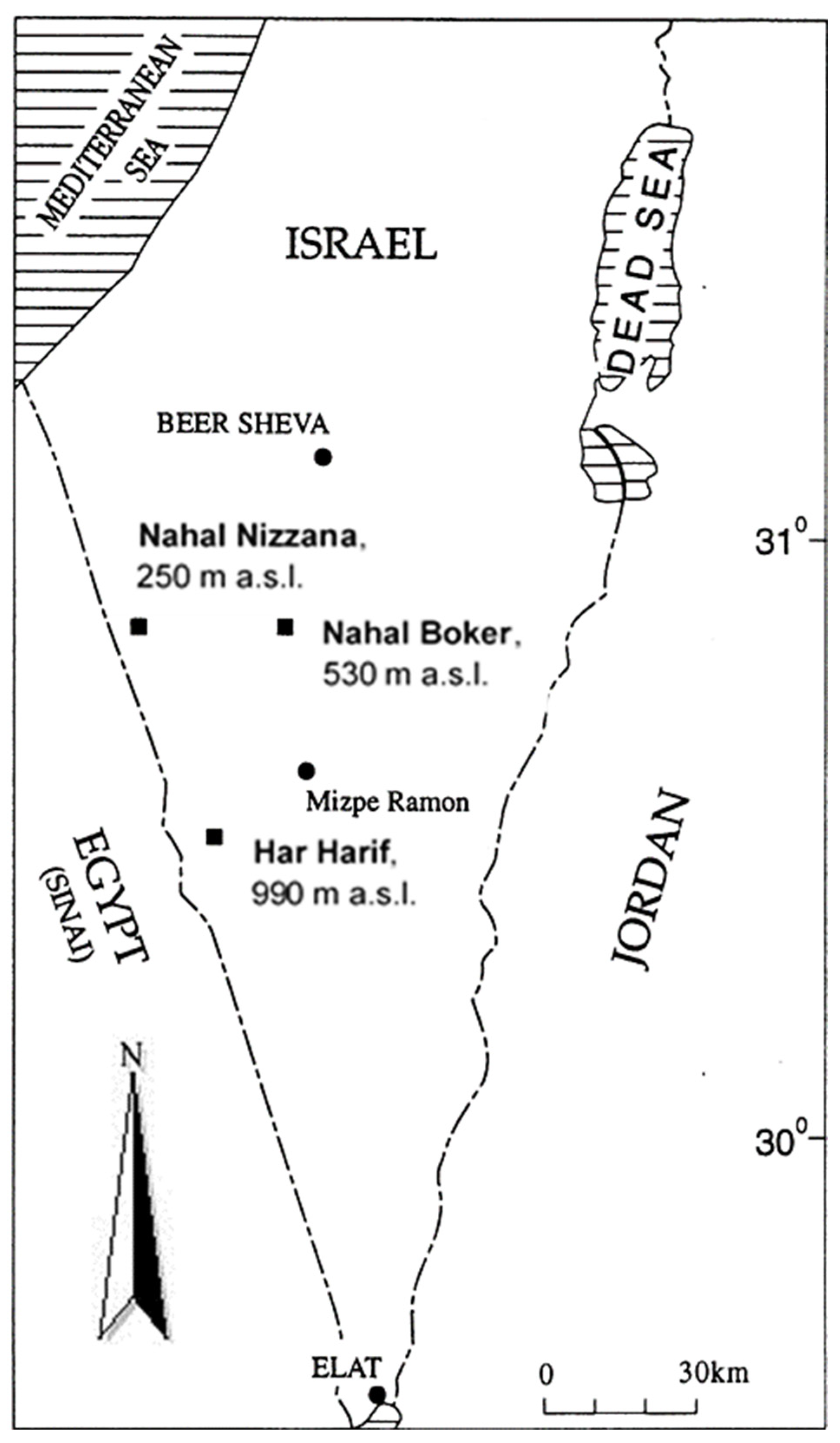
2.1. Site Description
2.2. Sampling Design
2.3. Characterization of Fungal Communities
2.3.1. Culture-Dependent Approach
2.3.2. Culture-Independent Approach Based on Next-Generation Sequencing (NGS)
2.4. Data Analyses
3. Results
3.1. Culturable Fungal Communities
3.1.1. Composition and Diversity
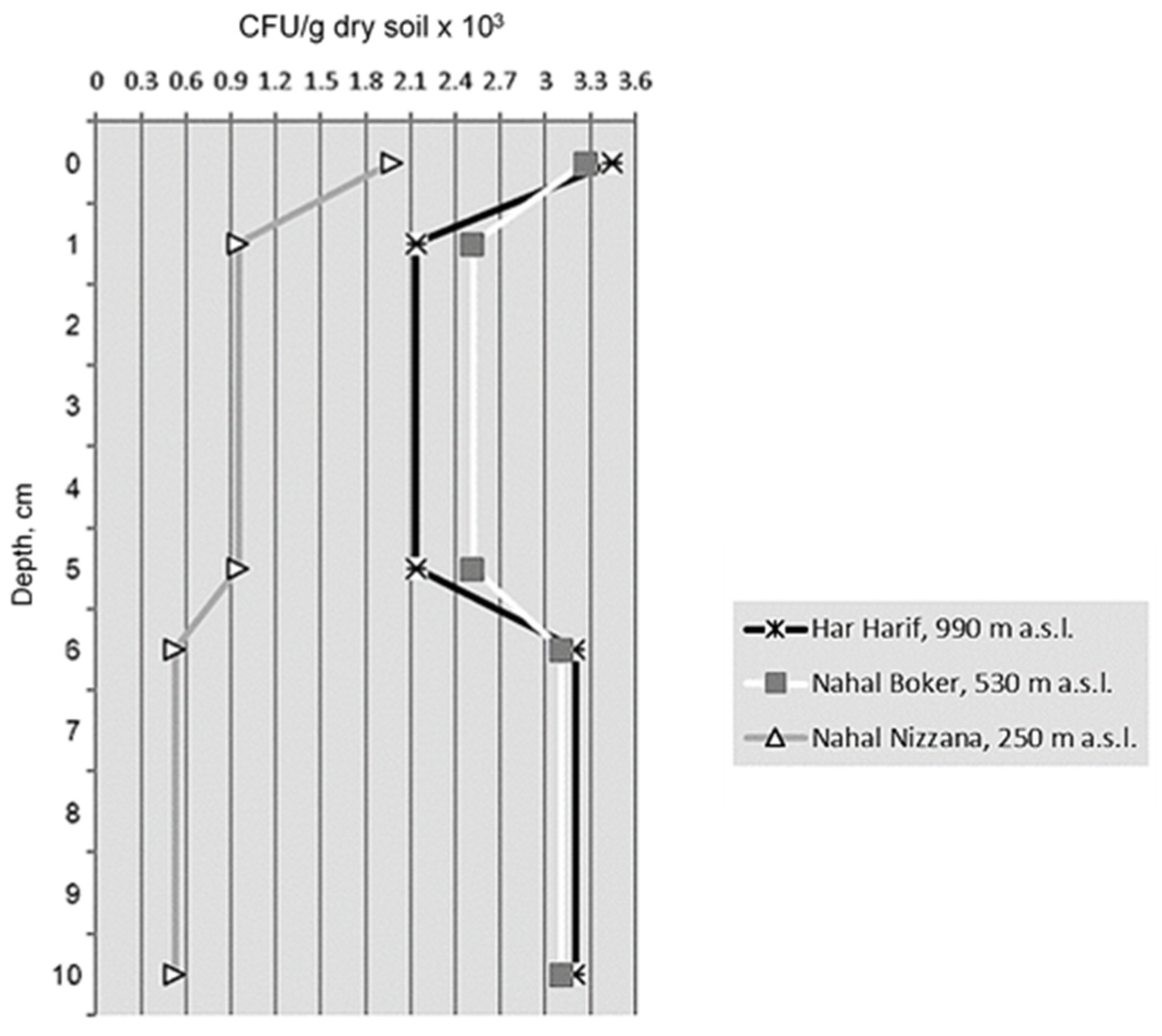
3.1.2. Density of Microfungal Isolates
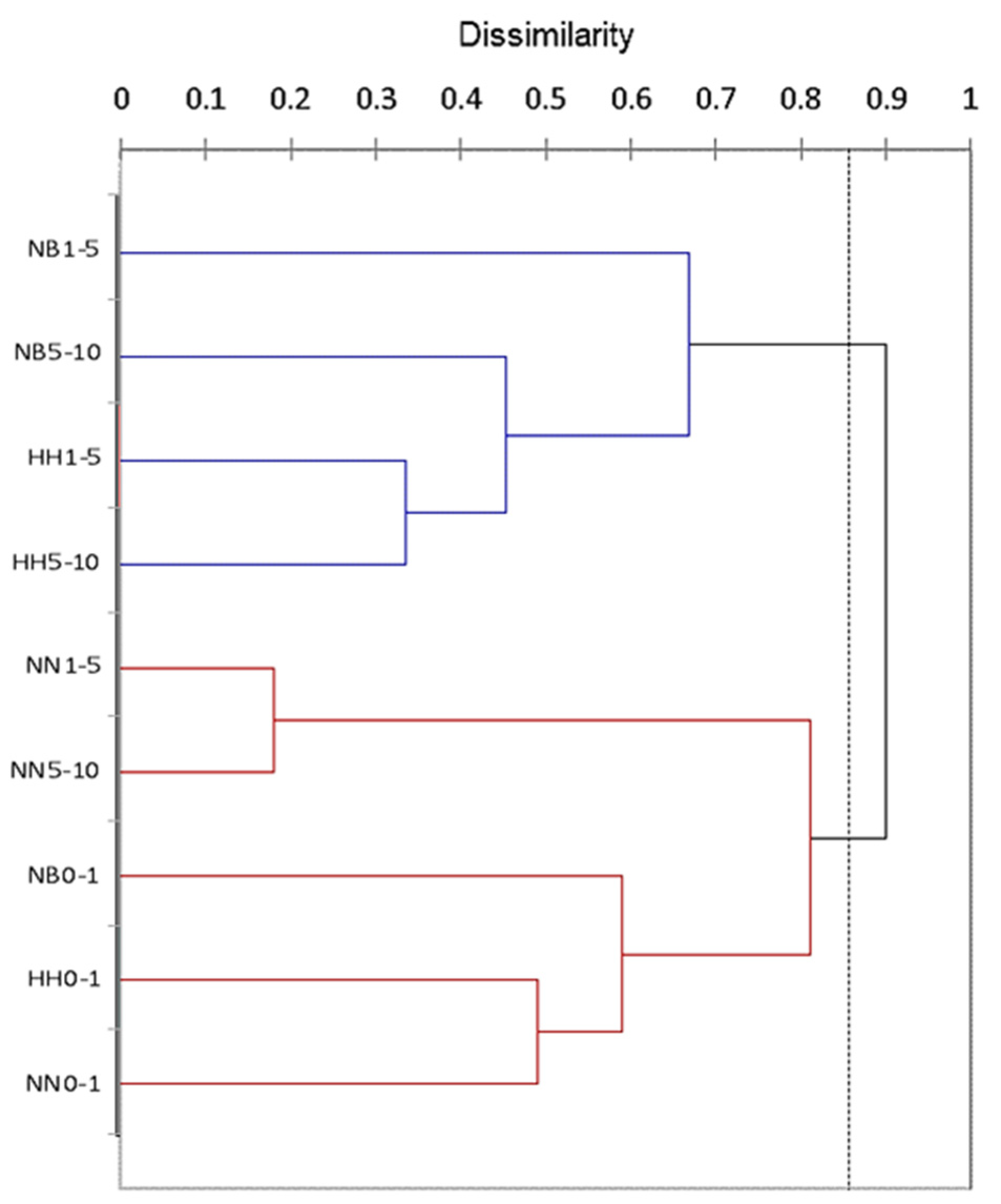
3.2. Effect of Altitude and Soil Depth on the Characteristics of Microfungal Communities
3.3. Fungal Communities Revealed by DNA Extraction
Composition and Diversity
4. Discussion
4.1. Effect of Altitude and Soil Depth on Fungal Communities
4.2. Comparison of Fungal Communities Revealed by Culture-Dependent and Culture-Independent Approaches
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caruso, T.; Chan, Y.; Lacap, D.C.; Lau, M.C.Y.; McKay, C.P.; Pointing, S.P. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011, 5, 1406–1413. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Tesei, D.; Zakharova, K. Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol. 2012, 5, 453–462. [Google Scholar] [CrossRef]
- Amiran, D.H.; Kadmon, N. Atlas of Israel, 3d ed.; Surveys of Israel: Tel Aviv, Israel, 1985. [Google Scholar]
- Grishkan, I.; Zaady, E.; Nevo, E. Soil crust microfungi along a southward rainfall aridity gradient in the Negev desert, Israel. Eur. J. Soil Biol. 2006, 42, 33–42. [Google Scholar] [CrossRef]
- Grishkan, I.; Beharav, A.; Kirzhner, V.; Nevo, E. Adaptive spatiotemporal distribution of soil microfungi in ‘‘Evolution Canyon’’ III, Nahal Shaharut, extreme Southern Negev desert, Israel. Biol. J. Linn. Soc. 2007, 90, 263–277. [Google Scholar] [CrossRef]
- Grishkan, I.; Kidron, G.J.; Wei, X. Microclimatic gradient as a source of variations in cultivable soil microfungal communities at the Negev Desert, Israel. Geomicrobio. J. 2021, 38, 829–841. [Google Scholar] [CrossRef]
- Grishkan, I.; Nevo, E. Spatiotemporal distribution of soil microfungi in the Makhtesh Ramon area, Central Negev Desert, Israel. Fungal Ecol. 2010, 3, 326–337. [Google Scholar] [CrossRef]
- Grishkan, I.; Kidron, G.J. Vertical divergence of microfungal communities through the depth in different soil formations at Nahal Nizzana, western Negev Desert, Israel. Geomicrobiol. J. 2016, 33, 564–577. [Google Scholar] [CrossRef]
