Potential of Microbial Communities to Perform Dehalogenation Processes in Natural and Anthropogenically Modified Environments—A Metagenomic Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Site and Sample Description
2.2. Physicochemical Analyses of Ground Samples
2.3. Extraction and Gas Chromatography Analyses
2.4. Isolation of DNA
2.5. Metagenome Sequencing and Analysis
3. Results
3.1. Physical and Chemical Analyses of Studied Samples
3.2. General Characteristics of Metagenomes of Natural and Anthropogenically Modified Environments
3.3. Taxonomic Composition and Diversity of Microbial Communities
3.4. Occurrence of Dehalogenase Genes in Natural and Anthropogenically Modified Environments
3.5. Correlations Analyses
4. Discussion
4.1. The Importance of Microbial Dehalogenation for the Environment
4.2. An Overview of the Studied Environments
4.3. The Characteristics of Identified Dehalogenases Genes
4.4. The Contribution of Microorganisms in HOC Dehalogenation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dolfing, J. Energetic considerations in organohalide respiration. In Organohalide-Respiring Bacteria; Adrian, L., Löffler, F.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 31–48. [Google Scholar]
- Häggblom, M.M.; Bossert, I.D. Dehalogenation: Microbial Processes and Environmental Applications; Kluwer Academic Publisher Group: Dordrecht, The Netherlands, 2003. [Google Scholar]
- Khan, M.F.S.; Wu, J.; Cheng, C.; Akbar, M.; Liu, B.; Liu, C.; Shen, Y.; Xin, Y. Insight into fluorescence properties of 14 selected toxic single-ring aromatic compounds in water: Experimental and DFT study. Front. Environ. Sci. Eng. 2020, 14, 42. [Google Scholar] [CrossRef]
- Sun, R.X.; Luo, X.J.; Tan, X.X.; Tang, B.; Li, Z.R.; Mai, B.X. Legacy and emerging halogenated organic pollutants in marine organisms from the Pearl River Estuary, South China. Chemosphere 2015, 139, 565–571. [Google Scholar] [CrossRef]
- Cui, J.; Yu, Z.; Mi, M.; He, L.; Sha, Z.; Yao, P.; Feng, J.; Sun, W. Occurrence of halogenated organic pollutants in hadal trenches of the Western Pacific Ocean. Environ. Sci. Technol. 2020, 54, 15821–15828. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, G.; Di Filippo, P.; Riccardi, C.; Pomata, D.; Sonego, E.; Buiarelli, F. Occurrence of halogenated pollutants in domestic and occupational indoor dust. Int. J. Environ. Res. Public Health 2020, 17, 3813. [Google Scholar] [CrossRef] [PubMed]
- Nsibande, S.A.; Forbes, P.B.C. Fluorescence detection of pesticides using quantum dot materials—A review. Anal. Chim. Acta 2016, 945, 9–22. [Google Scholar] [CrossRef]
- Lamb, C.; Watts, J. Dehalogenation of brominated phenolic compounds by environmental microorganisms. Access Microbiol. 2020, 2, 319. [Google Scholar] [CrossRef]
- Häggblom, M.M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol. Lett. 1992, 103, 29–71. [Google Scholar] [CrossRef]
- Chandra, R.; Kumar, V. Biotransformation and biodegradation of organophosphates and organohalides. In Environmental Waste Management; Chandra, R., Ed.; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Atashgahi, S.; Häggblom, M.H.; Smidt, H. Organohalide respiration in pristine environments: Implications for the natural halogen cycle. Environ. Microbiol. 2018, 20, 934–948. [Google Scholar] [CrossRef]
- Kameswaran, S.; Ramesh, B.; Pitchika, G.K.; Chandra, M.S.; Srinivasulu, M. Microbial Reductive Dehalogenation and Its Role in Bioremediation. In Innovations in Biotechnology for a Sustainable Future; Springer: Berlin/Heidelberg, Germany, 2021; pp. 205–226. [Google Scholar]
- Fetzner, S.; Lingens, F. Bacterial Dehalogenases: Biochemistry, Genetics, and Biotechnological Applications. Microbiol. Mol. Biol. Rev. 1994, 58, 641–685. [Google Scholar] [CrossRef]
- Arp, D.J.; Yeager, C.M.; Hyman, M.R. Molecular and Cellular Fundamentals of Aerobic Cometabolism of Trichloroethylene. Biodegradation 2001, 12, 81–103. [Google Scholar] [CrossRef]
- Hug, L.A. Diversity evolution and environmental distribution of reductive dehalogenase genes. In Organohalide-Respiring Bacteria; Adrian, L., Loffler, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 377–393. [Google Scholar]
- Agarwal, V.; Miles, Z.D.; Winter, J.M.; Eustáquio, A.S.; El Gamal, A.A.; Moore, B.S. Enzymatic halogenation and dehalogenation reactions: Pervasive and mechanistically diverse. Chem. Rev. 2017, 117, 5619–5674. [Google Scholar] [CrossRef] [PubMed]
- Bin Hudari, M.S.; Richnow, H.; Vogt, C.; Nijenhuis, I. Effect of temperature on microbial reductive dehalogenation of chlorinated ethenes: A review. FEMS Microbiol. Ecol. 2022, 98, fiac081. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 214–2120. [Google Scholar] [CrossRef] [PubMed]
- Rotmistrovsky, K.; Agarwala, R. BMTagger: Best Match Tagger for Removing Human Reads from Metagenomics Datasets. 2011. Available online: ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/ (accessed on 2 July 2022).
