Roles of the Crp/Fnr Family Regulator ArcR in the Hemolysis and Biofilm of Staphylococcus aureus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Primers, and Growth Conditions
2.2. Construction of the S. aureus Mutant and Complementary Strains
2.3. Detection of Biofilm Production
2.4. Confocal Laser Scanning Microscopy (CLSM) for the Observation of Biofilm Production
2.5. Determination of Hemolytic Activity
2.6. Total RNA Extraction, cDNA Generation, qRT-PCR, and RNA-seq
2.7. Construction of LacZ Reporter Vectors
2.8. β-Galactosidase Activity Assay
2.9. PIA Detection
2.10. ArcR Protein Expression and Purification
2.11. Electrophoretic Mobility Shift Assay (EMSA)
2.12. Statistical Analysis
3. Results
3.1. Deletion of arcR Increased the Hemolytic Activity in S. aureus
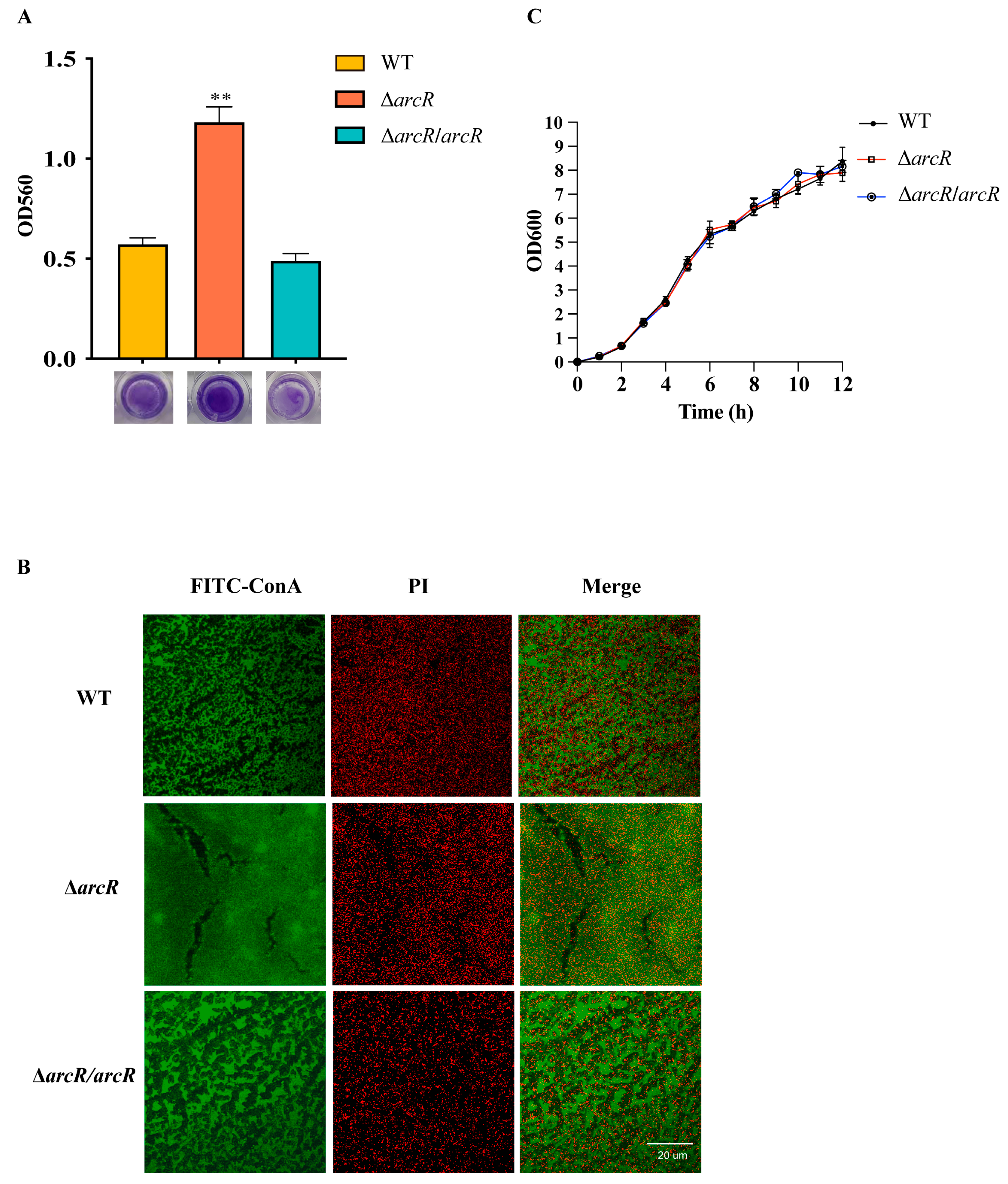
3.2. Deletion of arcR Increased Biofilm Formation in S. aureus
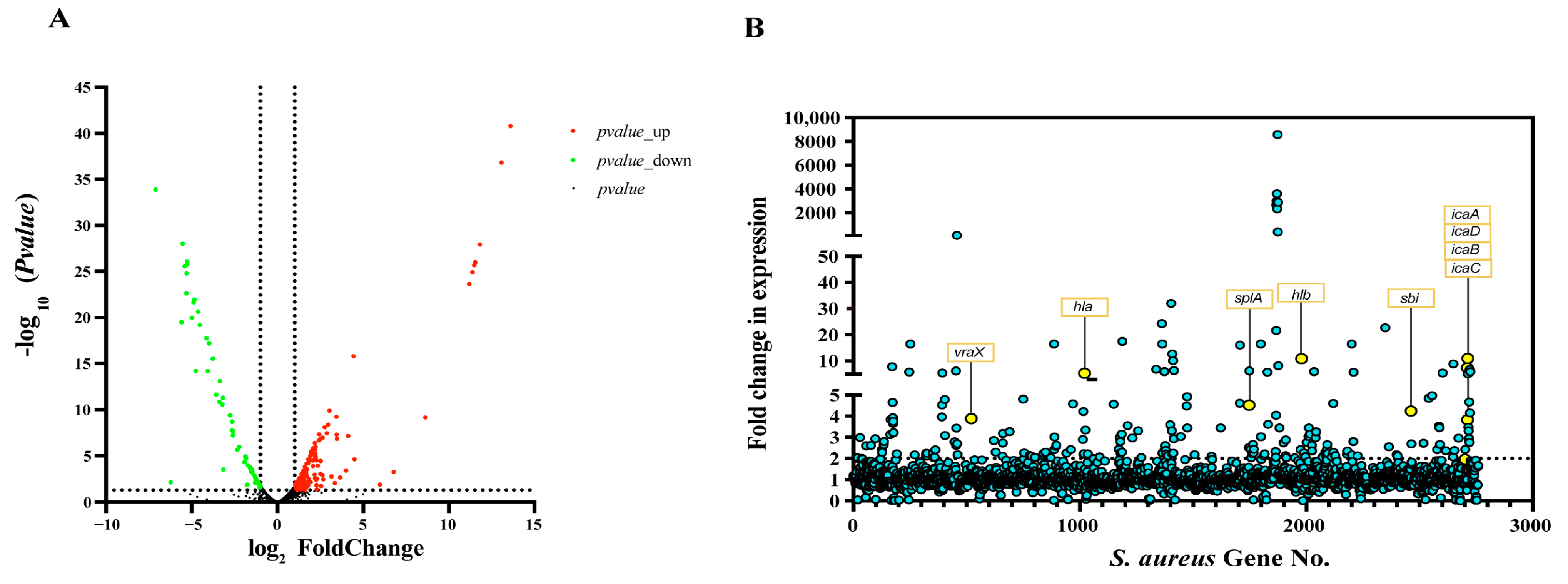
3.3. Gene Expression Profiling of the ΔarcR Mutant
3.4. ArcR Regulates the Transcription of HEMOLYTIC Genes
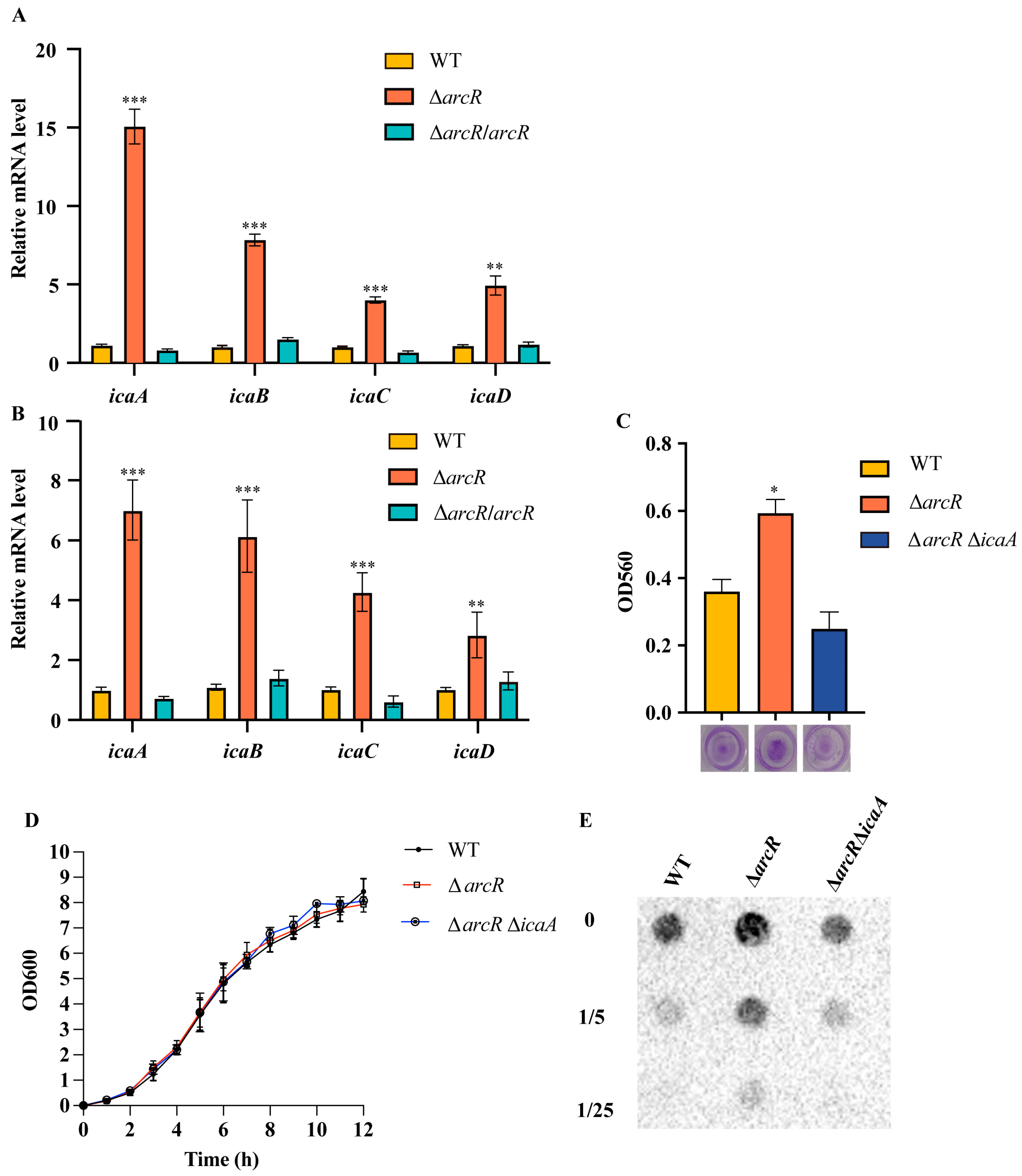
3.5. ArcR Influences Biofilm Formation through ica Operon
3.6. ArcR Could Bind to the Promoter Regions of the ica Operon and hla
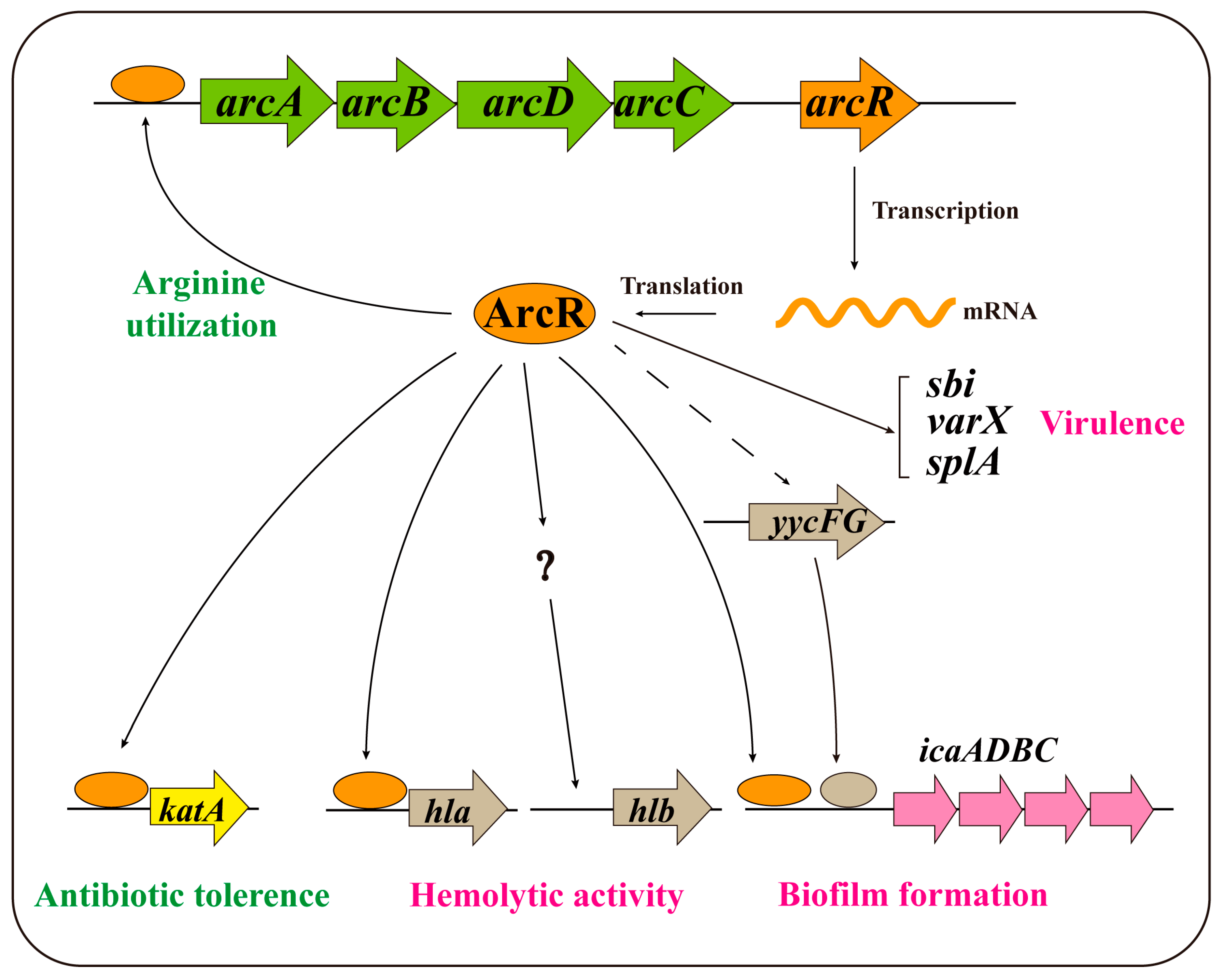
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Stobberingh, E.E. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 2008, 8, 747–763. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhou, X.; Li, W.; Zhang, H.; Zhang, B.; Li, G.; Liu, B.; Deng, X.; Peng, L. Diosmetin inhibits the expression of alpha-hemolysin in Staphylococcus aureus. Antonie Van Leeuwenhoek 2015, 108, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Gudeta, D.D.; Lei, M.G.; Lee, C.Y. Contribution of hla Regulation by SaeR to Staphylococcus aureus USA300 Pathogenesis. Infect. Immun. 