- Grishkan, I.; Kidron, G.J. Vertical divergence of cultured microfungal communities at the Hallamish dune field, Western Negev Desert, Israel. Geomicrobiol. J. 2017, 34, 706–721. [Google Scholar] [CrossRef]
- Grishkan, I. Spatiotemporal variations in soil cultivable mycobiota at the Arava desert (Israel) along latitudinal and elevational gradients. AIMS Microbiol. 2018, 4, 502–521. [Google Scholar] [CrossRef] [PubMed]
- Grishkan, I. Soil microfungi of Israeli deserts: Adaptations to environmental stress. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 97–117. [Google Scholar] [CrossRef]
- Ni, Y.; Yang, T.; Zhang, K.; Shen, C.; Chu, H. Fungal communities along a small-scale elevational gradient in an alpine tundra are determined by soil carbon nitrogen ratios. Front. Microbiol. 2018, 9, 1815. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Liu, H.; Liu, S.; Yin, Y.; Yuan, Z.; Zhao, Y.; Cao, H. Distribution and assembly processes of soil fungal communities along an altitudinal gradient in Tibetan Plateau. JoF 2021, 7, 1082. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in Alpine forest soils: What are the driving factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Yoo, S.; Park, M.S.; Kim, C.S.; Lim, Y.W. Different patterns of belowground fungal diversity along altitudinal gradients with respect to microhabitat and guild types. Environ. Microbiol. Rep. 2021, 13, 649–658. [Google Scholar] [CrossRef]
- Chen, J.; Shi, Z.; Liu, S.; Zhang, M.; Cao, X.; Chen, M.; Xu, G.; Xing, H.; Li, F.; Feng, Q. Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau. JoF 2022, 8, 807. [Google Scholar] [CrossRef]
- Kidron, G.J.; Temina, M. The effect of dew and fog on lithic lichens along an altitudinal gradient in the Negev Desert. Geomicrobiol. J. 2013, 30, 281–290. [Google Scholar] [CrossRef]
- Kidron, G.J.; Kronenfeld, R. Lithic cyanobacteria as bioindicators for dewless habitats within a dew desert. Flora 2022, 288, 152027. [Google Scholar] [CrossRef]
- Kidron, G.J.; Kronenfeld, R. Assessing the likelihood of the soil surface to condense vapor: The Negev experience. Ecohydrology 2020, 13, e2200. [Google Scholar] [CrossRef]
- Rosenan, N.; Gilad, M. Meteorological Data; Atlas of Israel; Carta Jerusalem: Jerusalem, Israel, 1985. [Google Scholar]