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- NCBI RefSeq Database. Available online: https://ftp.ncbi.nlm.nih.gov/refseq/ (accessed on 20 July 2022).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Picard Toolkit. Broad Institute, GitHub Repository. 2019. Available online: https://broadinstitute.github.io/picard/ (accessed on 31 July 2022).
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Dröge, J.; Gregor, I.; McHardy, A.C. Taxator-tk: Precise taxonomic assignment of metagenomes by fast approximation of evolutionary neighborhoods. Bioinformatics 2015, 31, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Kirk, K.L. Biochemistry of the Elemental Halogens and Inorganic Halides; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Fuge, R. Fluorine in the environment, a review of its sources and geochemistry. Appl. Geochem. 2019, 100, 393–406. [Google Scholar] [CrossRef]
- Geilfus, C.M. Chloride in soil: From nutrient to soil pollutant. Environ. Exp. Bot. 2019, 157, 299–309. [Google Scholar] [CrossRef]
- Vassilev, S.V.; Eskenazy, G.M.; Vassileva, C.G. Contents, modes of occurrence and origin of chlorine and bromine in coal. Fuel 2000, 79, 903–921. [Google Scholar] [CrossRef]
- Cox, E.M.; Arai, Y. Environmental chemistry and toxicology of iodine. Adv. Agron. 2014, 128, 47–96. [Google Scholar]
- de Souza, R.M.; Seibert, D.; Quesada, H.B.; de Jesus Bassetti, F.; Fagundes-Klen, M.R.; Bergamasco, R. Occurrence, impacts and general aspects of pesticides in surface water: A review. Process Saf. Environ. Prot. 2020, 135, 22–37. [Google Scholar] [CrossRef]
- Gribble, G.W. Naturally occurring organohalogen compounds a survay. J. Nat. Prod. 1992, 55, 1353–1395. [Google Scholar] [CrossRef]
- Gribble, G.W. Naturally Occurring Organohalogen Compounds—A Comprehensive Update; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; Volume 91, pp. 12–13, 21–23. [Google Scholar]
- Gribble, G.W. A recent survey of naturally occurring organohalogen compounds. Environ. Chem. 2015, 12, 396–405. [Google Scholar] [CrossRef]
- Gribble, G.W.; Scheuer, P.J.; Moore, R.E.; Faulkner, D.J. Newly discovered naturally occurring organohalogens. Org. Chem. 2018, vi, 372–410. [Google Scholar] [CrossRef]
- Oberg, G. Chloride and organic chlorine in soil. Acta Hydrochem. Hydrobiol. 1998, 26, 137–144. [Google Scholar] [CrossRef]
- Hoekstra, E.J.; de Weerd, H.; de Leer, E.W.B.; Brinkman, U.A.T. Natural formation of chlorinated phenols, dibenzo-p-dioxins and dibenzo furans in soil of Douglas fir forest. Environ. Sci. Technol. 1999, 33, 2543–2549. [Google Scholar] [CrossRef]