2019, 87, e00231-19. [Google Scholar] [CrossRef]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995, 14, 4569–4577. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.Q.; Willard, J.; Yeaman, M.R.; Cheung, A.L.; Bayer, A.S. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 2006, 194, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.L.; Jefferson, K.K. Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog. 2014, 10, e1004292. [Google Scholar] [CrossRef] [PubMed]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Débarbouillé, M.; Penadés, J.R.; Lasa, I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef]
- Beenken, K.E.; Blevins, J.S.; Smeltzer, M.S. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 2003, 71, 4206–4211. [Google Scholar] [CrossRef]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef]
- Ma, R.; Qiu, S.; Jiang, Q.; Sun, H.; Xue, T.; Cai, G.; Sun, B. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 257–267. [Google Scholar] [CrossRef]
- Tormo, M.A.; Martí, M.; Valle, J.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 2005, 187, 2348–2356. [Google Scholar] [CrossRef]
- Conlon, K.M.; Humphreys, H.; O’Gara, J.P. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 2004, 186, 6208–6219. [Google Scholar] [CrossRef]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The Toxin-Antitoxin MazEF Drives Staphylococcus aureus Biofilm Formation, Antibiotic Tolerance, and Chronic Infection. mBio 2019, 10, e01658-19. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, J.; Peng, Q.; Liu, Y.; Lei, L.; Zhang, H. The Role of Staphylococcus aureus YycFG in Gene Regulation, Biofilm Organization and Drug Resistance. Antibiotics 2021, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, F.; Zhang, H.; Lei, L. Staphylococcus aureus biofilm organization modulated by YycFG two-component regulatory pathway. J. Orthop. Surg. Res. 2019, 14, 10. [Google Scholar] [CrossRef]
