Abstract
Bifidobacterium longum is considered a microorganism with probiotic potential, which has been extensively studied, but these probiotic effects are strain dependent. This work aims to characterize the probiotic potential, based on the biochemical and genomic functionality, of B. longum LBUX23, isolated from neonates’ feces. B. longum LBUX23 contains one circular genome of 2,287,838 bp with a G+C content of 60.05%, no plasmids, no CRISPR-Cas operon, possesses 56 tRNAs, 9 rRNAs, 1 tmRNA and 1776 coding sequences (CDSs). It has chromosomally encoded resistance genes to ampicillin and dicloxacillin, non-hemolytic activity, and moderate inhibition of Escherichia coli ATCC 25922 and to some emergent pathogen’s clinical strains. B. longum LBUX23 was able to utilize lactose, sucrose, fructooligosaccharides (FOS), and lactulose. The maximum peak of bacterial growth was observed in sucrose and FOS at 6 h; in lactose and lactulose, it was shown at 8 h. B. longum LBUX23 can survive in gastrointestinal conditions (pH 4 to 7). A decrease in survival (96.5 and 93.8%) was observed at pH 3 and 3.5 during 120 min. argC, argH, and dapA genes could be involved in this tolerance. B. longum LBUX23 can also survive under primary and secondary glyco- or tauro-conjugated bile salts, and a mixture of bile salts due to the high extracellular bile salt hydrolase (BSH) activity (67.3 %), in taurocholic acid followed by taurodeoxycholic acid (48.5%), glycocholic acid (47.1%), oxgall (44.3%), and glycodeoxycholic acid (29.7%) probably due to the presence of the cbh and gnlE genes which form an operon (start: 119573 and end: 123812). Low BSH activity was determined intracellularly (<7%), particularly in glycocholic acid; no intracellular activity was shown. B. longum LBUX23 showed antioxidant effects in DPPH radical, mainly in intact cells (27.4%). In the case of hydroxyl radical scavenging capacity, cell debris showed the highest reduction (72.5%). In the cell-free extract, superoxide anion radical scavenging capacity was higher (90.5%). The genome of B. longum LBUX23 contains PNPOx, AhpC, Bcp, trxA, and trxB genes, which could be involved in this activity. Regarding adherence, it showed adherence up to 5% to Caco-2 cells. B. longum LBUX23 showed in vitro potential probiotic properties, mainly in BSH activity and antioxidant capacity, which indicates that it could be a good candidate for antioxidant or anti-cholesterol tests using in vivo models.
1. Introduction
Bifidobacterium longum is a human symbiont bacterium considered one of the most common and prevalent species in the healthy human gut, whose richness changes according to age [1]. Three subspecies have been identified: B. longum, Bifidobacterium infantis, and B. longum subsp. suis, commonly found in the porcine gut, while B. longum subsp. infantis and longum have been isolated from infant and adult intestines, respectively [2,3]. Several probiotic properties have been described for B. longum strains in both in vitro experiments and in vivo animal studies. These have included antiviral, metabolic, antioxidant, immunomodulatory, and neuromodulating effects, among others. However, these effects are strain dependent [4].
The scientific evidence suggests that B. longum has antioxidant properties and can reduce oxidative stress in the intestine. Some bifidobacteria possess peroxide reductase (PrxR) and NADPH oxidases (NOXs), which are capable of efficiently eliminating reactive oxygen species (ROS) and enhancing the tolerance of bifidobacteria to oxygen. When bifidobacteria exhibit higher resistance to oxygen, their capacity for scavenging free radicals becomes more potent [5]. An excess of ROS interacts with several biomolecules, resulting in dysfunction and cellular damage, leading to structural and functional disorders, including cancer, neurodegenerative diseases, diabetes mellitus, atherosclerosis, heart and/or brain failure, rheumatoid arthritis, inflammatory bowel diseases, colon cancer, Alzheimer’s disease, and aging among others [6,7]. The antioxidant properties of probiotics include their ability to generate different antioxidant metabolites such as butyrate, glutathione (GSH), and folate. Probiotics can also hinder the growth of harmful intestinal pathogens and lower post-meal lipid levels, which contribute to oxidative damage. Since appropriate antioxidant mechanisms play a vital role in maintaining our body’s proper functioning, it is important to extensively study probiotics for their potential antioxidant benefits [7]. In this context, the plasma lipid profile is another important factor modulated by the probiotic strains through antioxidant and enzyme activities. According to Jiang et al. (2021), it was proposed that B. longum strains possess a unique capacity to individually influence cholesterol reduction. These effects are influenced by specific bacterial traits, such as their ability to deconjugate bile salts, assimilate cholesterol, and express genes involved in cholesterol metabolism [8]. The hypocholesterolemic activity of B. longum can be exerted through two different mechanisms: i) bile salt deconjugation by bile salt hydrolase and ii) cholesterol assimilation by specific absorption [9]. BSH is an enzyme responsible for breaking down glycine- and/or taurine-conjugated bile salts into amino acid residues and liberating bile acids through hydrolysis. Conjugated bile salts usually move through enterohepatic circulation; due to deconjugation by BSH, the bile salts become less soluble and are excreted by the feces, which generates a replacement of the excreted bile salts with new synthesized bile salts at the expense of stored cholesterol [10,11,12,13].
Among the criteria for selecting probiotics or candidate strains are the identification, safety, functionality characterization, and healthy benefits. Currently, the most suitable method for identifying probiotics and attributing their effects to specific microbial strains appears to be through the analysis of phenotypic and genotypic data. Therefore, properly identifying a probiotic strain requires a comprehensive assessment of its morphological, physiological, and biochemical characteristics, along with considerations of its genetic profile [14]. The functionality characterization involves assessing the ability to withstand various stress conditions, such as salivary/gastric enzymes, low pH, gastric juice, and bile. This evaluation helps to elucidate the potential health-promoting effects of the tested substances. The aim of this study was to characterize the probiotic potential, based on the biochemical and genomic functionality, of B. longum LBUX23, a strain isolated from neonates’ feces. To assign probiotic effects in vitro and to generate evidence so that in the future, it can be tested in an in vivo model and/or clinical trial and could be used in biotechnological applications, human health, and food science.
2. Materials and Methods
2.1. Isolation and Strain Propagation
This study was performed using stool samples from clinically healthy neonates delivered vaginally at Obstetrics Gynecology Hospital No. 4, "Luis Castelazo Ayala". The first sample was collected from birth to hospital discharge. This protocol was accepted by the Ethics Commission of the Mexican Social Security Institute (IMSS), document number 36068. Prior to inclusion, written informed consent was obtained from each donor sample’s parent or guardian, ensuring that fecal samples were collected under ethical conditions during that time. The samples (1 g) were diluted in 1 mL of reduced-PBS (phosphate-buffered saline) and kept at −80 °C until analyses were conducted. Subsequently, the samples were plated in triplicate onto TPY medium agar plates, which were incubated anaerobically in an anaerobic chamber (85% nitrogen, 10% hydrogen, 5% carbon dioxide) (Forma anaerobic system Model 1025, Thermo Scientific, Waltham, MA, USA) at 37 °C for 48 h. The chosen isolates were subjected to optical microscopy for the examination of their morphology and Gram staining outcomes. Furthermore, their catalase activity was assessed. The Gram-positive and catalase-negative isolates with typical bifidobacteria shapes were genotypically identified [15]. The propagation of the Bifidobacterium strains was performed in TPY medium in an anaerobic chamber at 37 °C for 24–48 h.
2.2. DNA Extraction and Genotypic Identification
The Wizard® Genomic DNA Purification Kit (Promega, Tokyo, Japan) was used to isolate total genomic DNA according to the manufacturer’s instructions. The 16S rRNA sequencing was conducted at the Divisional Molecular Biology Laboratory of Universidad Autonoma Metropolitana campus Iztapalapa, Mexico City, using the Bif 164 (GGGTGGTAATGCCGGATG) and Bif 662 (CCAC-CGTTACACCGGGAA) primers [16]. Comparisons and sequence alignments were performed using MEGA5 and NCBI’s basic local alignment search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 16 January 2023)).
2.3. Multilocus Sequence Analysis (MLSA)
A comparison of constitutive genes in different strains of B. longum strains was made: tlyC, cbh, glnE, pNPOx, ahpC, trxA, trxB [11,17,18]. The MLSA approach has been used to generate a robust and highly discriminatory supertree to infer phylogeny in the genus Bifidobacterium. MLSA was performed according to González–Vázquez et al., 2022 [18].
2.4. Genomic analysis and annotation
The sequencing was conducted at the Integrated Microbiome Resource (IMR) located t Dalhousie University in Nova Scotia, Halifax, NS, Canada [17]. Assembling was carried out by IMR using SMRT analysis software v12.0. Data processing, plot construction, and genome annotation were performed according to González–Vázquez et al., 2022 [18].