- Loffler, F.E.; Cole, J.R.; Ritalahti, K.M.; Tiedje, J.M. Diversity of dechlorinating bacteria. In Dehalogenation Microbial Processes and Environmental Applications; Häggblom, M.M., Bossert, I.D., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherland, 2003; pp. 53–87. [Google Scholar]
- Fetzner, S. Mini-Review Bacterial Dehalogenation. Appl. Microbiol. Biotechnol. 1998, 50, 633–657. [Google Scholar] [CrossRef]
- Janssen, D.B.; Pries, F.; Van Der Ploeg, J.R. Genetics and biochemistry of dehalogenating enzymes. Annu. Rev. Microbiol. 1994, 48, 163–191. [Google Scholar] [CrossRef] [PubMed]
- Dunaway-Mariano, D.; Babbitt, P.C. On the Origins and Functions of the Enzymes of the 4-Chlorobenzoate to 4-Hydroxybenzoate Converting Pathway. Biodegradation 1994, 5, 259–276. [Google Scholar] [CrossRef] [PubMed]
- Slater, J.H.; Bull, A.T.; Hardman, D.J. Microbial Dehalogenation. Biodegradation 1995, 6, 181–189. [Google Scholar] [CrossRef]
- Reineke, W.; Knackmuss, H.J. Chemical Structure and Biodegradability of Halogenated Aromatic Compounds Substituent Effects on 1,2-Dioxygenation of Benzoic Acid. BBA-Gen. Subj. 1978, 542, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Bartels, I.; Knackmuss, H.J.; Reineke, W. Suicide Inactivation of Catechol 2,3-Dioxygenase from Pseudomonas Putida Mt-2 by 3-Halocatechols Appl. Environ. Microbiol. 1984, 47, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Vannelli, T.; Hooper, A.B. Reductive Dehalogenation of the Trichloromethyl Group of Nitrapyrin by the Ammonia-Oxidizing Bacterium Nitrosomonas Europaea Appl. Environ. Microbiol. 1993, 59, 3597–3601. [Google Scholar] [CrossRef]
- Matturro, B.; Tandoi, V.; Rossetti, S. Different Activity Levels of Dehalococcoides Mccartyi Revealed by FISH and CARD-FISH under Non-Steady and Pseudo-Steady State Conditions. New Biotechnol. 2013, 30, 756–762. [Google Scholar] [CrossRef]
- Huang, Y.; Wen, L.; Zhang, L.; Xu, J.; Wang, W.; Hu, H.; Xu, P.; Li, Z.; Tang, H. Community-integrated multi-omics facilitates screening and isolation of the organohalide dehalogenation microorganism. Innovation 2023, 4, 100355. [Google Scholar]
- Oyewusi, H.A.; Wahab, R.A.; Huyop, F. Whole genome strategies and bioremediation insight into dehalogenase-producing bacteria. Mol. Biol. Rep. 2021, 48, 2687–2701. [Google Scholar] [CrossRef]
- Wan, J.; Chen, K.; Chen, J.; Qin, Z.; Adrian, L.; Shen, C. Enhanced perchloroethene dechlorination by humic acids via increasing the dehalogenase activity of Dehalococcoides strains. FEMS Microbiol. Ecol. 2022, 98, fiac034. [Google Scholar] [CrossRef]
- Matturro, B.; Majone, M.; Aulenta, F.; Rossetti, S. Correlations between maximum reductive dechlorination rates and specific biomass parameters in Dehalococcoides mccartyi consortia enriched on chloroethenes PCE, TCE and cis-1, 2-DCE. FEMS Microbiol. Ecol. 2021, 97, fiab064. [Google Scholar] [CrossRef]