- Bellucci, M.; Ofiteru, I.D.; Graham, D.W.; Head, I.M.; Curtis, T.P. Low-dissolved-oxygen nitrifying systems exploit ammonia-oxidizing bacteria with unusually high yields. Appl. Environ. Microbiol. 2011, 77, 7787–7796. [Google Scholar] [CrossRef]
- Kiley, P.J.; Beinert, H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 2003, 6, 181–185. [Google Scholar] [CrossRef]
- Stelling, C.R.; Orsi, R.H.; Wiedmann, M. Complementation of Listeria monocytogenes null mutants with selected Listeria seeligeri virulence genes suggests functional adaptation of Hly and PrfA and considerable diversification of prfA regulation in L. seeligeri. Appl. Environ. Microbiol. 2010, 76, 5124–5139. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.J.; Bochkareva, A.; Rolfe, M.D.; Hunt, D.M.; Kahramanoglou, C.; Braun, Y.; Rodgers, A.; Blockley, A.; Coade, S.; Lougheed, K.E.A.; et al. Cmr is a redox-responsive regulator of DosR that contributes to M. tuberculosis virulence. Nucleic Acids Res. 2017, 45, 6600–6612. [Google Scholar] [CrossRef] [PubMed]
- Akhter, Y.; Tundup, S.; Hasnain, S.E. Novel biochemical properties of a CRP/FNR family transcription factor from Mycobacterium tuberculosis. Int. J. Med. Microbiol. 2007, 297, 451–457. [Google Scholar] [CrossRef]
- Fazli, M.; O’Connell, A.; Nilsson, M.; Niehaus, K.; Dow, J.M.; Givskov, M.; Ryan, R.P.; Tolker-Nielsen, T. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 2011, 82, 327–341. [Google Scholar] [CrossRef]