2.5. Effect of Carbon Source on the Growth of B. longum LBUX23
The growth kinetics were monitored for a duration of 8 h of fermentation, representing the time required for the bacteria to reach the stationary phase. TPY medium was utilized, and various substances, including lactose, sucrose, fructooligosaccharides, and lactulose, were independently added at a concentration of 1% w/v. All media were adjusted to a pH of 7 using 0.1 M NaOH (Thermo Scientific Orion 410A+, USA). Prior to sterilization, the medium was purged of oxygen by bubbling CO2. Samples were collected at regular intervals of 2 h from time zero to 8 h. The bacterial concentration was determined by the plate count method [19]. The pH of the supernatant was measured according to González–Vázquez et al., 2022 [18]. All experiments were completed in triplicate.
2.6. Antibiotic Profile
Multidisc PT-34 Multibac I.D. (Investigación Diagnostica, Ciudad de Mexico, Mexico) were used to determine the ability to resist antibiotics following the supplier’s instructions. The antibiotics used in the study were as follows: β-lactam antibiotics, including vancomycin (30 μg), ampicillin (10 μg), dicloxacillin (1 μg), cephalothin (30 μg), penicillin (10 U), and cefotaxime (30 μg); inhibitors of protein synthesis such as gentamicin (10 μg), clindamycin (30 μg), erythromycin (15 μg), and tetracycline (30 μg); inhibitors of nucleic acid synthesis such as ciprofloxacin (5 μg) and other compounds such as trimethoprim-sulfamethoxazole (25 μg). The antibiotic profile of Bifidobacterium animalis subsp. lactis Bb-12 (CHR Hansen, Hørsholm, Denmark) was also determined for comparison. All experiments were conducted in triplicate [18].
2.7. Hemolysis Test
Prior to the experiment, B. longum LBUX23 was cultured overnight in MRS broth supplemented with 0.5% (w/v) L-cysteine. A concentration of 1 × 109 CFU/mL of the bacterial sample was inoculated onto blood agar plates and then incubated for 48 h at 37 °C in an anaerobic chamber. The test was performed in triplicate for each strain. Staphylococcus aureus ATCC 6538 served as the positive control, while Bifidobacterium animalis subsp. lactis Bb-12 (CHR Hansen) was used as the negative control. Results were reported as follows: positive indicating total hemolysis, and negative as non-hemolytic.
2.8. Antimicrobial Activity
The strain’s capacity to inhibit various bacterial strains, including Escherichia coli ATCC 25922, Escherichia coli O157:H7, Salmonella typhi ATCC 14028, Staphylococcus aureus ATCC 6538, as well as emergent pathogens obtained from clinical samples such as Klebsiella pneumoniae, Proteus mirabilis, and Pseudomonas aeruginosa, was assessed using the agar well diffusion assay. According to antibiotic susceptibility, these strains were classified as multi-drug resistant (MDR) and extremely resistant (XDR), following the general criteria of Magiorakos et al. (2012) [20]. XDR strains were categorized as strains that exhibited resistance to at least one agent in all or two therapeutic election categories (referred to as groups A and B) [20]. B. longum LBUX23 and B. animalis Bb-12 (control) were cultivated on TPY agar at a concentration of 1 × 107 CFU/mL and incubated at 37 °C for 24 h. Following this, warm, soft tryptic soy agar (8 g/L) was poured over each Bifidobacterium culture. Once solidified, other bacteria were individually plated onto the agar at a concentration of 1 × 108 CFU/mL. The plates were then incubated in anaerobic conditions at 37 °C for 24 h. The results were interpreted as follows: absence of inhibition (−), weak inhibition (+), and strong inhibition (++).
2.9. Bile Salt Tolerance Assay
Fresh bacterial cultures were inoculated onto TPY agar supplemented with different concentrations (0.1%, 0.2%, 0.3%, and 0.5% w/v) of glycocholic acid, glycodeoxycholic acid, taurocholic acid, taurodeoxycholic acid, or oxgall (obtained from Sigma Aldrich, St. Louis, MO, USA). The agar plates were then incubated anaerobically at 37 °C for 48 h. Strains exhibiting bile salt hydrolase activity were characterized by the presence of a halo surrounding them, indicating the precipitation of deconjugated bile salts [21] and demonstrating their resistance. B. animalis subsp. lactis Bb-12 was employed as the positive control in this experiment.
2.10. Bile Salt Hydrolase Activity
This activity was determined in intracellular cell-free extracts and cell debris. The method described by González–Vázquez et al. (2015) [22] was used to quantify BSH activity. First, B. longum LBUX23 was inoculated at a concentration of 1 × 109 CFU/mL in sodium phosphate buffer (PBS) supplemented with 0.5% w/v of glycocholic acid, glycodeoxycholic acid, taurocholic acid, taurodeoxycholic acid oxgall or any bile salt as control and incubated for 48 h. Following incubation, a 1 mL portion of the culture was subjected to centrifugation at 29,286× g for 5 min at 4 °C. Subsequently, 50 μL of the resulting supernatant was combined with 50 μL of sodium PBS (0.1 M, pH 6.0), 100 μL of a solution containing each bile salt or glycine (used as a control group) at a concentration of 0.5% w/v, and 10 μL of dithiothreitol (DTT). The mixture was incubated at 37 °C for 30 min to stop the enzyme activity, and 100 μL of trichloroacetic acid (15% w/v) was added. Afterward, the mixture was subjected to centrifugation for 5 min at 29,286× g and 4 °C. Then, 50 μL of the resulting supernatant was combined with 50 μL of distilled water and 1.9 mL of ninhydrin reagent. The ninhydrin reagent consisted of 0.5 mL of 1% w/v ninhydrin in citrate buffer (0.5 M, pH 5.5), 1.2 mL of glycerol, and 0.2 mL of buffer solution (0.5 M citrate, pH 5.5). The mixture was boiled for 14 min, followed by cooling for 3 min in an ice bath. The absorbance at 570 nm was measured. Prior to each assay, a standard curve was prepared using glycine and 10 μL of DTT to determine the concentration of released glycine. The percentage of BSH activity was calculated with the following:
2.11. pH Tolerance Assay
B. longum LBUX23 was cultured in TPY medium and incubated at 37 °C for 48 h. The culture was centrifuged at 10,000× g for 5 min, and the cells were washed twice using PBS buffer. The cells (1 × 109 CFU/mL) were inoculated into TPY broth adjusted to different pH 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, and 7 (control) with HCL 1N or NaOH 1N, respectively (Thermo Scientific Orion 410A+, USA). The media was incubated at 37 °C for 120 min and the bacterial concentration was determined by the plate count method [19]. The growth in CFU/mL was determined at 5, 10, 15, 20, 30, 60, and 120 min, respectively. All experiments were completed in triplicate. The survival rate of the strains at different pH and times was calculated as described above using the following expression according to Buntin et al. (2008) [23]:
2.12. DPPH Radical Scavenging Activity Assay
The scavenging activity of the 2,2-diphenyl-1 picrylhydrazyl (DPPH) radical was assessed utilizing the procedure outlined by Su et al. (2015) [24]. In summary, a mixture was prepared by combining 1 mL of freshly prepared DPPH solution (0.2 mmol/L in ethanol) with 1 mL of the sample, which included intact cells, intracellular cell-free extracts, or cell debris. Subsequently, the mixture was incubated in darkness for 30 min. The absorbance of the solution was then determined at a wavelength of 517 nm. The DPPH scavenging ability was calculated as follows:
2.13. Hydroxyl Radical Scavenging Activity Assay
The hydroxyl radical scavenging ability was analyzed according to Yan et al. (2020) [25]. Intact cells, intracellular cell-free extracts, or cell debris (1 mL per sample) were mixed with 1.0 mL of 1,10-phenanthroline solution (2.5 mM), 1 mL of PBS (pH 7.4), and 1 mL of FeSO4 (2.5 mM). To inhibit the reaction, 1 mL of 20 mM H2O2 was added, and the reaction was incubated at 37 °C for 90 min. The absorbance of the solution was measured at 517 nm. The scavenging ability of hydroxyl radicals was determined as follows:
2.14. Superoxide Anion Radical Scavenging Activity Assay
The superoxide anion radical scavenging ability was evaluated following the method of Yan et al. (2020) [25], with some modifications. Intact cells, intracellular cell-free extracts, or cell debris (1mL per sample) were added to 3 mL of Tris–HCl solution (pH 8.2) and incubated at 25 °C for 20 min. Then, 0.4 mL of pyrogallol (25 mM) was added at room temperature for 4 min. The reactions were stopped by adding 0.5 mL of HCl, and the absorbance was measured at 325 nm. The superoxide anion radical scavenging activity was determined as follows:
2.15. Adhesion Assay
Caco-2 cells (ATCC® HTB-37™, Sigma Aldrich, USA) were used to determine the cell adhesion ability of B. longum LBUX23 by utilizing the methodology reported in Gonzalez-Vazquez et al. (2022) [18]. Caco-2 cells were cultured in Eagle’s minimal essential medium (obtained from Gibco Invitrogen, Carlsbad, CA, USA) supplemented with 20% fetal bovine serum (FBS; Gibco Invitrogen, Carlsbad, CA, USA), 1% penicillin/streptomycin, 1% L-glutamine, and 1% (v/v) nonessential amino acids solution (Gibco). The cells, with a density of 1 × 105, were seeded into 24-well culture plates and incubated at 37 °C with 5% CO2. After 24 h of incubation, a monolayer of cells was formed, and the plates were further incubated for a period of 15 days. To prepare the bacterial samples, an overnight culture of either B. longum LBUX23 or B. animalis subsp. lactis Bb-12 (utilized as a control) was subjected to centrifugation at 6000× g for 5 min at 4 °C. The resulting pellet was then washed twice with PBS (pH 7.4) and suspended in Dulbecco’s Modified Eagle Medium without antibiotics and fetal bovine serum. In each well of the 24-well plates, a bacterial suspension of 1 mL containing 1 × 108 CFU/mL was introduced and incubated for 1 h at 37 °C in a 5% CO2 environment. Subsequently, each well was subjected to three washes with PBS (pH 7.4) to eliminate non-adherent bacteria. The bacteria that adhered to the monolayer were detached using a solution of trypsin-EDTA (obtained from Gibco, Carlsbad, California, USA) in PBS. The enzymatic activity was deactivated by adding a culture medium. The bacterial samples were subjected to serial dilutions, and the resulting dilutions were plated onto MRS agar plates containing 0.5% (w/v) L-cysteine hydrochloride (obtained from Merck, Rahway, NJ, USA). After 48 h of incubation in anaerobic conditions, the bacterial colonies were counted. All experiments were performed in triplicate.