- Hlouchova, K.; Rudolph, J.; Pietari, J.M.H.; Behlen, L.S.; Copley, S.D. Pentachlorophenol Hydroxylase, a Poorly Functioning Enzyme Required for Degradation of Pentachlorophenol by Sphingobium Chlorophenolicum. Biochemistry 2012, 51, 3848–3860. [Google Scholar] [CrossRef]
- Rudolph, J.; Erbse, A.H.; Behlen, L.S.; Copley, S.D. A Radical Intermediate in the Conversion of Pentachlorophenol to Tetrachlorohydroquinone by Sphingobium Chlorophenolicum. Biochemistry 2014, 53, 6539–6549. [Google Scholar] [CrossRef] [PubMed]
- Xun, L.; Bohuslavek, J.; Cai, M. Characterization of 2,6-Dichloro-p-Hydroquinone 1,2-Dioxygenase (PcpA) of Sphingomonas Chlorophenolica ATCC 39723. Biochem. Biophys. Res. Commun. 1999, 266, 322–325. [Google Scholar] [CrossRef]
- Hayes, R.P.; Green, A.R.; Nissen, M.S.; Lewis, K.M.; Xun, L.; Kang, C. Structural Characterization of 2,6-Dichloro-p-Hydroquinone 1,2-Dioxygenase (PcpA) from Sphingobium Chlorophenolicum, a New Type of Aromatic Ring-Cleavage Enzyme. Mol. Microbiol. 2013, 88, 523–536. [Google Scholar] [CrossRef]
- Hasan, Q.A.K.M.; Takada, H.; Esaki, N.; Soda, K. Catalytic Action of L-2-halo Acid Dehalogenase on Long-chain L-2-haloalkanoic Acids in Organic Solvents. Biotechnol. Bioeng. 1991, 38, 1114–1117. [Google Scholar] [CrossRef]
- Beadle, C.A.; Smith, A.R.W. The Purification and Properties of 2,4-Dichlorophenol Hydroxylase from a Strain of Acinetobacter Species. Eur. J. Biochem. 1982, 123, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Nariya, T.; Ohtomo, R.; Fukuda, M.; Yano, K.; Takagi, M. Cloning and Sequencing of a Dehalogenase Gene Encoding an Enzyme with Hydrolase Activity Involved in the Degradation of γ-Hexachlorocyclohexane in Pseudomonas Paucimobilis. J. Bacteriol. 1993, 175, 6403–6410. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Miyauchi, K.; Damborsky, J.; Manova, K.; Ansorgova, A.; Takagi, M. Purification and Characterization of a Haloalkane Dehalogenase of a New Substrate Class from a Gamma-Hexachlorocyclohexane-Degrading Bacterium, Sphingomonas Paucimobilis UT26. Appl. Environ. Microbiol. 1997, 63, 3707–3710. [Google Scholar] [CrossRef] [PubMed]
- Hynková, K.; Nagata, Y.; Takagi, M.; Damborský, J. Identification of the Catalytic Triad in the Haloalkane Dehalogenase from Sphingomonas Paucimobilis UT26. FEBS Lett. 1999, 446, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Goldman, P. The Enzymatic Cleavage of the Carbon-Fluorine Bond in Fluoroacetate. J. Biol. Chem. 1965, 240, 3434–3438. [Google Scholar] [CrossRef] [PubMed]