- Kidron, G.J. Altitude dependent dew and fog in the Negev desert, Israel. AFM 1999, 96, 1–8. [Google Scholar] [CrossRef]
- Kidron, G.J. Measurements of evaporation with a novel mini atmometer in the Negev. Weather 2005, 60, 268–272. [Google Scholar] [CrossRef]
- Evenari, M. Ecology of the Negev Desert, a critical review of our knowledge. In Development in Arid Zone Ecology and Environmental Quality; Shuval, H., Ed.; Balaban International Science Services: Glenside, PA, USA, 1981; pp. 1–33. [Google Scholar]
- Bitan, A.; Rubin, S. Climatic Atlas of Israel for Physical and Environmental Planning and Design. Minist. Transp. Jerus. 1991. [Google Scholar]
- Davet, R.; Rouxel, F. Detection and Isolation of Soil Microfungi; Science Publisher Inc.: Enfield, NH, USA; Plymouth, UK, 2000. [Google Scholar]
- Bills, G.F.; Christensen, M.; Powell, M.; Thorn, G. Saprobic soil microfungi. In Biodiversity of Microfungi. Inventory and Monitoring Methods; Mueller, G.M., Bills, G.F., Foster, M.S., Eds.; Elsevier Academic Press: San-Diego, CA, USA, 2004; pp. 271–302. [Google Scholar]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Krebs, C.J. Ecological Methodology; Addison Wesley: San Francisco, CA, USA; Longman: Harlow, UK, 1999. [Google Scholar]
- Adams, J.E. Influence of mulches on runoff, erosion, and soil water depletion. SSSAJ 1966, 30, 110–114. [Google Scholar] [CrossRef]
- Cerdà, A. Effects of rock fragment cover on soil infiltration, interrill runoff and erosion. Eur. J. Soil Sci. 2001, 52, 59–68. [Google Scholar] [CrossRef]
- Grishkan, I.; Lazaro, R.; Kidron, G.J. Weak effect of plant canopy but strong impact of depth on variation of cultivable microfungal communities through soil profiles in semiarid Spain. Pedobiologia 2021, 85–86, 150710. [Google Scholar] [CrossRef]
- Grishkan, I. Chapter 10.2. Ecological stress: Melanization as a response in fungi to radiation. In Extremophiles Handbook; Horikoshi., K., Antranikian, G., Bull, A., Robb, F., Stetter, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1136–1148. [Google Scholar]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. MMBR 2015, 79, 243–262. [Google Scholar] [CrossRef]