- Salazar, J.K.; Wu, Z.; McMullen, P.D.; Luo, Q.; Freitag, N.E.; Tortorello, M.L.; Hu, S.; Zhang, W. PrfA-like transcription factor gene lmo0753 contributes to L-rhamnose utilization in Listeria monocytogenes strains associated with human food-borne infections. Appl. Environ. Microbiol. 2013, 79, 5584–5592. [Google Scholar] [CrossRef]
- Śmiga, M.; Olczak, T. PgRsp Is a Novel Redox-Sensing Transcription Regulator Essential for Porphyromonas gingivalis Virulence. Microorganisms 2019, 7, 623. [Google Scholar] [CrossRef] [PubMed]
- Makhlin, J.; Kofman, T.; Borovok, I.; Kohler, C.; Engelmann, S.; Cohen, G.; Aharonowitz, Y. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 2007, 189, 5976–5986. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Fan, Z.; Li, Y.; Li, Z.; Du, B.; Liu, S.; Cui, X.; Zhang, R.; Zhao, H.; Feng, Y.; et al. ArcR contributes to tolerance to fluoroquinolone antibiotics by regulating katA in Staphylococcus aureus. Front. Microbiol. 2023, 14, 1106340. [Google Scholar] [CrossRef]
- Kreiswirth, B.N.; Löfdahl, S.; Betley, M.J.; O’Reilly, M.; Schlievert, P.M.; Bergdoll, M.S.; Novick, R.P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983, 305, 709–712. [Google Scholar] [CrossRef]
- Bae, T.; Schneewind, O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006, 55, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Anton, A.I.; Barry, P.; Alfonso, B.; Fang, Y.; Novick, R.P. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70, 6076–6085. [Google Scholar] [CrossRef] [PubMed]
- Schneewind, O.; Mihaylova-Petkov, D.; Model, P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993, 12, 4803–4811. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, X.; Liu, X.; Chen, C.; Sun, B. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob. Agents Chemother. 2015, 59, 1352–1355. [Google Scholar] [CrossRef]