- Goldman, P.; Milne, G.W. Carbon-Fluorine Bond Cleavage. II. Studies on the Mechanism of the Defluorination of Fluoroacetate. J. Biol. Chem. 1966, 241, 5557–5559. [Google Scholar] [CrossRef] [PubMed]
- Leisinger, T.; Bader, R. Microbial Dehalogenation of Synthetic Organohalogen Compounds: Hydrolytic Dehalogenases. Chimia 1993, 47, 116–121. [Google Scholar] [CrossRef]
- Kayser, M.F.; Vuilleumier, S. Dehalogenation of dichloromethane by dichloromethane dehalogenase/glutathione S-transferase leads to the formation of DNA adducts. J. Bacteriol. 2001, 183, 5209–5212. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shi, J.; Zhang, Y.; Yu, Z. Molecular docking and site-directed mutagenesis of dichloromethane dehalogenase to improve enzyme activity for dichloromethane degradation. Appl. Biochem. Biotechnol. 2020, 190, 487–505. [Google Scholar] [CrossRef]
- Suwa, Y.; Wright, A.D.; Fukimori, F.; Nummy, K.A.; Hausinger, R.P.; Holben, W.E.; Forney, L.J. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/alphaketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 1996, 62, 2464–2469. [Google Scholar] [CrossRef]
- Kitagawa, W.; Takami, S.; Miyauchi, K.; Masai, E.; Kamagata, Y.; Tiedje, J.M.; Fukuda, M. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 2002, 184, 509–518. [Google Scholar] [CrossRef]
- Poelarends, G.J.; Serrano, H.; Person, M.D.; Johnson, W.H., Jr.; Murzin, A.G.; Whitman, C.P. Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: Insights into the mechanistic, structural, and evolutionary relationship between isomer-specific 3-chloroacrylic acid dehalogenases. Biochemistry 2004, 43, 759–772. [Google Scholar] [CrossRef]
- Cai, M.; Xun, L. Organization and Regulation of Pentachlorophenol-Degrading Genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 2002, 184, 4672–4680. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.; Wohlfarth, G.; Diekert, G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: Cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 1998, 180, 4140–4145. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, J.; Reineke, W.; Knackmuss, H.J. Metabolism of 3-Chloro-, 4-Chloro-, and 3,5-Dichlorobenzoate by a Pseudomonad. Appl. Environ. Microbiol. 1979, 37, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Vaillancourt, F.; Bolin, J.; Eltis, L. The Ins and Outs of Ring-Cleaving Dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 241–267. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszyńska, D.; Hupert-Kocurek, K.; Guzik, U. Factors Affecting Activity of Catechol 2,3-Dioxygenase from 2-Chlorophenol-Degrading Stenotrophomonas Maltophilia Strain KB2. Biocatal. Biotransform. 2013, 31, 141–147. [Google Scholar] [CrossRef]