2.16. Statistical Analysis
Significant differences in growth by using different carbon sources and bile salt in regard to DPPH scavenging ability, hydroxyl scavenging ability, and anion superoxide scavenging were determined by ANOVA (p ≤ 0.05). In the case of adhesion assay, differences were determined by t-student (p ≤ 0.05) using GraphPad Prism software version 5.01 (GraphPad Software, Inc., San Diego, CA, USA).
3. Results
3.1. Isolation and Genotypic Identification
In this study, we isolated one-Gram-positive rod-shaped bacterium; according to the BLAST analysis, those strains showed 99% of identity with the Bifidobacterium genus. The multilocus analysis showed that it belongs to longum subspecies (Figure 1) and was named B. longum LBUX23.
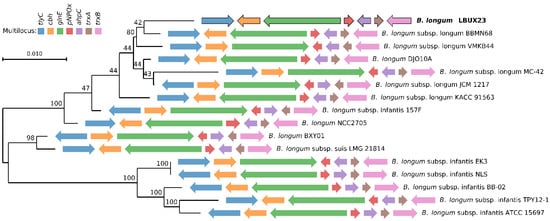
Figure 1.
Multilocus sequence analysis (MLSA) and the maximum likelihood phylogenetic tree were constructed using genomic sequences of bifidobacterial strains obtained from the GenBank database. The B. longum LBUX23 strains identified and characterized in this study are indicated in bold font. The solid arrows represent the housekeeping genes utilized for constructing the multilocus (tlyC, cbh, glnE, pNPOx, ahpC, trxA, and trxB). The direction of the arrows reflects the orientation of these genes within the genome of each Bifidobacterium strain.
The complete genome was registered at GenBank CP116390 as B. longum LBUX23. The BioProject accession number is PRJNA924960.
3.2. Genome Analysis
The genomic analysis showed that B. longum LBUX23 has a plasmid-free genome with a single 2,287,838 bp circular chromosome with a G+C content of 60.1%. The genomic annotations illustrated a total of 1853 coding sequences. The genome of B. longum LBUX23 possesses 56 tRNAs, 9 rRNAs, 1 tmRNA, and 1776 CDSs (95.84%). These last were grouped according to their functionality in cellular processes and signaling: (17.54%), information storage and processing (23.98%), metabolism (36.19%), and poorly characterized (22.29%). After COG categories were assigned, 22.29% were genes of unknown function, 9.51 % belonged to the carbohydrate transport and metabolism categories, 9.14% to amino acid transport and metabolism, 8.56 % to transcription, 7.82% to replication, recombination, and repair, 7.55% to translation, ribosomal structure, and biogenesis, 4.38% inorganic ion transport and metabolism, 4.23% nucleotide transport and metabolism, 3.96% cell wall, membrane, envelope biogenesis, 3.38% coenzyme transport and metabolism, 3.28% signal transduction mechanisms, 3.12% defense mechanisms, 2.96% energy production and conversion, 2.54% intracellular trafficking, secretion, and vesicular transport, 2.27% posttranslational modifications, protein turnover and chaperones, 2.01% lipid transport and metabolism, 1.95% cell cycle control, cell division, and chromosome partitioning, 0.58% secondary metabolites biosynthesis, transport, and catabolism, 0.42% cell motility and 0.05% chromatin structure and dynamics. B. longum LBUX23 did not contain clustered regularly interspaced short palindromic repeats (CRISPR).
3.3. Safety and Functional Characterization of B. longum LBUX23
3.3.1. Effect of Carbon Source on Growth
The B. longum LBUX23 grew in all carbon sources (Figure 2), initiating the exponential phase between 4-8 h. The maximum peak of bacterial growth was observed in sucrose and FOS at 6 h and at 8 h in lactose and lactulose (Figure 2). The pH decreased to 1.27 in the sucrose medium, followed by FOS (0.86), lactulose (0.65), and lactose (0.49), with respect to the initial pH of each substrate (Figure 2). However, no significant differences were found between them.
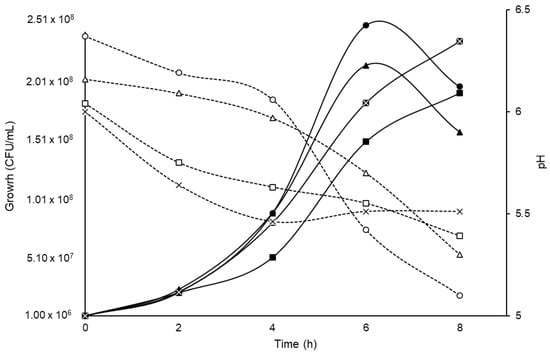
Figure 2.
Effect of different carbon sources on the growth of B. longum LBUX23. Continuous lines in black correspond to the growth (CFU/mL). Black dotted lines indicate changes in pH over time regarding different carbon sources:  sucrose,
sucrose,  FOS,
FOS,  lactulose, and
lactulose, and  lactose. In the case of pH:
lactose. In the case of pH:  sucrose,
sucrose,  FOS,
FOS,  lactulose, and
lactulose, and  lactose.
lactose.
 sucrose,
sucrose,  FOS,
FOS,  lactulose, and
lactulose, and  lactose. In the case of pH:
lactose. In the case of pH:  sucrose,
sucrose,  FOS,
FOS,  lactulose, and
lactulose, and  lactose.
lactose.
3.3.2. Antibiotic Profile
In terms of the antibiotic profile, both B. longum LBUX23 and B. animalis Bb-12 exhibited resistance to ampicillin and dicloxacillin. However, B. animalis Bb12 also showed resistance to cephalothin tetracycline and ciprofloxacin (Table 1a).

Table 1.
Characterization of B. longum LBUX23.
3.3.3. Hemolysis Test
Table 1b shows the results of the hemolysis activity. B. longum LBUX23 and B. animalis Bb-12 demonstrated negative hemolysis activity.
3.3.4. Antimicrobial Activity
Table 1c presents the results of the antimicrobial activity testing of B. longum LBUX23 and B. animalis Bb-12 against various strains.
3.3.5. Bile Salt Tolerance Assay and Activity
When B. longum LBUX23 and B. animalis Bb-12 grew in different concentrations and types of bile salts (Table 1d), a halo of precipitation was observed. B. longum LBUX23 showed significant differences in extracellular bile salt hydrolase activity between taurocholic acid (67.3%) and glycocholic acid (47.8%), oxgall (44.3%), and glycodeoxycholic acid (29.7%) (p = 0.0494, p = 0.0247, and p < 0.001, respectively) (Figure 3a). In addition, a higher percentage of extracellular BSH activity in taurocholic acid to B. longum LBUX23 (67.3%) in comparison to B. animalis Bb-12 (13.9%) (p < 0.0001) was determined (Figure 3a). In the case of intracellular BSH activity, no activity in glycocholic acid was shown by B. longum LBUX23 and B. animalis Bb-12 (Figure 3a). B. longum LBUX23 showed the highest BSH activity in taurodeoxycholic acid (8.2%), followed by taurocholic acid (6.3%) in comparison to B. animalis Bb-12, which showed the highest BSH activity in glycodeoxycholic acid (4.2%) acid and oxgall (2.9%). No significant difference in intracellular activity between B. longum LBUX23 and B. animalis Bb-12 was shown (Figure 3b). The genomic analysis showed the presence of the cbh gene, which codify to BSH (EC 3.5.1.24), as a part of an operon containing two genes, cbh, and glnE, which in turn codify to a glutamine synthetase adenylyltransferase (E.C. 2.7.7.42 2.7.7.89); this operon might be responsible for BSH activity.