- Balkwill, D.I.; Murphy, E.M.; Fair, D.M.; Ringelberg, D.B.; White, D.C. Microbial communities in high and low recharge environments: Implication for microbial transport in the vadose zone. Micro. Ecol. 1998, 35, 156–171. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Detection and diversity of fungi from environmental samples: Traditional versus molecular approaches. In Soil Biology, Advanced Techniques in Soil Microbiology; Varma, A., Oelmuller, R., Eds.; Springler: Berlin/Heidelberg, Germany, 2007; Volume 11, pp. 1–15. [Google Scholar]
- Willoughby, L.G. The activity of Rhizophlyctis rosea in soil: Some deductions from laboratory observations. Mycologist 2001, 15, 113–117. [Google Scholar] [CrossRef]
- Nguyen, H.D.T.; Nickerson, N.L.; Seifert, K.A. Basidioascus and Geminibasidium: A new lineage of heat-resistant and xerotolerant basidiomycetes. Mycologia 2013, 105, 1231–1250. [Google Scholar] [CrossRef]
- Gorbushina, A. Microcolonial fungi: Survival potential of terrestrial vegetative structures. Astrobiology 2003, 3, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Adamo, M.; Voyron, S.; Chialva, M.; Marmeisse, R.; Girlanda, M. Metabarcoding on both environmental DNA and RNA highlights differences between fungal communities sampled in different habitats. PLoS ONE 2020, 15, e0244682. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.W. Aspergillus and Penicillium identification using DNA sequences: Barcode or MLST? Appl. Microbiol. Biotechnol. 2012, 95, 339–344. [Google Scholar] [CrossRef]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 2020, 11, 14. [Google Scholar] [CrossRef]
 —melanin-containing spp.;
—melanin-containing spp.;  —light-colored spp. with small one-celled spores; area left from the line on the bars of melanin-containing spp. indicates contributions of species with large multicellular spores.
—light-colored spp. with small one-celled spores; area left from the line on the bars of melanin-containing spp. indicates contributions of species with large multicellular spores.
 —melanin-containing spp.;
—melanin-containing spp.;  —light-colored spp. with small one-celled spores; area left from the line on the bars of melanin-containing spp. indicates contributions of species with large multicellular spores.
—light-colored spp. with small one-celled spores; area left from the line on the bars of melanin-containing spp. indicates contributions of species with large multicellular spores.