- Brückner, R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 151, 1–8. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Lin, M.H.; Shu, J.C.; Huang, H.Y.; Cheng, Y.C. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE 2012, 7, e34388. [Google Scholar] [CrossRef] [PubMed]
- Koppenhöfer, S.; Lang, A.S. Interactions among Redox Regulators and the CtrA Phosphorelay in Dinoroseobacter shibae and Rhodobacter capsulatus. Microorganisms 2020, 8, 562. [Google Scholar] [CrossRef] [PubMed]
- Ke, N.; Kumka, J.E.; Fang, M.; Weaver, B.; Burstyn, J.N.; Bauer, C.E. Redox Brake Regulator RedB and FnrL Function as Yin-Yang Regulators of Anaerobic-Aerobic Metabolism in Rhodobacter capsulatus. Microbiol. Spectr. 2022, 10, e0235422. [Google Scholar] [CrossRef]
- Giachino, P.; Engelmann, S.; Bischoff, M. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 2001, 183, 1843–1852. [Google Scholar] [CrossRef]
- Wu, S.; Liu, Y.; Lei, L.; Zhang, H. Antisense yycG modulates the susceptibility of Staphylococcus aureus to hydrogen peroxide via the sarA. BMC Microbiol. 2021, 21, 160. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Chong, A.S.; Montgomery, C.P. Importance of B Lymphocytes and the IgG-Binding Protein Sbi in Staphylococcus aureus Skin Infection. Pathogens 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.K.; Reuter, M.; Barthel, D.; Böhm, S.; van den Elsen, J.; Kraiczy, P.; Zipfel, P.F.; Skerka, C. Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PLoS ONE 2012, 7, e47638. [Google Scholar] [CrossRef]