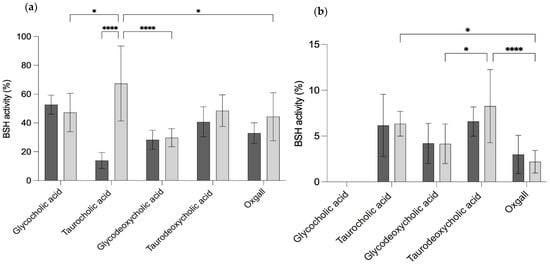
Figure 3.
BSH activity (a) extracellular and (b) intracellular using glycol, and tauro cholic acids (primary bile salts), glycol, and tauro deoxycholic acids (secondary bile salts), and oxgall to  B. animalis Bb-12 and
B. animalis Bb-12 and  B. longum LBUX23. The ANOVA analysis **** means significant to p < 0.0001 and * to p = 0.01 between different Bifidobacterium species, while ns means no significant difference.
B. longum LBUX23. The ANOVA analysis **** means significant to p < 0.0001 and * to p = 0.01 between different Bifidobacterium species, while ns means no significant difference.
 B. animalis Bb-12 and
B. animalis Bb-12 and  B. longum LBUX23. The ANOVA analysis **** means significant to p < 0.0001 and * to p = 0.01 between different Bifidobacterium species, while ns means no significant difference.
B. longum LBUX23. The ANOVA analysis **** means significant to p < 0.0001 and * to p = 0.01 between different Bifidobacterium species, while ns means no significant difference.
3.3.6. pH Tolerance Assay
The survival rate of B. longum LBUX23 at different pH is shown in Figure 4a. B. longum LBUX23, did not survive to pH 1-2.5. In the case of pH 3 and 3.5, a decrease in survival rates to 96.5 and 93.8% were observed. An increase of 20.1%, 22.8%, 30.8%, 50.8%, 77.8%, 89.6%, and 117.8% in the percentage of survival with respect to pH 3.5 was shown to pH 4, 4.5, 5, 5.5, 6, 6.5, and 7, respectively (Figure 4a). When survival was tested at pH 1 after 5 min, a 99.3% decrease was observed for B. longum LBUX23, while at pH 1.5, the same percentage of reduction was observed but after 10 min. In the case of pH 2, a decrease in survival of 98.8% was shown after 30 min. At pH 2.5, we observed a decrease of 99% after 120 min of submission (Figure 4b). The genomic analysis of B. longum LBUX23 showed the presence of argC (NCBI Locus tag: PIB40_02085), argH (PIB40_02045), and dapA (PIB40_02760) genes, which have been reported as responsible for the ability to tolerate acid environments in other species of B. longum [26].
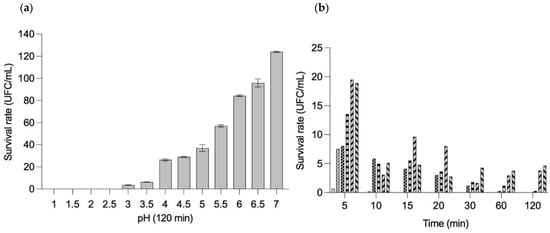
Figure 4.
The survival rate of the percentage of survivors of B. longum LBUX23 (a) in different pH’s for 120 min (b) at different pH’s:  pH 1,
pH 1,  pH 1.5,
pH 1.5,  pH 2,
pH 2,  pH 2.5,
pH 2.5,  pH 3, and
pH 3, and  pH 3.5.
pH 3.5.
 pH 1,
pH 1,  pH 1.5,
pH 1.5,  pH 2,
pH 2,  pH 2.5,
pH 2.5,  pH 3, and
pH 3, and  pH 3.5.
pH 3.5.
3.3.7. Antioxidant Activity
B. longum LBUX23 showed more DPPH radical scavenging activity than B. animalis Bb-12 in cell-free extracts (5.2 and 2.3%, respectively, p < 0.0095) and intact cells (27.4 and 24.2%, respectively, p < 0.0038). In contrast, cell debris of B. animalis Bb-12 showed higher antioxidant ability (17.9%) than B. longum LBUX23 (13.5%) p < 0.0003 (Figure 5a). By searching in the genome deposited at GenBank, we found nrdH (PIB40_05210), msrAB (PIB40_04685), PNPOx (PIB40_09440), AhpC (PIB40_00775), Bcp (PIB40_00775), trxA (PIB40_02940), and trxB (PIB40_04850) genes, which have been previously reported as possibly responsible of antioxidant activity involved in antioxidant activity (Table 2) [5,27,28].
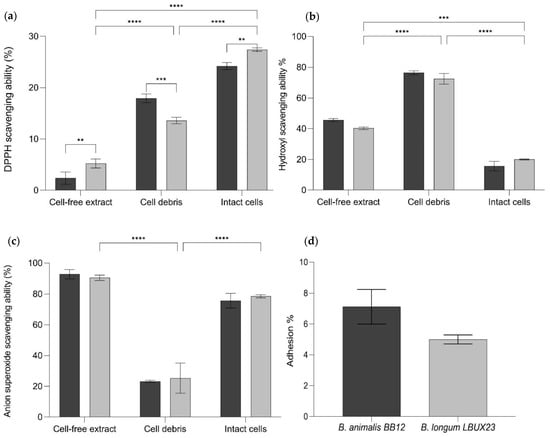
Figure 5.
(a) DPPH scavenging ability (b) Hydroxyl scavenging ability (c) Anion superoxide scavenging ability in cell-free extract, cell debris, and intact cells. (d) Percentage of adhesion to Caco-2 cells. For all test to  B. animalis Bb-12 and
B. animalis Bb-12 and  B. longum LBUX23. **** means significant p < 0.0001. The ANOVA analysis *** to p = 0.0003 and ** to 0.0095 between different Bifidobacterium species.
B. longum LBUX23. **** means significant p < 0.0001. The ANOVA analysis *** to p = 0.0003 and ** to 0.0095 between different Bifidobacterium species.
 B. animalis Bb-12 and
B. animalis Bb-12 and  B. longum LBUX23. **** means significant p < 0.0001. The ANOVA analysis *** to p = 0.0003 and ** to 0.0095 between different Bifidobacterium species.
B. longum LBUX23. **** means significant p < 0.0001. The ANOVA analysis *** to p = 0.0003 and ** to 0.0095 between different Bifidobacterium species.

Table 2.
Genes associated with antioxidant activity identified in B. longum LBUX23.
The ability to eliminate the hydroxyl radical was determined for B. longum LBUX23 and B. animalis Bb-12 (Figure 5b). We observed that intact cells of B. longum LBUX23 showed more hydroxyl activity (19.9%) than B. animalis Bb-12 (15.6%). Regarding cell-free extract and cell debris, hydroxyl radical scavenging activity was higher in B. animalis Bb-12 (45.6 and 76.3%, respectively) than in B. longum LBUX23 (40.2 and 72.5%, respectively). However, no significant differences among strains (Figure 5b) were shown. Cell debris showed major hydroxyl activity followed by cell-free extract and intact cell (p < 0.0001) in B. longum LBUX23 and B. animalis Bb-12 (Figure 5b). In the genome, we found some genes related to this activity, which are described in Table 2.
In this study, we showed that B. longum LBUX23 and B. animalis Bb-12 could inhibit superoxide anion radicals (Figure 5c). B. longum LBUX23 showed more activity in cell debris (25.3%) and intact cells (78.6%) in comparison to B. animalis Bb-12 (23.2 and 75.6%, respectively). In the case of cell-free extract, higher activity was found in B. animalis Bb-12 (92.8%) than in B. longum LBUX23 (90.5%). However, no significant differences were found for both strains in the case of cell-free extract, cell debris, and intact cells of both strains (Figure 5c). In addition, we observed that B. longum LBUX23 and B. animalis Bb-12 had major activity in cell-free extracts, followed by activity in cells and cell debris. Significant differences were found to cell debris in comparison to cell-free extracts (p < 0.0001) and intact cells (p < 0.0001) in both strains (Figure 5c). Genome analysis showed the presence of some genes involved in antioxidant activity (Table 2).