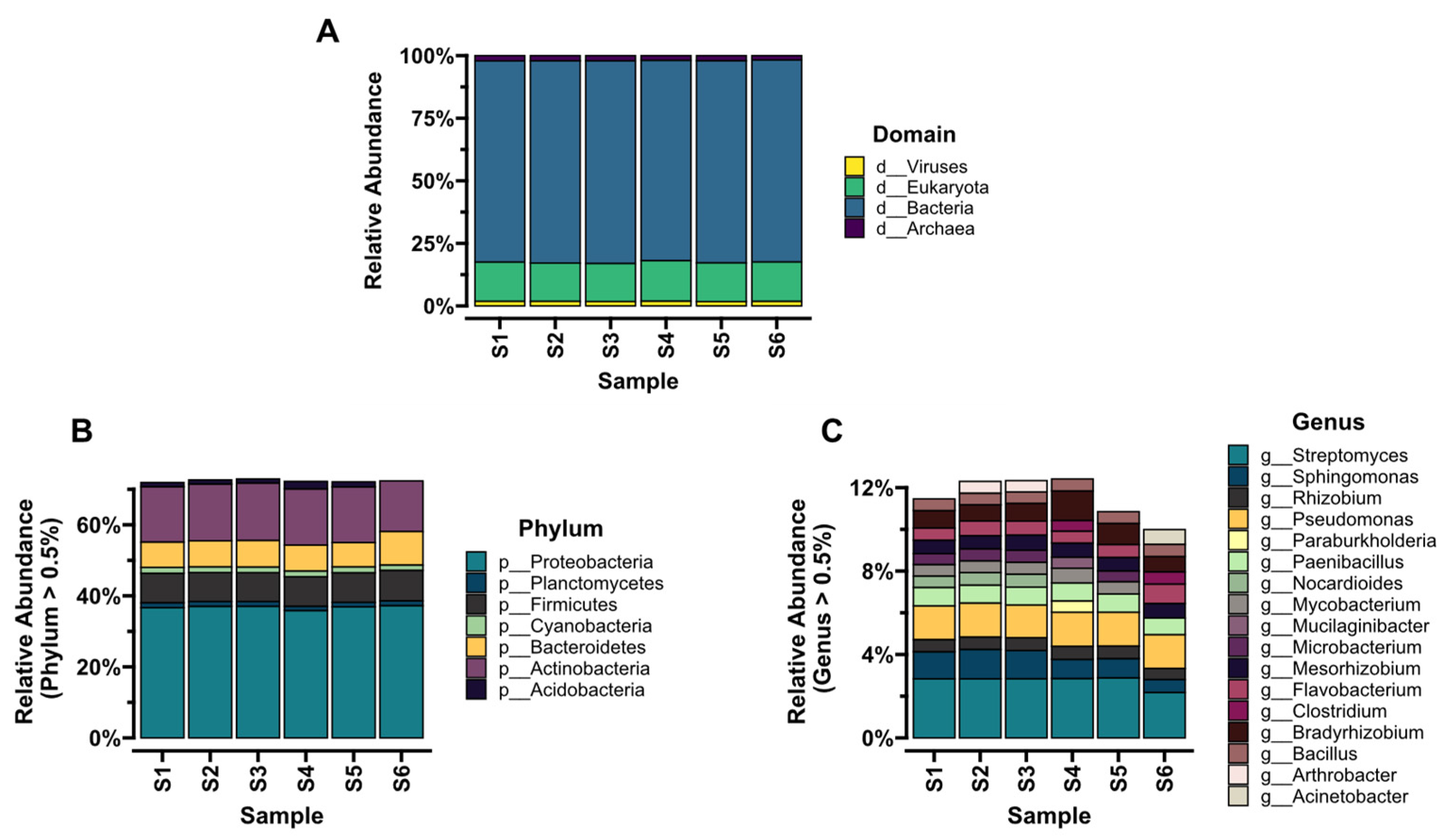

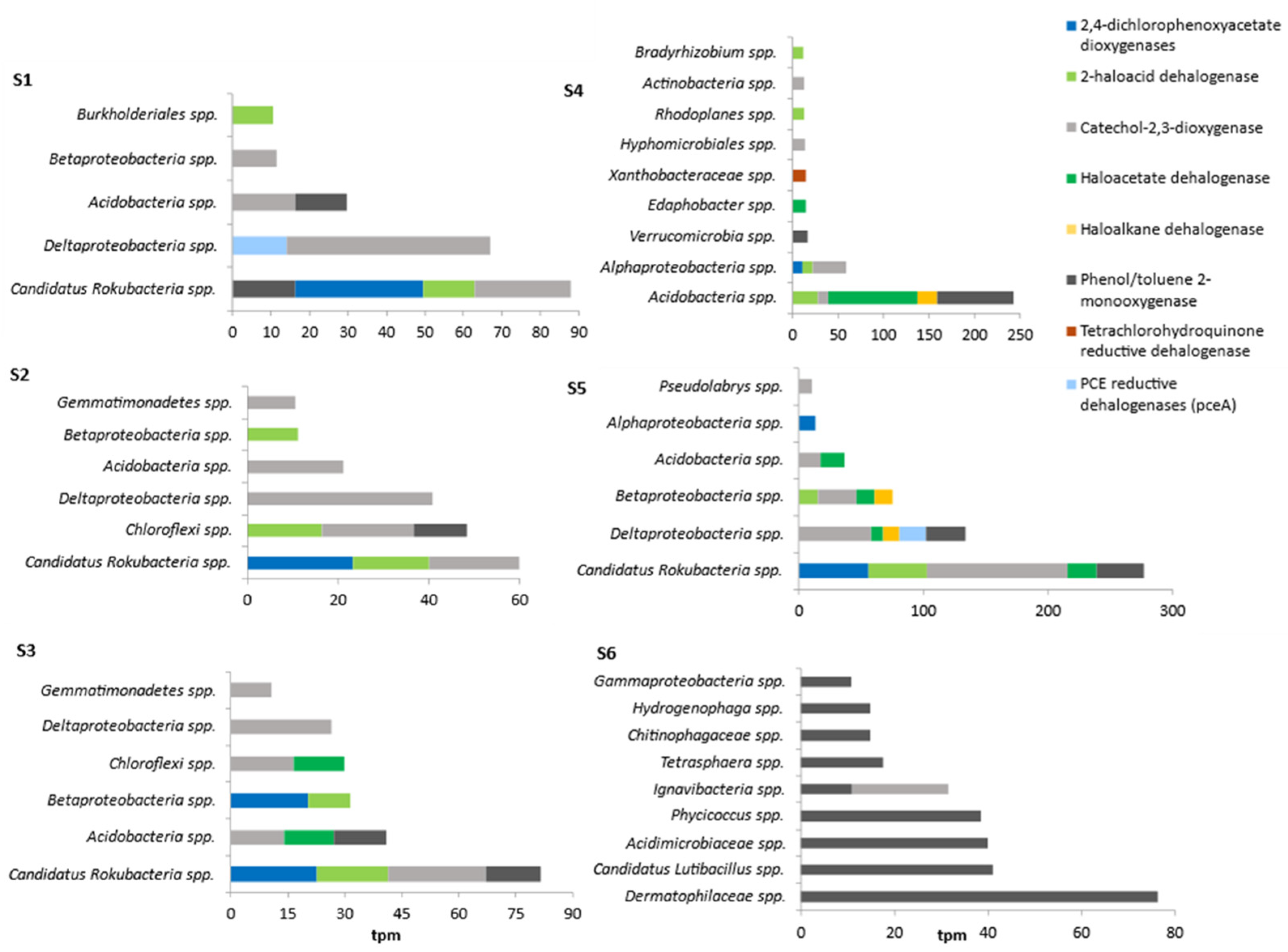
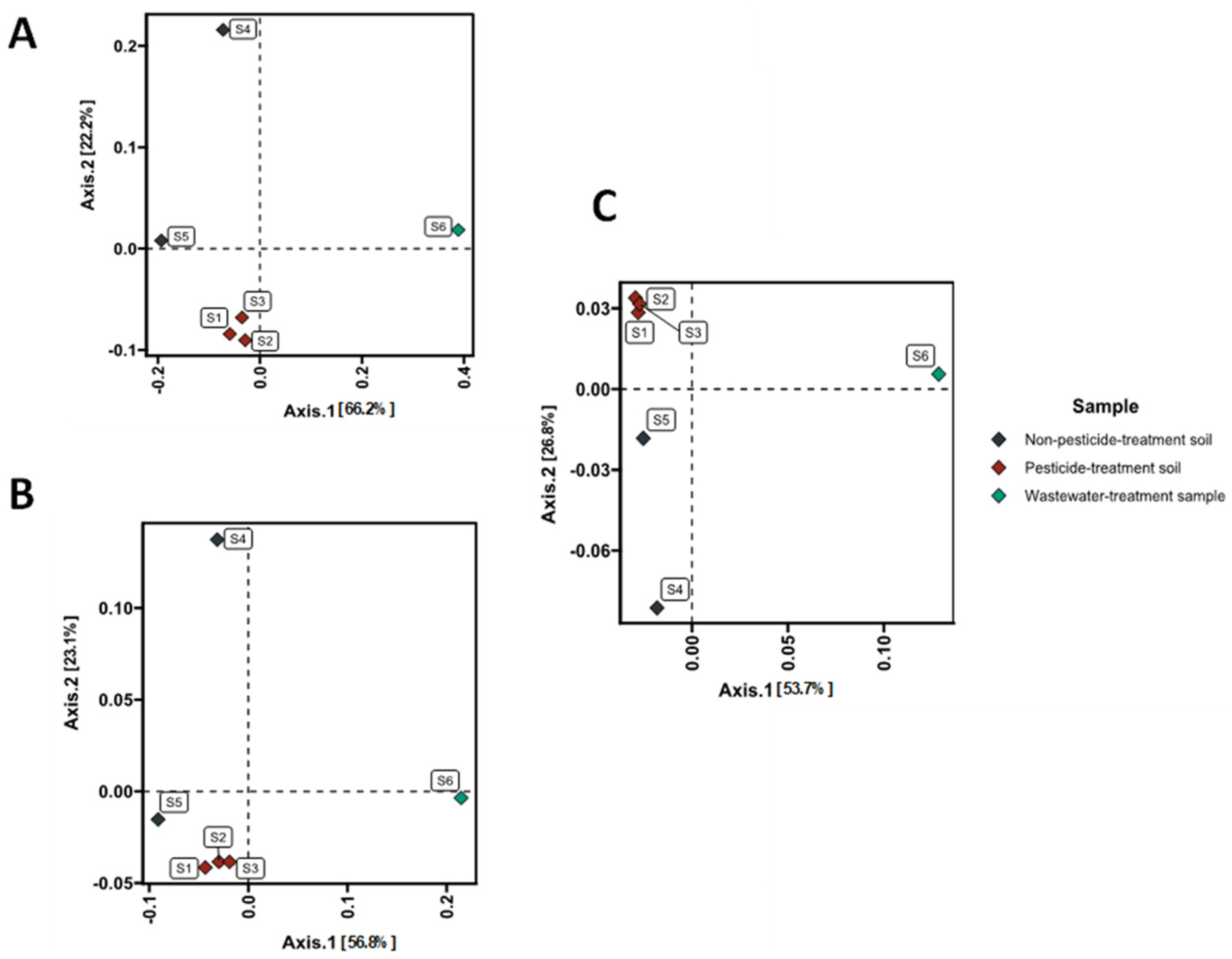
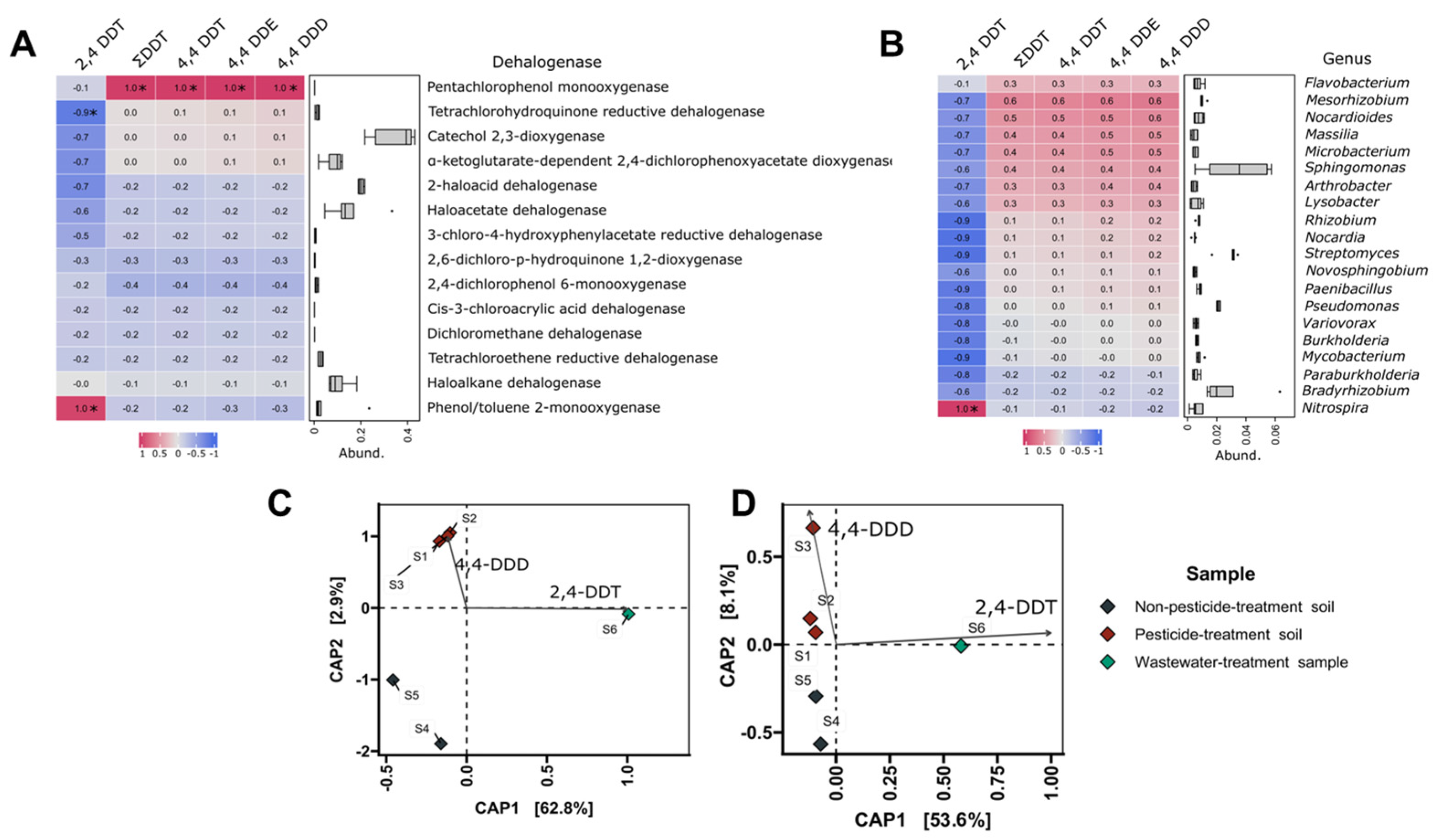

| Sample | As | Ba | Cr | Sn | Zn | Cd | Co | Cu | Mo | Ni | Pb | Fe |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mg/kg d.w. | % | |||||||||||
| S1 | <3 | 33 | 10 | <2 | 19 | <0.5 | 3 | 7 | <0.5 | 7 | 11 | 0.71 |
| S2 | <3 | 68 | 11 | <2 | 39 | <0.5 | 3 | 22 | <0.5 | 13 | 14 | 0.50 |
| S3 | 4 | 91 | 14 | 2 | 58 | <0.5 | 3 | 21 | <0.5 | 9 | 21 | 1.08 |
| S4 | 1.8 | 44 | 15 | <1 | 21 | <0.5 | 4 | 7 | <0.5 | 9 | 13 | 0.99 |
| S5 | <0.5 | 76 | 16 | <1 | 38 | <0.5 | 3 | 21 | <0.5 | 15 | 16 | 0.77 |
| S6 | 2.6 | 132 | 22 | 12 | 470 | 0.8 | 3 | 199 | 4 | 16 | 11 | 1.13 |
| Sample | F | Cl | Br | I |
|---|---|---|---|---|
| mg/kg | mg/kg | mg/kg | mg/kg | |
| S1 | <5.00 | 37.5 | <5.00 | <5.00 |
| S2 | 31.5 | 45.5 | <5.00 | 14.5 |
| S3 | 13.5 | 78.0 | <5.00 | <5.00 |
| S4 | 13.5 | 127 | <5.00 | <5.00 |
| S5 | 13.5 | 127 | <5.00 | <5.00 |
| Sample | 2,4- DDD | 2,4- DDT | 4,4′- DDD | 4,4′- DDE | 4,4′- DDT | Sum of 4 Isomers of DDT | Sum of 6 Isomers of DDT |
|---|---|---|---|---|---|---|---|
| mg/kg d.w. | |||||||
| S1 | <0.010 | <0.010 | <0.010 | <0.010 | <0.010 | <0.040 | <0.060 |
| S2 | <0.010 | 0.012 | 0.014 | 0.171 | 0.062 | 0.259 | 0.259 |
| S3 | <0.010 | <0.010 | <0.010 | <0.010 | <0.010 | <0.040 | <0.060 |
| S4 | <0.010 | <0.010 | <0.010 | <0.010 | <0.010 | <0.040 | <0.060 |
| S5 | <0.010 | <0.010 | <0.010 | <0.010 | <0.010 | <0.040 | <0.060 |
| S6 | 0.011 | 0.039 | <0.010 | 0.016 | 0.014 | 0.069 | 0.080 |
| Type of Enzyme | Name of Enzyme | Reaction | Characteristic of Dehalogenation Mechanism | References |
|---|---|---|---|---|
| Metabolic dehalogenases | pentachlorophenol monooxygenase | H+ + NADPH + O2 + pentachlorophenol = 2,3,5,6-tetrachloro-1,4-benzoquinone + chloride + H2O + NADP+ | Pentachlorophenol monooxygenase belongs to oxidoreductases (monooxygenases) responsible for dechlorination of pentachlorophenol to tetrachlorobenzoquinone. | [55,56] |
| 2,6-dichloro-p-hydroquinone 1,2-dioxygenase | 2,6-dichlorohydroquinone + O2 + H2O = 2-chloromaleylacetate + hydrochloric acid | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase is an oxidoreductase (dioxygenase) and is responsible for opening the aromatic ring of 2,6-dichloro-p-hydroquinone (cleaves the aromatic ring between a hydroxyl group and a chlorine group) to produce 2-chloromaleylacetate. | [57,58] | |
| 2-haloacid dehalogenase | 2-haloacid + H2O = 2-hydroxyacid + halide | Dehalogenates 2-haloalkanoic acids to the hydroxyalkanoic acids, with inversion of configuration at C-2. Acts on 2-haloalkanoic acids whose carbon chain lengths are five or less. | [46,59] | |