| Species | HH 0–1 cm | HH 1–5 | HH 5–10 | NB 0–1 | NB 1–5 | NB 5–10 | NN 0–1 | NN 1–5 | NN 5–10 |
|---|---|---|---|---|---|---|---|---|---|
| Mortierellomycota | |||||||||
| * Mortierella humilis | - | 5.8 | 4.6 | - | 1.7 | 0.8 | 0.5 | - | - |
| Ascomycota | |||||||||
| * Acremonium alternatum | - | - | - | - | - | - | - | - | 0.24 |
| Acrospeira mirabilis | - | - | - | - | - | - | 0.7 | - | - |
| Alternaria alternata | 20.6 | - | 0.3 | 4.7 | 0.15 | - | 14.6 | 1.3 | 2.9 |
| A. atra | 17.4 | - | - | 35.5 | 3.8 | 0.8 | 11.7 | - | 1.2 |
| A. botrytis | - | - | - | - | - | - | - | - | 0.24 |
| A. chartarum | - | - | - | - | - | - | - | - | 0.24 |
| A. chlamydospora | - | - | - | - | - | 1.6 | 0.25 | - | 0.5 |
| A. chlamydosporigena | 5.8 | 1.9 | 2.6 | 1.1 | 0.5 | 0.5 | 4.1 | 0.16 | 1 |
| A. raphani | 0.2 | - | - | 1.2 | - | - | 0.25 | 0.16 | - |
| * Aphanocladium album | 1.7 | 3.0 | 0.8 | 0.3 | 0.5 | - | - | 1.7 | 0.5 |
| Arthrinium phaeospermum | 0.2 | - | - | - | - | - | - | - | 0.24 |
| Ascohyta medicaginicola | 0.9 | - | - | - | - | - | - | - | - |
| * Aspergillus crustosum | - | - | - | - | - | 0.13 | 0.5 | 0.3 | - |
| * A. fumigatus | - | - | - | - | - | - | - | 0.5 | - |
| A. niger | 0.4 | - | - | 0.3 | - | - | 0.25 | - | - |
| * Auxarthron umbrinum | - | - | - | - | 0.5 | - | - | 0.16 | - |
| Boeremia exigua | 3.2 | - | - | 0.4 | - | - | - | - | - |
| Camarosporium aequivocum | 1.9 | - | - | 0.3 | - | 0.13 | - | - | - |
| Chaetomium elatum | 1.1 | 3.0 | 0.9 | 0.3 | - | 1.7 | - | - | - |
| Ch. globosum | - | 2.8 | 2.6 | 1.5 | 1.2 | 0.5 | 0.5 | - | - |
| Ch. hispanicum | - | - | - | 1.7 | 1.3 | - | - | - | - |
| Chaetomium piluliferum | - | 16.7 | 9.8 | 2.7 | 4.5 | 6.8 | - | 0.16 | 1 |
| Ch. strumarium | - | - | - | - | - | - | 0.25 | - | - |
| Ch. cf. subspirilliferum 1 | - | - | - | - | - | - | - | 3.7 | 2.4 |
| Chaetomium sp. | - | - | - | 1.1 | 5.9 | 0.5 | - | - | - |
| * Chrysosporium merdarium | - | 0.9 | - | - | - | - | - | - | - |
| Cladosporium cladosporioides | 8.8 | 0.23 | 0.5 | 4.7 | 0.7 | - | 26.8 | 0.5 | 5.9 |
| C. herbarum | 0.2 | - | - | - | - | - | - | - | - |
| C. sphaerospermum | 1.5 | - | - | 0.3 | - | - | 0.25 | - | - |
| Collariella bostrychodes | - | 8.8 | 0.15 | - | - | - | - | - | - |
| Cordyceps farinosa | - | 0.23 | - | - | - | - | - | - | - |
| Cylindrocarpon didymum | - | - | 0.3 | - | - | - | - | - | - |
| Epicoccum nigrum | - | - | - | - | - | - | 0.25 | - | - |
| Fusarium equiseti | 1.7 | - | 2.3 | 1.4 | 3.3 | 1.5 | 0.25 | - | 0.7 |
| F. oxysporum | - | - | - | - | 1.8 | 1.7 | 0.5 | - | - |
| F. sporotrichioides | - | - | - | - | 0.5 | - | - | - | - |
| F. tricinctum2 | 1.9 | 1.2 | 1.2 | 1.1 | 3.6 | 0.3 | 0.5 | - | - |
| * Gymnoascus reessii | - | - | 5.2 | 5 | 42.8 | 20.6 | 1 | - | - |
| Helicodendron articulatum | - | - | - | 1.5 | - | - | - | - | - |
| Juxtiphoma eupyrena | 0.9 | 0.7 | - | - | - | - | - | - | - |
| Lasiodiplodia theobromae | - | - | - | 0.6 | - | - | - | - | - |
| * Lecanicillium psalliotae | - | - | - | 0.3 | - | - | - | - | - |
| Massarina igniaria | - | - | - | - | - | - | 0.25 | - | 0.5 |
| Metulocladosporiella musae | - | - | - | - | - | 0.4 | - | - | - |
| * Metarhizium marquandii | 1.5 | - | - | - | - | - | - | - | - |
| Microascus cirrosus | - | - | - | - | 12.2 | - | - | - | - |
| M. trigonosporus | - | - | - | - | - | 8.9 | 1.2 | - | - |
| Microsphaeropsis olivacea | 6.6 | - | - | - | - | - | - | - | - |