3.3.8. Adhesion Assay
It was observed that B. longum LBUX23 could adhere up to 5% to the human epithelial cells Caco-2, whereas B. animalis subsp. lactis Bb-12 was able to adhere up to 7.12%. However, neither strain showed significant differences (p > 0.05) (Figure 5d). In addition, genomic annotation of B. longum LBUX23 strain showed the presence of lspA (PIB40_08330), dnaK (PIB40_04505), grpE (PIB40_00120), aprE (PIB40_04245), dppB3 (PIB40_02590), sortase family (PIB40_04400 and PIB40_04760) and fibronectin type III-like domain (PIB40_05180 and PIB40_07405) genes, might be responsible for adhesion as have been previously reported [29].
4. Discussion
The population of Bifidobacterium is a common inhabitant of healthy humans. The alteration in the number and composition of Bifidobacterium population present in the human microbiome has been associated with several diseases, such as irritable bowel syndrome, inflammatory bowel disease (IBD), obesity, and allergy, among others. The consumption of bifidobacteria in various clinical trials has demonstrated promising outcomes in several clinical aspects, including the prevention of diarrhea, alleviation of symptoms in ulcerative colitis and irritable bowel syndrome (IBS), and the prevention of necrotizing enterocolitis [30,31]. Bifidobacterium genus is present in early life and plays an important role in newborns and infant development, which will determine an individual’s health at later stages of life [32]; Bifidobacterium species are most abundant in infants from the first week to the sixth month of life. However, it should be noted that not all fecal samples contain isolates of Bifidobacterium strains [33]. In this study, we found one Bifidobacterium bacteria (Figure 1) belonging to longum species from a fecal sample of a newborn. The genomic size and G+C content were similar to the one reported to B. longum LTBL16 [5], B. longum subsp. longum 35624 [34], B. longum GT15 [35], B. longum subsp. longum D4, M12, E1, S3, [3] and another strain reported by Arboleya et al. (2018) [36]. Research on bifidobacteria has utilized various bioinformatic tools to examine their complete genomes, providing valuable insights into the mechanisms through which these bacteria adapt to the unique conditions of the gastrointestinal tract. These genome characterizations have also revealed probiotic functions that facilitate specific interactions between the host and microorganisms [37]. Therefore, the genomic characterization of bacteria with probiotic potential is necessary and important, including newly isolated probiotics [38]. According to the genomic analysis, we showed that no CRISPR system in the B. longum LBUX23 was found, in comparison with other strains of B. longum (9, 17-1B, 105-A, BG7, and GT15) [39]. In 2015, Briner et al. (2015) [40] reported several strains of the Bifidobacterium that contained CRISPR sequences and concluded that the presence of CRISPR systems might be strain- rather than species-dependent [40]. The CRISPR system represents the strain’s record of immunity and the environmental challenges suffered by invasive DNA [31]. In the case of Bifidobacterium strains, the CRISPR targeting prophages are present in the genome of several bifidobacterial species (bifidophages). These findings indicate that these species reside in the same ecological habitat, where coevolution between CRISPR immune systems and prophages has likely taken place [31]. We hypothesize that B. longum LBUX23 has not been exposed to invasive DNA or infection and interaction with bifidophages. In addition, it has been suggested that the occurrence of CRISPR systems in the Bifidobacterium genus was 77.2% [40]. In the case of B. longum subsp. longum strains, the occurrence of CRISPR has been around 44.2% [41]. Bifidobacteria usually have diverse and extensive CRISPR systems. A relationship was noted between strains without CRISPR systems and the frequency of targeting of prophages in their chromosome by other CRISPR spacers [40].
The Bifidobacterium genus can metabolize a wide variety of mono-, di-, and oligosaccharides that are present in the intestinal environment. These carbohydrates are imported into their cytoplasm through ABC transporters [42]. In this study, we found that 9.51% of COG, are involved in metabolizing and transporting carbohydrates. In addition to B. longum, LBUX23 could be grown on different carbon sources, including glucose (data not shown), sucrose, lactose, and lactulose (Figure 2), and likely, this ability to metabolize carbohydrates is via bifid-shunt, as has been reported in B. longum NCC2705 [18,43]. Additionally, B. longum LBUX23 was grown in fructooligosaccharides (FOS), which are non-digestible carbohydrates, that can be found in various plant-based sources such as onions, asparagus, artichokes, garlic, wheat, bananas, tomatoes, and honey [44]. FOS are one of the significant classes of bifidogenic oligosaccharides and is one of the established prebiotics, defined as "a substrate that is selectively utilized by Bifidobacterium conferring a health benefit" [45,46]. Additionally, in the genomic analysis, we found some genes involved in carbohydrates metabolism, such as GalA1 (PIB40_05300), Ldh (PIB40_00085 and PIB40_08485), Ppc (PIB40_04825), GlGP (PIB40_04785), Fba (PIB40_04610), TreY (PIB40_04580), AmyA (PIB40_01230), MalQ1 (PIB40_04535), LeuA (PIB40_04350), and several genes to code by glycosyl hydrolase. These results showed a great diversity of genes and glycosyl-hydrolases, which confer the ability to metabolize a wide range of sugars [18,36,42]. These results were similar to those reported by González–Vázquez et al. (2022) [18], who grew B. pseudocatenulatum JCLA3 on glucose, lactose, sucrose, or inulin and found 44 predicted glycosyl hydrolases genes, which can act over carbohydrates metabolism [18]. In addition, Blanco et al. (2020) [47] found the presence of genes coding glycosyl hydrolases in B. longum subsp. longum [47].
The European Food Safety Authority (EFSA) suggests that assessing hemolytic activity is important when choosing probiotic strains. These strains are considered non-virulent, making them suitable for use in food products [48]. In this study, the hemolytic activity of B. longum LBUX23 was evaluated. Neither α-hemolytic nor β-hemolytic activity was shown in blood agar plates (Table 1). Our results agreed with the findings of Kim et al. (2018) [49] as they did not show hemolysis in B. bifidum BGN4 and B. longum BORI [49]; and Yasmin et al. (2020) [48], who also evaluated hemolytic activity as sensitive to antibiotics [48]. The B. longum LBUX23 has been resistant to β-lactam antibiotics, like other Bifidobacterium species such as B. pseudocatenulatum B700 and B. pseudocatenulatum JCLA3 [18,50]. However, in the genome of B. longum LBUX23, we found that resistance to antibiotics is chromosomal since the genome did not present plasmids, then β-lactam resistance cannot be transferred by conjugation to other bacteria. However, the possibility exists that B. longum LBUX23 and other generally recognized as safety (GRAS) bacteria could transfer the genomic material by transformation mechanism; therefore, it is necessary to define the GRAS status again from a molecular point of view.
In addition, oppA (PIB40_00680 and PIB40_008910), oppB (PIB40_08915), oppC (PIB40_08920), and oppF (PIB40_08925) are found in its genome, which are involved in Quorum sensing (Qs) and resistance to β-lactam, as reported by González–Vázquez et al. (2022) [18], who found intrinsic resistance to β-lactam genes in B. pseudocatenulatum JCLA3 [18,50].
In the current study, the antimicrobial activity of B. longum LBUX23 was investigated against ATCC strains and clinical isolates (Table 1). These last were categorized according to their antibiotic resistance pattern [20]. We found weak inhibition of K. pneumoniae MDR, P. mirabilis MDR, P. aeruginosa XDR, and E. coli ATCC 25922. The antimicrobial effects observed are likely attributed to the synthesis of antimicrobial compounds, including organic acids, hydrogen peroxide, ethanol, diacetyl, acetaldehyde, saturated or unsaturated free fatty acids, and other substances such as peptides and bacteriocins [51,52,53,54]. Additionally, we hypothesized that the possible advantage of this strain is the activity against emerging pathogens shown, not in the regard of being used during the infection, but instead in the sense of being used as a probiotic favoring self-defense from the gastrointestinal tract, decreasing the probability of an emerging pathogen attacking the host; however, this would have to be demonstrated in an in vivo study. Furthermore, a metabolomic study is required when the B. longum LBUX23 grows in a coculture with the pathogen to find postbiotics. Postbiotics have been demonstrated to possess various biological activities, including antimicrobial, antioxidant, anti-inflammatory, antiproliferative, and immunomodulatory effects [55].