| 2,4-dichlorophenol 6-monooxygenase | 2,4-dichlorophenol + H+ + NADPH + O2 = 3,5-dichlorocatechol + H2O + NADP+ | 2,4-dichlorophenol 6-monooxygenase is an oxidoreductase (monooxygenase). Transforms 2,4-dichlorophenol (2,4-DCP) into 3,5-dichlorocatechol, using NADH or NADPH as one donor and incorporating an atom of oxygen into the other donor. | [60] | |
| haloalkane dehalogenase | 1-haloalkane + H2O = a primary alcohol + halide | Haloalkane dehalogenase is a hydrolase, has a broad substrate specificity, and catalyzes hydrolytic cleavage of carbon–halogen bonds in halogenated aliphatic compounds. Acts on a wide range of 1-haloalkanes, haloalcohols, haloalkenes, and some haloaromatic compounds, leading to the formation of the corresponding primary alcohols, halide ions, and protons. | [61,62,63] | |
| haloacetate dehalogenase | a haloacetate + H2O = a halide anion + glycolate + H+ | Haloacetate dehalogenase is a hydrolase and catalyzes hydrolytic cleavage of carbon–halogen bonds in fluoroacetate (haloacetate dehalogenase DehH1), and chloro-, bromo-, and iodoacetate, but not fluoroacetate (haloacetate dehalogenase DehH2), yielding halide and glycolate. | [13,64,65,66] | |
| dichloromethane dehalogenase | dichloromethane + H2O = 2 chloride + formaldehyde + 2 H+ | Dichloromethane dehalogenase is a lyase enzyme that generates formaldehyde and belongs to the family of glutathione S-transferases. Glutathione is required for its activity. | [67,68] | |
| alpha-ketoglutarate-dependent 2,4-dichlorophenoxyacetate dioxygenase | 2,4-dichlorophenoxyacetate + 2-oxoglutarate + O2 = 2,4-dichlorophenol + CO2 + glyoxylate + succinate | Alpha-ketoglutarate-dependent 2,4-dichlorophenoxyacetate dioxygenase is an oxidoreductase (dioxygenase). Involved in degradation of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D). Is also able to degrade 2-methyl-4-chlorophenoxyacetic acid and 3-chlorobenzoic acid. | [69,70] | |
| cis-3-chloroacrylic acid dehalogenase | Cis-3-chloroacrylic acid dehalogenase is a hydrolase, which is involved in the hydrolytic cleavage of the vinylic carbon–halogen bond in 3-haloacrylates to afford malonate semialdehyde. | [71] | ||
| tetrachlorohydroquinone reductive dehalogenase | 2,6-dichlorohydroquinone + chloride + glutathione disulfide + H+ = 2,3,6-trichlorohydroquinone + 2 glutathione | Tetrachlorohydroquinone reductive dehalogenase is an oxidoreductase and is responsible for sequential reduction of tetrachloro-p-hydroquinone to monochlorophenol, using glutathione as the reducing agent. | [72] | |