| Neocamarosporium chichastianum 2 | 0.2 | 0.23 | - | 2.1 | 0.7 | - | - | - | 0.7 |
| Neocucurbitaria cava | 0.2 | 0.5 | - | - | - | - | - | 1.8 | 0.7 |
| Nigrospora oryzae | 0.4 | - | - | - | - | - | - | 0.16 | - |
| * Parengyodontium album | 3.2 | 4.4 | 3.2 | 0.8 | 0.8 | 0.4 | 23.7 | 87.2 | 75.4 |
| Papulaspora pannosa | 1.3 | - | - | - | - | 0.13 | 2.4 | 1.2 | - |
| * Penicillium simplicissimum | 0.6 | 34.0 | 43.9 | 0.3 | 10.9 | 49.4 | 0.5 | 0.16 | 3.3 |
| * Pseudogymnoascus pannorum | - | 3.7 | 5.7 | - | 1.7 | 1.3 | - | - | - |
| Pseudothielavia terricola | - | - | - | - | - | - | 0.25 | - | - |
| * Sarocladium strictum | - | 0.7 | - | - | - | 0.3 | - | - | - |
| Scolecobasidium humicola | - | - | - | - | - | - | - | 0.16 | - |
| Sordaria fimicola | 0.2 | - | - | - | - | - | - | - | - |
| Sporormiella australis | 2.6 | 0.23 | - | 11.8 | 0.8 | 0.4 | 0.7 | 0.16 | 0.5 |
| S. minimoides | - | - | - | 1.8 | - | - | 0.25 | - | - |
| Stachybotrys chartarum | 1.5 | - | 0.15 | 1.2 | - | 0.13 | 2.9 | - | 0.7 |
| Stemphylium botryosum | 12.3 | - | 0.3 | 11.9 | - | 0.5 | 3.2 | 0.5 | 0.5 |
| S. vesicarium | - | - | - | 2.7 | - | - | - | - | - |
| * Talaromyces variabilis | - | 1.9 | 13.7 | - | - | - | - | - | - |
| Trichocladium acropullum 2 | - | 5.1 | 1.4 | - | - | - | - | - | 0.5 |
| Venustampulla parva | 0.2 | - | - | - | - | - | - | - | - |
| Immature fruit bodies | 0.2 | - | - | - | 0.15 | - | - | - | - |
| Basidiomycota | |||||||||
| Cutaneotrichosporon mucoides | - | 0.5 | - | - | - | - | - | - | - |
| Locality | S | H | J | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Depth | 0–1 cm | 1–5 cm | 5–10 cm | 0–1 cm | 1–5 cm | 5–10 cm | 0–1 cm | 1–5 cm | 5–10 cm | |
| a. Culturable communities | ||||||||||
| Har Harif, 990 m.a.s.l. | 32 | 23 | 21 | 2.54 | 2.18 | 2.01 | 0.73 | 0.69 | 0.63 | |
| Nahal Boker, 530 m.a.s.l. | 32 | 23 | 25 | 2.38 | 2.12 | 1.71 | 0.69 | 0.68 | 0.53 | |
| Nahal Nizzana, 250 m.a.s.l. | 31 | 18 | 23 | 2.14 | 0.67 | 1.15 | 0.62 | 0.23 | 0.37 | |
| b. DNA-based communities | ||||||||||
| Har Harif | 46 | - | 37 | 2.86 | - | 2.45 | 0.64 | - | 0.68 | |
| Nahal Boker | 20 | - | 28 | 2.50 | - | 2.34 | 0.83 | - | 0.70 | |
| Nahal Nizzana | 52 | - | 43 | 2.58 | - | 2.82 | 0.72 | - | 0.75 | |
| Parameter | R2 | Adjusted R2 | FPr>F | |||
|---|---|---|---|---|---|---|
| Source | Altitude (DF = 2) 1 | Soil Depth (DF = 2) | Altitude × Depth (DF = 4) | |||
| Species richness (S) | 0.547 | 0.412 | 2.42 ns | 12.43 **** | 0.72 ns | |
| Shannon index (H) | 0.732 | 0.653 | 14.18 **** | 16.97 **** | 2.88 @ | |
| Evenness (J) | 0.654 | 0.551 | 12.02 **** | 7.48 ** | 2.99 @ | |
| Melanin-containing (MC) spp. | 0.884 | 0.85 | 5.13 * | 95.02 **** | 1.26 ns | |
| MC spp. with large multicellular spores | 0.918 | 0.893 | 2.74 ns | 137.01 **** | 5.25 ** | |
| Light-colored spp. with small one-celled spores | 0.886 | 0.852 | 8.36 *** | 93.40 **** | 1.58 ns | |
| Isolate density | 0.458 | 0.298 | 7.53 ** | 1.61 ns | 1.14 ns | |
| Species | HH 0–1 cm | HH 5–10 cm | NB 0–1 cm | NB 5–10 cm | NN 0–1 cm | NN 5–10 cm |
|---|---|---|---|---|---|---|
| Chytridiomycota | ||||||
| Powellomyces sp. | 0.3 | - | - | - | - | - |
| Rhizophlyctis rosea | 0.13 | - | - | - | 0.5 | 0.22 |
| Spizellomyces sp. | - | - | - | - | - | 0.44 |
| Chytridiomycota_sp. | - | - | - | - | - | 0.44 |
| Glomeromycota | ||||||
| Claroideoglomus drummondii | - | - | - | - | - | 0.44 |
| Rhizophagus sp. | - | - | - | - | - | 0.11 |
| Mortierellomycota | ||||||