The potential of the isolate as a probiotic was evaluated by assessing its ability to survive under simulated conditions mimicking the digestive tract [48,56]. Strain selection typically relies on assessing the tolerance of probiotics to physiologically relevant stresses, including low pH and bile [57]. The pH level is a critical factor that affects the growth and viability of probiotics during their journey through the stomach [58]. Bifidobacteria, being moderately acid-tolerant microorganisms, exhibit optimal growth and viability at pH levels ranging from 6.5 to 7.0. However, these commensal microbes can grow at a pH higher than 4.5 [59] since they are natural inhabitants of the large intestine, where the pH begins to be more basic with respect to the stomach and small intestine. We showed that B. longum LBUX23 survived optimally in a range of pH 4 to 7 (Figure 4). Therefore, in the case of using this bacterium as a supplement, it should be administered after consuming food to previously increase gastric pH or by using a matrix that protects it from stomach acid to ensure its viability until it reaches the large intestine [60,61]. In the case of pH 3 and 3.5, B. longum LBUX23 is capable of surviving < 10%, probably due to the presence of argC (PIB40_02085), argH (PIB40_02045), and dapA (PIB40_02760) genes. These genes have been previously reported by Sundararaman et al. 2021 [26] as those that confer to B. longum NCIM 5672 a survival advantage in an acid environment (pH 3). In the case of B. Longum LBUX23, those genes could be responsible for acid tolerance; nevertheless, more research is needed to prove their expression and to elucidate how the proteins codified by those genes act against acidity.
The ability of probiotics to tolerate bile salts has often been included among the criteria for probiotic strain selection [57]. Bile salt hydrolase catalyzes the hydrolysis of the amide bond in conjugated bile salts by choloylglycine hydrolase (EC 3.5.1.24.), resulting in the release of free amino acids, which makes the bile salt insoluble and is finally secreted through stools. Bile salts are primarily produced by combining cholesterol with amino acids glycine or taurine in the liver. These synthesized bile salts are then stored in the gallbladder until they are released into the duodenum following the consumption of fatty foods [62]. The identification of BSH activity has been included as a criterion for probiotic strain selection. Currently, there is a growing focus on investigating the bacterial conversion of bile in the human gastrointestinal tract due to its potential role in the development or prevention of metabolic and inflammatory conditions [63]. Our study shows that B. longum LBUX23 has the presence of 2.01 % of COG assigned to lipid transport of metabolism and includes the choloylglycine hydrolase gene. This gene can metabolize primary and secondary biliary salts (Figure 3), such as another Bifidobacterium species [10,18,64,65]. In addition, Begley et al. (2006) [57] suggest that the ratio of glycoconjugated to tauroconjugated bile salts is 3:1 in enterohepatic circulation [57]. In our study, B. longum LBUX23 had high extracellular BSH activity (67.3%) in taurocholic acid, followed by taurodeoxycholic acid (48.5 %), glycocholic acid (47.1%), oxgall (44.3 %), and glycodeoxycholic acid (29.7 %). Likely, the high activity in a taurocholic acid or taurodeoxycholic acid was due to tlyC1 ( PIB40_08320) gene present in B. longum LBUX23. The gene in question encodes a protein like hemolysin, which enhances the resistance of B. longum BBMN68 to bile acids conjugated with taurine, as documented in previous studies [66]. The intracellular activity was not found for glycocholic acid and less than 7 % for the other bile salts. These results suggest that BSH activity is extracellular, such as the results shown by Morinaga et al. (2022) [67,68]. Due to the high concentration of bile salts present in the intestine, some bacteria synthesize the BSH enzyme, and the BSH activity can decrease the toxicity of conjugated bile acids for them. The deconjugation of bile acids results in a reduction in their solubility and detergent properties, which can potentially decrease their toxicity to intestinal bacteria [68,69]. Bifidobacteria was able to produce precipitates in agar plates supplemented with biliary salt; due to the deconjugation of biliary salts by the BSH enzyme [67]. This effect is associated with probiotic cholesterol-lowering properties [70]. Another reason why B. longum LBUX23 has high BSH activity could be due to a previous adaptation since the source of its isolation was the newborn’s stools. Jarocki et al. (2014) [65] and Tanaka et al. (1999) [71] independently suggested that the activity of BSH is closely associated with the natural habitat of bacteria. Strains that exhibit BSH activity are typically found in the intestinal environment, where they are exposed to bile salts [65,71]. Additionally, Bordoni et al. (2013) [72] conducted a study evaluating cholesterol assimilation and BSH activity in 34 Bifidobacterium strains of human origin. They observed that strains belonging to B. animalis, B. breve, B. longum, and B. pseudocatenulatum species showed higher levels of BSH activity. Notably, B. longum subsp. longum MB 214 demonstrated the highest activity, followed by B. breve MB 11. These results are like the activity shown by B. longum LBUX23; however, it is necessary to homogenize the determination of BSH activity among the different probiotic strains in order to really be able to compare among them. Finally, high levels of activity of BSH are interesting as correlated with the ability to lower serum cholesterol levels in hypercholesterolemic [12]. Therefore, B. longum LBUX23 may be a candidate strain to test this effect in an in vivo model.
The presence of an excess of ROS significantly contributes to cell damage, including cell membrane injury, protein denaturation, and erroneous DNA replications. These effects can induce aging and many diseases, including cancer, diabetes, and rheumatoid arthritis, among others [27]. Several studies have been conducted on the antioxidant activity of bifidobacteria [5,27,28]. DPPH, hydroxyl, and superoxide radicals were used in this study to evaluate the antioxidant capacity of B. longum LBUX23. We observed that B. longum LBUX23 reduced DPPH radicals, mainly in intact cells following the cell debris and cell-free extract (Figure 5). In the case of hydroxyl radical scavenging capacity, it was shown that cell debris had major reductions in comparison with cell extract-free and intact cells, respectively, which suggests that it is not necessary that the bacteria be alive to have an antioxidant effect. In the case of superoxide anion radical scavenging capacity B. longum LBUX23 showed higher activity in cell-free extract, followed by intact cells and cell debris (Figure 5). Therefore, B. longum LBUX23 has a low effect over anion radical by itself. We showed a high anion radical effect in cell-free extract; therefore, it is suggested that this effect is due to a metabolite produced by the bacteria. B. longum LBUX23 did not contain coding genes related to catalase and superoxide dismutase. Additionally, it contains genes associated with ABC transporter ATP-binding protein, ferredoxin, thioredoxin, NADH oxidase, NADH peroxidase, and genes such as nrdH (PIB40_05210), msrAB (PIB40_04685), PNPOx (PIB40_09440), AhpC (PIB40_00775), Bcp (PIB40_00775), trxA (PIB40_02940), and trxB (PIB40_04850), which are related to such antioxidant activity. However, more research is needed on the expression of these genes and the mechanism that the produced proteins use to show the effect. This result is similar to B. longum LTBL16 [5], B. Longum BBMN68 [73], and B. Longum NCC2705 [74], which present different genes in response to ROS. In general, Bifidobacterium longum can decline the production of ROS, suppress oxidative stress, and reduce damage to the intestinal tract as a way of protecting intestinal epithelial cells [75]. Finally, B. longum LBUX23 is a candidate strain to test the antioxidant effect in an in vivo model.
5. Conclusions
In summary, the complete genome of B. longum LBUX23 is an effective tool for the discovery and identification of possible host-interaction and capacities of new probiotic strains. Among them, B. longum LBUX23 is grown in different carbon sources; apparently, it is considered a safe bacterium and shows adhesion ability, antimicrobial activity over emerging pathogens, tolerance of bile salts, high BSH activity, and antioxidant capacity. Our study indicates that B. longum LBUX23 has excellent probiotic properties, mainly in BSH activity and antioxidant capacity, and can be used as a candidate for antioxidant or anti-cholesterol effects in vivo models, among others. All the information generated in this study serves as evidence of the safety and functionality of this microorganism so that in the future, if any of these benefits are demonstrated in an in vivo model, B. longum LBUX23 could be used in biotechnological applications, human healthcare, and food science.
Author Contributions
Conceptualization, P.A.R.-C., R.G.-V., E.T.-M. and L.M.-R.; methodology, P.A.R.-C., J.I.B.-H., E.Z.-L., and R.G.-V.; validation, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; formal analysis, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; investigation, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; resources, R.G.-V. and L.M.-R.; data curation, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; writing—original draft preparation, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; writing—review and editing, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; visualization, P.A.R.-C., R.G.-V., E.T.-M., J.I.B.-H., E.Z.-L., M.L.-L., L.M.-S., F.M.-P., M.A.G.-N., D.R.-P., A.A.-E., and L.M.-R.; supervision, R.G.-V., E.T.-M., E.Z.-L., and L.M.-R.; project administration, R.G.-V., E.T.-M., E.Z.-L., and L.M.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are openly available from the NCBI in the BioProject PRJNA924960.