| tetrachloroethene reductive dehalogenase | AH2 + tetrachloroethene = A + chloride + H+ + trichloroethene | Tetrachloroethene reductive dehalogenase is an oxidoreductase and a member of the reductive dehalogenase enzyme family. Catalyzes the reductive dechlorination of tetrachloroethene (PCE) to trichloroethene (TCE) and of trichloroethene to cis-1,2-dichloroethene (DCE). Can also use various chlorinated ethanes such as tetrachloroethane, pentachloroethane, and hexachloroethane. A—hydrogen acceptor; AH2—hydrogen donor. | [73] | |
| Cometabolic dehalogenases | phenol monooxygenase | phenol + NADPH + H+ + O2 = catechol + NADP+ + H2O | Phenol monooxygenase has a broad substrate specificity and is an oxidoreductase (monooxygenase). It hydroxylates a variety of substituted phenols, such as fluoro- and chloro-phenols. | [13,43] |
| toluene monooxygenase | H+ + NADH + O2 + toluene = 4-methylphenol + H2O + NAD+ | Toluene monooxygenase is an oxidoreductase (monooxygenase) and uses the reducing power of NADH to split dioxygen and incorporate a single oxygen atom in the form of a hydroxyl group into the halogenated substrate, creating an unstable halohydrin. Subsequently, the unstable halohydrin spontaneously eliminates a halide ion. | [13,14,43] | |
| catechol 2,3-dioxygenase | catechol + O2 = 2-hydroxy-6-oxohexa-2,4-dienoate + H+ | Catechol 2,3-dioxygenase belongs to the extradiol dioxygenase family and oxidoreductase class, and catalyzes ring cleavage of catechol and chloro-, methyl-, and ethyl-substituted catechols in a meta fashion. | [47,48,74,75,76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łomża, P.; Krucoń, T.; Tabernacka, A. Potential of Microbial Communities to Perform Dehalogenation Processes in Natural and Anthropogenically Modified Environments—A Metagenomic Study. Microorganisms 2023, 11, 1702. https://doi.org/10.3390/microorganisms11071702
Łomża P, Krucoń T, Tabernacka A. Potential of Microbial Communities to Perform Dehalogenation Processes in Natural and Anthropogenically Modified Environments—A Metagenomic Study. Microorganisms. 2023; 11(7):1702. https://doi.org/10.3390/microorganisms11071702
Chicago/Turabian StyleŁomża, Pola, Tomasz Krucoń, and Agnieszka Tabernacka. 2023. "Potential of Microbial Communities to Perform Dehalogenation Processes in Natural and Anthropogenically Modified Environments—A Metagenomic Study" Microorganisms 11, no. 7: 1702. https://doi.org/10.3390/microorganisms11071702
APA StyleŁomża, P., Krucoń, T., & Tabernacka, A. (2023). Potential of Microbial Communities to Perform Dehalogenation Processes in Natural and Anthropogenically Modified Environments—A Metagenomic Study. Microorganisms, 11(7), 1702. https://doi.org/10.3390/microorganisms11071702