| Mortierella alpina | - | - | - | - | 0.1 | 0.6 |
| Ascomycota | ||||||
| Acremonium alternatum | - | - | - | 0.8 | - | - |
| A. rutilum | 7.8 | - | - | - | - | - |
| Acremonium sp. | - | - | - | - | 2.0 | - |
| Alternaria sp. | 37.6 | 8.5 | 6.5 | 20.8 | 13.9 | 1.5 |
| A. chlamydospora | 4.8 | - | 1.3 | 4.3 | 0.1 | 0.7 |
| A. chlamydosporigena | - | - | - | - | 5.4 | 0.44 |
| A. hungarica | 1.0 | - | - | - | 2.1 | - |
| A. obovoidea | 1.8 | - | - | 0.33 | 0.1 | - |
| A. oregonensis | 0.9 | - | - | 0.44 | 0.4 | - |
| Ascobolus sp. | 0.5 | - | - | - | - | - |
| Auxarthron alboluteum | - | - | - | - | - | 16.8 |
| Auxarthron sp. | - | - | - | - | - | 1.1 |
| Botryotrichum spirotrichum | 0.7 | 5.5 | 2.4 | 0.3 | 2.1 | |
| Botryotrichum sp. | - | - | - | - | 0.6 | - |
| Camarosporidiella sp. | - | 1.0 | - | - | - | - |
| Cephaliophora sp. | - | 1.3 | - | 0.11 | - | - |
| Chaetomium acropullum | - | 0.3 | - | - | - | 1.1 |
| Ch. elatum | - | - | - | - | - | 2.9 |
| Ch. iranianum | - | - | - | - | 0.3 | - |
| Ch. subspirilliferum | - | - | - | - | 0.1 | - |
| Chaetomium sp. | - | 3.8 | - | - | - | 1.2 |
| Chrysosporium carmichaelii | - | 21.1 | - | 1.3 | - | - |
| Cladosporium ossifragi | 0.3 | - | - | - | 1.0 | - |
| C. sphaerospermum | 0.6 | 0.14 | 1.3 | 0.11 | 0.4 | - |
| C. tenuissimum | - | 0.14 | - | - | - | - |
| Cladosporium sp. | - | - | - | 0.22 | - | - |
| Comoclathris sp. | - | 0.6 | 1.3 | - | 0.4 | - |
| Darksidea sp. | - | - | - | - | 0.1 | 1.5 |
| Dematiopleospora sp. | 0.13 | 0.14 | - | - | 0.1 | - |
| Foliophoma fallens | - | - | - | - | - | - |
| Fusarium oxysporum | - | - | - | - | 0.1 | - |
| F. redolens | - | - | - | - | - | 1.3 |
| F. tricinctum | 0.13 | 1.4 | 1.3 | 18.8 | 1.0 | 5.7 |
| Gymnoascus reessii | - | 0.14 | - | 5.3 | 0.1 | 6.3 |
| Knufia karalitana | 0.3 | - | - | 0.11 | - | - |
| Knufia sp. | 1.0 | - | - | - | 1.3 | - |
| Libertasomyces myopori | 0.13 | 1.0 | - | - | - | |
| Libertasomyces sp. | - | - | - | - | 0.7 | - |
| Mallochia reticulata | - | - | - | - | 0.2 | - |
| Monosporascus sp. | - | 1.3 | - | - | 0.2 | 3.4 |
| Monosporascus sp.1 | - | - | - | - | - | 0.22 |
| Neocamarosporium sp. | 4.3 | 2.4 | 6.5 | 3.7 | 0.2 | - |
| Neodevriesiasp. | 1.9 | - | - | 0.11 | - | - |
| Paraphaeosphaeria sporulosa | 0.13 | 1.4 | 1.3 | - | - | - |
| Parengyodontium album | 0.13 | 0.7 | - | 2.2 | - | - |
| Penicillium sp. | - | 0.6 | - | 2.2 | - | 0.6 |
| Phaeotheca triangularis | 0.5 | - | 1.3 | - | - | - |
| Preussia flanaganii | 1.6 | - | - | - | 0.8 | 0.22 |
| P. terricola | - | - | - | - | - | 2.4 |
| Preussia sp. | - | - | - | - | 3.1 | 0.11 |
| Pseudocoleophoma calamagrostidis | - | - | - | - | 0.1 | - |
| Pseudopithomyces chartarum | - | 1.7 | - | - | - | - |
| Saxophila tyrrhenica | 0.13 | - | - | 0.11 | - | - |
| Schizothecium carpinicola | - | - | - | - | 0.7 | - |
| S. inaequale | - | - | - | - | - | 0.33 |
| Scutellinia sp. | - | - | - | - | - | 11.2 |
| Sigarispora sp. | - | - | - | - | 0.5 | 0.11 |
| Sporormiella megalospora | 0.13 | - | - | - | 1.3 | - |
| S. minimoides | 0.13 | - | - | - | 0.1 | - |
| Stachybotrys chartarum | 0.13 | - | - | - | - | - |
| Stemphylium sp. | 0.4 | 5.6 | 2.6 | - | 1.2 | 0.11 |
| S. vesicarium | 0.13 | - | - | - | - | - |
| Subramaniula sp. | - | - | - | - | 0.4 | - |
| Toxicocladosporium sp. | - | - | - | - | 0.1 | - |
| Trichophaeopsis sp. | - | - | - | - | 15.7 | - |
| Trimmatostroma salinum | 1.3 | - | - | - | - | - |