Acknowledgments
This work was financially supported by the research strengthening program of the Rector of UAM Xochimilco, 2022.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Kato, K.; Odamaki, T.; Mitsuyama, E.; Sugahara, H.; Xiao, J.Z.; Osawa, R. Age-Related Changes in the Composition of Gut Bifidobacterium Species. Curr. Microbiol. 2017, 74, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Tarracchini, C.; Milani, C.; Lugli, G.A.; Mancabelli, L.; Fontana, F.; Alessandri, G.; Longhi, G.; Anzalone, R.; Viappiani, A.; Turroni, F.; et al. Phylogenomic disentangling of the Bifidobacterium longum subsp. infantis taxon. Microb. Genom. 2021, 7, 000609. [Google Scholar] [CrossRef]
- Díaz, R.; Torres-Miranda, A.; Orellana, G.; Garrido, D. Comparative Genomic Analysis of Novel Bifidobacterium longum subsp. longum Strains Reveals Functional Divergence in the Human Gut Microbiota. Microorganisms 2021, 9, 1906. [Google Scholar]
- Quigley, E.M.M. Chapter 16—Bifidobacterium longum. In The Microbiota in Gastrointestinal Pathophysiology; Floch, M.H., Ringel, Y., Allan Walker, W., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 139–141. [Google Scholar]
- Huang, G.; Pan, H.; Zhu, Z.; Li, Q. The complete genome sequence of Bifidobacterium longum LTBL16, a potential probiotic strain from healthy centenarians with strong antioxidant activity. Genomics 2020, 112, 769–773. [Google Scholar] [CrossRef]
- Preiser, J.-C. Oxidative Stress. J. Parenter. Enter. Nutr. 2012, 36, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Kleniewska, P.; Pawliczak, R. Antioxidative activity of probiotics. Arch. Med. Sci. 2021, 17, 792–804. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, C.; Zhang, C.; Zhang, Q.; Yu, L.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W.; Zhai, Q. Strain-Specific Effects of. Int. J. Mol. Sci. 2021, 22, 1305. [Google Scholar] [CrossRef]
- Kim, Y.-T.; Kim, C.-H.; Kwon, J.-G.; Cho, J.H.; Shin, Y.-S.; Kim, H.B.; Lee, J.-H. In vivo Trial of Bifidobacterium longum Revealed the Complex Network Correlations Between Gut Microbiota and Health Promotional Effects. Front. Microbiol. 2022, 13, 886934. [Google Scholar] [CrossRef]
- Tanaka, H.; Hashiba, H.; Kok, J.; Mierau, I. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Ghatani, K. Bile Salt Hydrolase Activity of Probiotics ans their Rople in hypolipidemia. J. Biol. Todays World 2023, 12, 1–4. [Google Scholar]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In Vitro Bile Salt Hydrolase (BSH) Activity Screening of Different Probiotic Microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, I.; Moussa, O.B.; Hassouna, M. Symbiotic, Hypocholesterolemic and Antioxidant Effects of Potential Probiotic Lactobacilli Strains Isolated from Tunisian Camel Milk. Adv. Microbiol. 2017, 7, 328–342. [Google Scholar] [CrossRef]
- Donelli, G.; Vuotto, C.; Mastromarino, P. Phenotyping and genotyping are both essential to identify and classify a probiotic microorganism. Microb. Ecol. Health Dis. 2013, 24, 20105. [Google Scholar] [CrossRef]
- Martín, R.; Jiménez, E.; Heilig, H.; Fernández, L.; Marín, M.L.; Zoetendal, E.G.; Rodríguez, J.M. Isolation of Bifidobacteria from Breast Milk and Assessment of the Bifidobacterial Population by PCR-Denaturing Gradient Gel Electrophoresis and Quantitative Real-Time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef]
- Satokari, R.M.; Vaughan, E.E.; Akkermans, A.D.; Saarela, M.; de Vos, W.M. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 504–513. [Google Scholar] [CrossRef]
- Comeau André, M.; Douglas Gavin, M.; Langille Morgan, G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- González–Vázquez, R.; Zúñiga-León, E.; Torres-Maravilla, E.; Leyte-Lugo, M.; Mendoza-Pérez, F.; Hernández-Delgado, N.C.; Pérez-Pastén-Borja, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L. Genomic and Biochemical Characterization of Bifidobacterium pseudocatenulatum JCLA3 Isolated from Human Intestine. Microorganisms 2022, 10, 2100. [Google Scholar] [CrossRef]
- Wendel, U. Assessing Viability and Stress Tolerance of Probiotics—A Review. Front. Microbiol. 2021, 12, 818468. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Zuo, F.; Yu, R.; Feng, X.; Chen, L.; Zeng, Z.; Khaskheli, G.B.; Ma, H.; Chen, S. Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast-fed infant feces. Ann. Microbiol. 2016, 66, 1027–1037. [Google Scholar] [CrossRef]
- González–Vázquez, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L.; Reyes-Nava, L.A.; Shah, N.P.; Rivera-Espinoza, Y. Isolation, Identification and Partial Characterization of a Lactobacillus casei Strain with Bile Salt Hydrolase Activity from Pulque. Probiotics Antimicrob. Proteins 2015, 7, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Buntin, N.; Chanthachum, S.; Hongpattarakere, T. Screening of lactic acid bacteria from gastrointestinal tracts of marine fish for their potential use as probiotics. Songklanakarin J. Sci. Technol. 2008, 30, 141–148. [Google Scholar]
- Su, J.; Wang, T.; Li, Y.Y.; Li, J.; Zhang, Y.; Wang, Y.; Wang, H.; Li, H. Antioxidant properties of wine lactic acid bacteria: Oenococcus oeni. Appl. Microbiol. Biotechnol. 2015, 99, 5189–5202. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Li, N.; Yue, Y.; Wang, C.; Zhao, L.; Evivie, S.E.; Li, B.; Huo, G. Screening for Potential Novel Probiotics With Dipeptidyl Peptidase IV-Inhibiting Activity for Type 2 Diabetes Attenuation in vitro and in vivo. Front. Microbiol. 2020, 10, 2855. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, A.; Bansal, K.; Sidhic, J.; Patil, P.; Halami, P.M. Genome of Bifidobacterium longum NCIM 5672 provides insights into its acid-tolerance mechanism and probiotic properties. Arch. Microbiol. 2021, 203, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Westermann, C.; Gleinser, M.; Corr, S.C.; Riedel, C.U. A Critical Evaluation of Bifidobacterial Adhesion to the Host Tissue. Front. Microbiol. 2016, 7, 1220. [Google Scholar] [CrossRef]
- Tojo, R.; Suárez, A.; Clemente, M.G.; de los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Crawley, A.B.; Sanchez, B.; Barrangou, R. Characterization and Exploitation of CRISPR Loci in Bifidobacterium longum. Front. Microbiol. 2017, 8, 1851. [Google Scholar] [CrossRef]
- Saturio, S.; Nogacka, A.M.; Suárez, M.; Fernández, N.; Mantecón, L.; Mancabelli, L.; Milani, C.; Ventura, M.; de Los Reyes-Gavilán, C.G.; Solís, G.; et al. Early-Life Development of the Bifidobacterial Community in the Infant Gut. Int. J. Mol. Sci. 2021, 22, 3382. [Google Scholar] [CrossRef]
- Tannock, G.W.; Lee, P.S.; Wong, K.H.; Lawley, B. Why Don’t All Infants Have Bifidobacteria in Their Stool? Front. Microbiol. 2016, 7, 834. [Google Scholar] [CrossRef]
- Altmann, F.; Kosma, P.; O’Callaghan, A.; Leahy, S.; Bottacini, F.; Molloy, E.; Plattner, S.; Schiavi, E.; Gleinser, M.; Groeger, D.; et al. Genome Analysis and Characterisation of the Exopolysaccharide Produced by Bifidobacterium longum subsp. longum 35624™. PLoS ONE 2016, 11, e0162983. [Google Scholar] [CrossRef]
- Zakharevich, N.V.; Averina, O.V.; Klimina, K.M.; Kudryavtseva, A.V.; Kasianov, A.S.; Makeev, V.J.; Danilenko, V.N. Complete Genome Sequence of Bifidobacterium longum GT15: Identification and Characterization of Unique and Global Regulatory Genes. Microb. Ecol. 2015, 70, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Bottacini, F.; O’Connell-Motherway, M.; Ryan, C.A.; Ross, R.P.; van Sinderen, D.; Stanton, C. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genom. 2018, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Turroni, F.; van Sinderen, D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng. Bugs 2012, 3, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Zawistowska-Rojek, A.; Kociszewska, A.; Zaręba, T.; Tyski, S. New Potentially Probiotic Strains Isolated from Humans—Comparison of Properties with Strains from Probiotic Products and ATCC Collection. Pol. J. Microbiol. 2022, 71, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Zarrinhaghighi, A.; Moradi, A.; Dehshahri, A. Bioinformatics investigation of CRISPR/Cas systems in Bifidobacterium longum. Trends Pharm. Sci. 2021, 7, 169–178. [Google Scholar] [CrossRef]