- Yan, J.; Han, D.; Liu, C.; Gao, Y.; Li, D.; Liu, Y.; Yang, G. Staphylococcus aureus VraX specifically inhibits the classical pathway of complement by binding to C1q. Mol. Immunol. 2017, 88, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Stec-Niemczyk, J.; Pustelny, K.; Kisielewska, M.; Bista, M.; Boulware, K.T.; Stennicke, H.R.; Thogersen, I.B.; Daugherty, P.S.; Enghild, J.J.; Baczynski, K.; et al. Structural and functional characterization of SplA, an exclusively specific protease of Staphylococcus aureus. Biochem. J. 2009, 419, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Salgado-Pabon, W.; Meyerholz, D.K.; White, M.J.; Schlievert, P.M.; Horswill, A.R. The Spl Serine Proteases Modulate Staphylococcus aureus Protein Production and Virulence in a Rabbit Model of Pneumonia. mSphere 2016, 1, e00208-16. [Google Scholar] [CrossRef]



| Strain or Plasmid | Relevant Characteristic | Source or Reference |
|---|---|---|
| S. aureus | ||
| RN4220 | 8325-4, r−, restriction-deficient mutagenized RN450 | [34] |
| NCTC8325 WT | NCTC8325 wild-type strain | NASA a |
| NCTC8325 ΔarcR | NCTC8325 arcR deletion mutant | [33] |
| NCTC8325 ΔarcR ΔicaA | NCTC8325 arcR icaA double mutant strain | This study |
| USA300 | CA-MRSA, wild-type | NARSA |
| USA300 ΔarcR | USA300 arcR deletion mutant | This study |
| E. coli | ||
| DH5α | E. coli host for cloning | Vazyme |
| BL21 (DE3) | Expression strain; F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Vazyme |
| Plasmid | ||
| pBTs | temp-sensitive plasmid, E. coli and S. aureus shutle vector, Apr, Cmr, for the construction of allelic-exchange mutants | [35] |
| pBTs-arcR | pBTs derivative, for arcR deletion | This study |
| pBTs-icaA | pBTs derivative, for icaA deletion | This study |
| pCN51 | E. coli/S. aureus expression vector, Apr, Emr | [36] |
| pCN51-arcR | Complementary arcR in S. aureus | This study |
| pET28a | Expression vector with His tag in E. coli, Kmr | Addgene |
| pET28a-arcR | His6-ArcR expression vector | This study |