| Vermiconia antarctica | 4.4 | 1.8 | 6.5 | 0.11 | 0.1 | - |
| Verrucocladosporium dirinae | - | - | - | - | 0.1 | - |
| Zopfiella sp. | - | - | - | - | - | 0.33 |
| Botryosphaeriales sp. | - | - | - | 0.11 | - | - |
| Capnodiales sp. | 1.6 | 0.3 | - | - | - | - |
| Capnodiales sp.1 | - | 0.4 | 7.8 | - | - | - |
| Chaetothyriales sp. | - | - | - | - | - | 2.3 |
| Hypocreales sp. | - | - | 25.9 | - | - | - |
| Pleosporales sp. | 1.7 | 2.5 | 3.9 | 2.0 | 26.0 | 0.11 |
| Pleosporales sp.1 | 5.7 | 0.6 | 11.7 | - | 3.9 | 0.11 |
| Xylariales sp. | - | 0.14 | - | 1.5 | - | 7.2 |
| Eurotiomycetes sp. | - | 6.5 | - | 9.6 | - | 0.8 |
| Ascomycota_ sp.1 | 0.13 | - | - | 20.2 | 3.8 | - |
| Ascomycota_ sp.2 | 0.13 | 2.3 | - | 0.11 | 1.4 | 1.9 |
| Ascomycota_ sp.3 | 4.7 | - | 1.3 | - | - | - |
| Ascomycota_ sp.4 | 1.3 | - | - | - | 1.00 | - |
| Ascomycota_ sp.5 | - | - | - | - | - | 2.0 |
| Ascomycota_ sp.6 | 1.2 | 1.0 | - | - | 0.1 | - |
| Ascomycota_ sp.7 | - | 1.3 | - | - | - | - |
| Ascomycota_ sp.8 | - | - | - | - | 0.6 | - |
| Ascomycota_ sp.9 | 0.5 | - | - | - | - | - |
| Ascomycota_ sp.10 | 0.13 | - | - | - | 0.2 | - |
| Ascomycota_ sp.11 | - | - | 3.9 | - | - | - |
| Ascomycota_ sp.12 | - | - | 1.3 | - | 0.1 | - |
| Ascomycota_ sp.13 | 2.3 | - | - | - | - | - |
| Ascomycota_ sp.14 | 3.8 | 4.3 | 11.7 | 1.0 | 3.3 | 1.2 |
| Ascomycota_ sp.15 | - | 0.14 | 1.3 | - | - | - |
| Basidiomycota | ||||||
| Agaricus gennadii | - | 6.7 | - | - | - | - |
| A. velutipes | 2.3 | - | - | - | - | - |
| Ceratobasidium sp. | - | - | - | - | - | 0.22 |
| Coniophora sp. | - | - | - | - | 0.1 | - |
| Geminibasidium sp. | - | - | - | 1.9 | - | 18.5 |
| Lyomyces sp. | - | - | - | - | 2.7 | - |
| Malassezia restricta | - | - | - | 0.11 | - | - |
| Naganishia sp. | - | - | - | - | 0.7 | - |
| Psathyrella arenosa | - | 10.5 | - | - | - | - |
| Thanatephorus sp. | 0.7 | - | - | - | - | - |
| Urocystis tritici | 0.2 | - | - | - | 0.1 | - |
| Basidiomycota_ sp.1 | - | - | - | - | - | 0.33 |
| Basidiomycota_ sp.2 | - | - | 1.3 | - | - | 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grishkan, I.; Kidron, G.J.; Rodriguez-Berbel, N.; Miralles, I.; Ortega, R. Altitudinal Gradient and Soil Depth as Sources of Variations in Fungal Communities Revealed by Culture-Dependent and Culture-Independent Methods in the Negev Desert, Israel. Microorganisms 2023, 11, 1761. https://doi.org/10.3390/microorganisms11071761
Grishkan I, Kidron GJ, Rodriguez-Berbel N, Miralles I, Ortega R. Altitudinal Gradient and Soil Depth as Sources of Variations in Fungal Communities Revealed by Culture-Dependent and Culture-Independent Methods in the Negev Desert, Israel. Microorganisms. 2023; 11(7):1761. https://doi.org/10.3390/microorganisms11071761
Chicago/Turabian StyleGrishkan, Isabella, Giora J. Kidron, Natalia Rodriguez-Berbel, Isabel Miralles, and Raúl Ortega. 2023. "Altitudinal Gradient and Soil Depth as Sources of Variations in Fungal Communities Revealed by Culture-Dependent and Culture-Independent Methods in the Negev Desert, Israel" Microorganisms 11, no. 7: 1761. https://doi.org/10.3390/microorganisms11071761
APA StyleGrishkan, I., Kidron, G. J., Rodriguez-Berbel, N., Miralles, I., & Ortega, R. (2023). Altitudinal Gradient and Soil Depth as Sources of Variations in Fungal Communities Revealed by Culture-Dependent and Culture-Independent Methods in the Negev Desert, Israel. Microorganisms, 11(7), 1761. https://doi.org/10.3390/microorganisms11071761