- Briner, A.E.; Lugli, G.A.; Milani, C.; Duranti, S.; Turroni, F.; Gueimonde, M.; Margolles, A.; van Sinderen, D.; Ventura, M.; Barrangou, R. Occurrence and Diversity of CRISPR-Cas Systems in the Genus Bifidobacterium. PLoS ONE 2015, 10, e0133661. [Google Scholar] [CrossRef]
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.-z.; van Sinderen, D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018, 8, 85. [Google Scholar] [CrossRef]
- Kelly, S.M.; Munoz-Munoz, J.; van Sinderen, D. Plant Glycan Metabolism by Bifidobacteria. Front. Microbiol. 2021, 12, 609418. [Google Scholar] [CrossRef]
- Liu, D.; Wang, S.; Xu, B.; Guo, Y.; Zhao, J.; Liu, W.; Sun, Z.; Shao, C.; Wei, X.; Jiang, Z.; et al. Proteomics analysis of Bifidobacterium longum NCC2705 growing on glucose, fructose, mannose, xylose, ribose, and galactose. Proteomics 2011, 11, 2628–2638. [Google Scholar] [CrossRef]
- Sabater-Molina, M.; Larqué, E.; Torrella, F.; Zamora, S. Dietary fructooligosaccharides and potential benefits on health. J. Physiol. Biochem. 2009, 65, 315–328. [Google Scholar] [CrossRef]
- Parhi, P.; Song, K.P.; Choo, W.S. Growth and survival of. J. Food Sci. Technol. 2022, 59, 3775–3786. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Blanco, G.; Ruiz, L.; Tamés, H.; Ruas-Madiedo, P.; Fdez-Riverola, F.; Sánchez, B.; Lourenço, A.; Margolles, A. Revisiting the Metabolic Capabilities of Bifidobacterium longum susbp. longum and Bifidobacterium longum subsp. infantis from a Glycoside Hydrolase Perspective. Microorganisms 2020, 8, 723. [Google Scholar] [PubMed]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In Vitro Probiotic Potential and Safety Evaluation (Hemolytic, Cytotoxic Activity) of Bifidobacterium Strains Isolated from Raw Camel Milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- Kim, M.J.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Johnston, T.V.; Park, M.S.; Ji, G.E. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. [Google Scholar] [CrossRef] [PubMed]
- Di Gioia, D.; Strahsburger, E.; Lopez de Lacey, A.M.; Bregola, V.; Marotti, I.; Aloisio, I.; Biavati, B.; Dinelli, G. Flavonoid bioconversion in Bifidobacterium pseudocatenulatum B7003: A potential probiotic strain for functional food development. J. Funct. Foods 2014, 7, 671–679. [Google Scholar] [CrossRef]
- De Keersmaecker, S.C.; Verhoeven, T.L.; Desair, J.; Marchal, K.; Vanderleyden, J.; Nagy, I. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 2006, 259, 89–96. [Google Scholar] [CrossRef]
- Inturri, R.; Trovato, L.; Volti, G.L.; Oliveri, S.; Blandino, G. In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates. Heliyon 2019, 5, e02891. [Google Scholar] [CrossRef]
- Tejero-Sariñena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Xing, D. The Current and Future Perspectives of Postbiotics. Probiotics Antimicrob. Proteins 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Marchwińska, K.; Gwiazdowska, D. Isolation and probiotic potential of lactic acid bacteria from swine feces for feed additive composition. Arch. Microbiol. 2021, 204, 61. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile Salt Hydrolase Activity in Probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Zhou, X.X.; Pan, Y.J.; Wang, Y.B.; Li, W.F. In vitro assessment of gastrointestinal viability of two photosynthetic bacteria, Rhodopseudomonas palustris and Rhodobacter sphaeroides. J. Zhejiang Univ. Sci. B 2007, 8, 686–692. [Google Scholar] [CrossRef]
- Grattepanche, F.; Lacroix, C. 13—Production of viable probiotic cells. In Microbial Production of Food Ingredients, Enzymes and Nutraceuticals; McNeil, B., Archer, D., Giavasis, I., Harvey, L., Eds.; Woodhead Publishing: Sawston, UK, 2013; pp. 321–352. [Google Scholar]
- Gunzburg, W.H.; Aung, M.M.; Toa, P.; Ng, S.; Read, E.; Tan, W.J.; Brandtner, E.M.; Dangerfield, J.; Salmons, B. Efficient protection of microorganisms for delivery to the intestinal tract by cellulose sulphate encapsulation. Microb. Cell Factories 2020, 19, 216. [Google Scholar] [CrossRef]
- Abd El-Salam, M.H.; El-Shibiny, S. Preparation and properties of milk proteins-based encapsulated probiotics: A review. Dairy Sci. Technol. 2015, 95, 393–412. [Google Scholar] [CrossRef]
- Kumar, R.S.; Brannigan, J.A.; Prabhune, A.A.; Pundle, A.V.; Dodson, G.G.; Dodson, E.J.; Suresh, C.G. Structural and Functional Analysis of a Conjugated Bile Salt Hydrolase from Bifidobacterium longum Reveals an Evolutionary Relationship with Penicillin V Acylase. J. Biol. Chem. 2006, 281, 32516–32525. [Google Scholar] [CrossRef]
- Ruiz, L.; Sánchez, B.; Margolles, A. Determination of Bile Salt Hydrolase Activity in Bifidobacteria. In Bifidobacteria: Methods and Protocols; van Sinderen, D., Ventura, M., Eds.; Springer US: New York, NY, USA, 2021; pp. 149–155. [Google Scholar]
- Grill, J.; Schneider, F.; Crociani, J.; Ballongue, J. Purification and Characterization of Conjugated Bile Salt Hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 1995, 61, 2577–2582. [Google Scholar] [CrossRef] [PubMed]
- Jarocki, P.; Podleśny, M.; Glibowski, P.; Targoński, Z. A New Insight into the Physiological Role of Bile Salt Hydrolase among Intestinal Bacteria from the Genus Bifidobacterium. PLoS ONE 2014, 9, e114379. [Google Scholar] [CrossRef]
- Liu, Y.; An, H.; Zhang, J.; Zhou, H.; Ren, F.; Hao, Y. Functional role of tlyC1 encoding a hemolysin-like protein from Bifidobacterium longum BBMN68 in bile tolerance. FEMS Microbiol. Lett. 2014, 360, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Kociubinski, G.; Pérez, P.; De Antoni, G. Screening of bile resistance and bile precipitation in lactic acid bacteria and bifidobacteria. J. Food Prot. 1999, 62, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, K.; Kusada, H.; Tamaki, H. Bile Salt Hydrolases with Extended Substrate Specificity Confer a High Level of Resistance to Bile Toxicity on Atopobiaceae Bacteria. Int. J. Mol. Sci. 2022, 23, 10980. [Google Scholar] [CrossRef] [PubMed]
- Moser, S.A.; Savage, D.C. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 2001, 67, 3476–3480. [Google Scholar] [CrossRef]
- Jones, M.L.; Tomaro-Duchesneau, C.; Martoni, C.J.; Prakash, S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin. Biol. Ther. 2013, 13, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Doesburg, K.; Iwasaki, T.; Mierau, I. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 1999, 82, 2530–2535. [Google Scholar] [CrossRef]
- Bordoni, A.; Amaretti, A.; Leonardi, A.; Boschetti, E.; Danesi, F.; Matteuzzi, D.; Roncaglia, L.; Raimondi, S.; Rossi, M. Cholesterol-lowering probiotics: In vitro selection and in vivo testing of bifidobacteria. Appl. Microbiol. Biotechnol. 2013, 97, 8273–8281. [Google Scholar] [CrossRef]
- Shen, Q.; Shang, N.; Li, P. In Vitro and In Vivo Antioxidant Activity of Bifidobacterium animalis 01 Isolated from Centenarians. Curr. Microbiol. 2011, 62, 1097–1103. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Dong, J.; Shi, J.; Guan, J.; Liu, D.; Liu, F.; Li, B.; Huo, G. Identification, Characterization, and Antioxidant Potential of Bifidobacterium longum subsp. longum Strains Isolated From Feces of Healthy Infants. Front. Microbiol. 2021, 12, 756519. [Google Scholar] [CrossRef]
- Zuo, F.; Yu, R.; Xiao, M.; Khaskheli, G.B.; Sun, X.; Ma, H.; Ren, F.; Zhang, B.; Chen, S. Transcriptomic analysis of Bifidobacterium longum subsp. longum BBMN68 in response to oxidative shock. Sci. Rep. 2018, 8, 17085. [Google Scholar] [CrossRef]
- Guo, Q.; Li, S.; Xie, Y.; Zhang, Q.; Liu, M.; Xu, Z.; Sun, H.; Yang, Y. The NAD+-dependent deacetylase, Bifidobacterium longum Sir2 in response to oxidative stress by deacetylating SigH (σH) and FOXO3a in Bifidobacterium longum and HEK293T cell respectively. Free Radic. Biol. Med. 2017, 108, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Zhao, Z.; Wang, W.; Liu, X. Bifidobacterium longum: Protection against Inflammatory Bowel Disease. J. Immunol. Res. 2021, 2021, 8030297. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).