| pOS1-lacZ | E. coli and S. aureus shuttle vector, with lacZ ORF lacking first 6 amino acids, Apr, Cmr. | [37] |
| pOS1hla | pOS1-lacZ plasimd derivative, including 546-bp sequence of hla | This study |
| pOS1hlb | pOS1-lacZ plasimd derivative, including 412-bp sequence of hlb | This study |
| pOS1sbi | pOS1-lacZ plasimd derivative, including 311-bp sequence of sbi | This study |
| pOS1vraX | pOS1-lacZ plasimd derivative, including 287-bp sequence of vraX | This study |
| pOS1splA | pOS1-lacZ plasimd derivative, including sequence of splA | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, T.; Fan, Z.; Li, Y.; Li, Z.; Zhao, H.; Feng, Y.; Xue, G.; Cui, J.; Yan, C.; Gan, L.; et al. Roles of the Crp/Fnr Family Regulator ArcR in the Hemolysis and Biofilm of Staphylococcus aureus. Microorganisms 2023, 11, 1656. https://doi.org/10.3390/microorganisms11071656
Fu T, Fan Z, Li Y, Li Z, Zhao H, Feng Y, Xue G, Cui J, Yan C, Gan L, et al. Roles of the Crp/Fnr Family Regulator ArcR in the Hemolysis and Biofilm of Staphylococcus aureus. Microorganisms. 2023; 11(7):1656. https://doi.org/10.3390/microorganisms11071656
Chicago/Turabian StyleFu, Tongtong, Zheng Fan, Yujie Li, Zhoufei Li, Hanqing Zhao, Yanling Feng, Guanhua Xue, Jinghua Cui, Chao Yan, Lin Gan, and et al. 2023. "Roles of the Crp/Fnr Family Regulator ArcR in the Hemolysis and Biofilm of Staphylococcus aureus" Microorganisms 11, no. 7: 1656. https://doi.org/10.3390/microorganisms11071656
APA StyleFu, T., Fan, Z., Li, Y., Li, Z., Zhao, H., Feng, Y., Xue, G., Cui, J., Yan, C., Gan, L., Feng, J., Yuan, J., & You, F. (2023). Roles of the Crp/Fnr Family Regulator ArcR in the Hemolysis and Biofilm of Staphylococcus aureus. Microorganisms, 11(7), 1656. https://doi.org/10.3390/microorganisms11071656






