The Mycobacterium smegmatis HesB Protein, MSMEG_4272, Is Required for In Vitro Growth and Iron Homeostasis
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. Generation of Mutant Strains of M. smegmatis
2.3. CRISPRi Mediated MSMEG_4272 Gene Silencing in M. smegmatis
2.4. Real-Time Quantitative PCR
2.5. Intracellular Iron Determination
2.6. Phenotypic Characterization of CRISPRi Mediated MSMEG_4272 Gene Silencing Strains
2.7. Statistical Analysis
3. Results
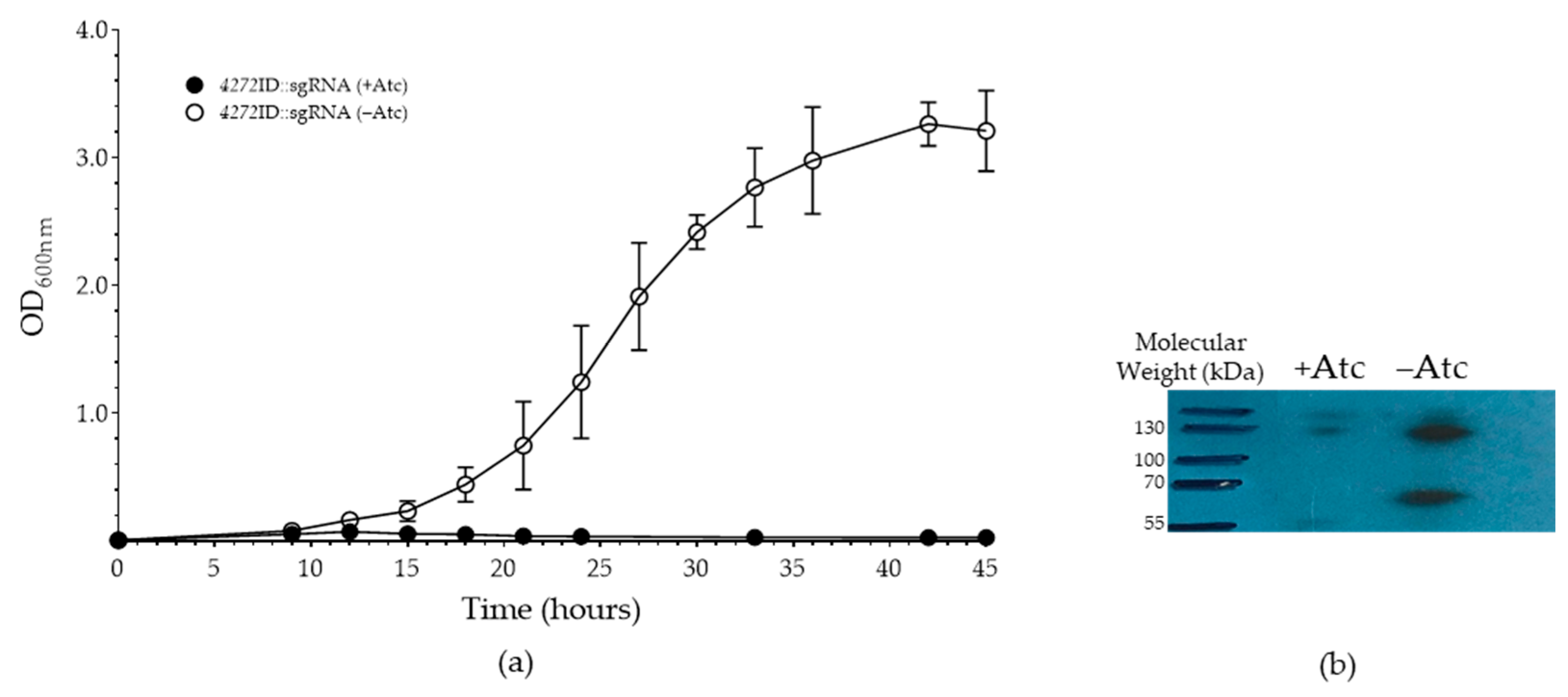
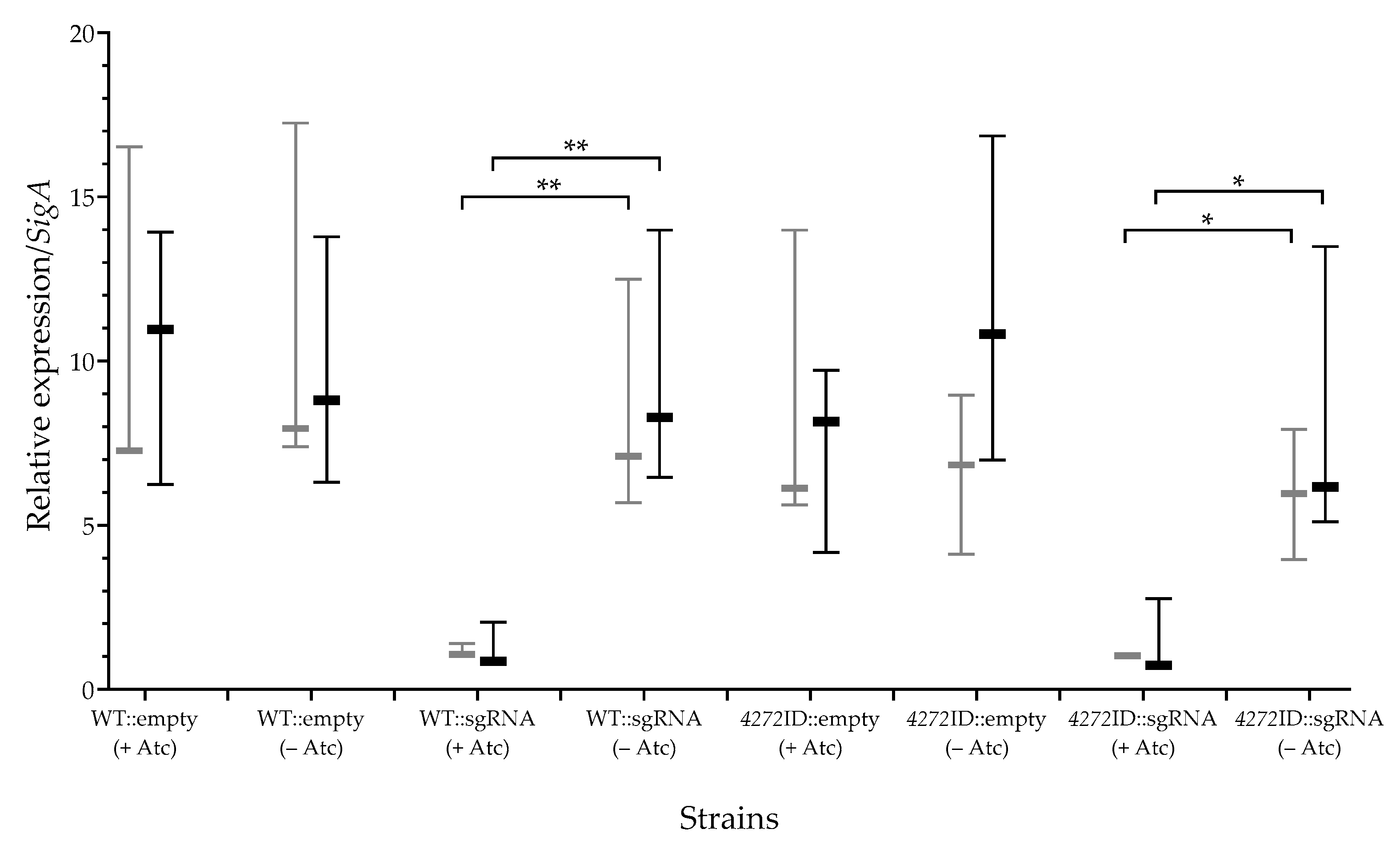
3.1. MSMEG_4272 Function Is Sensitive to the Modulation of Protein Levels
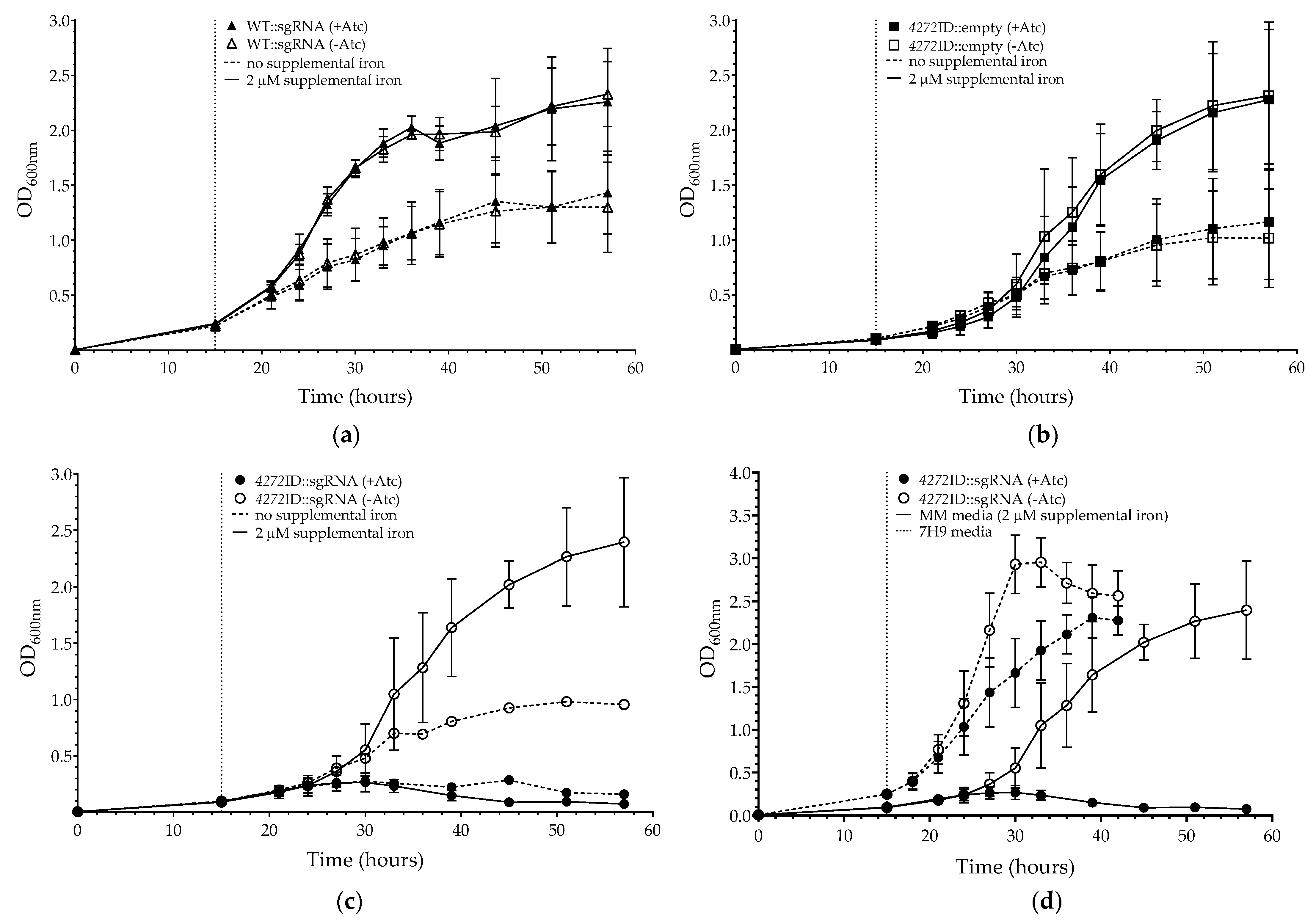
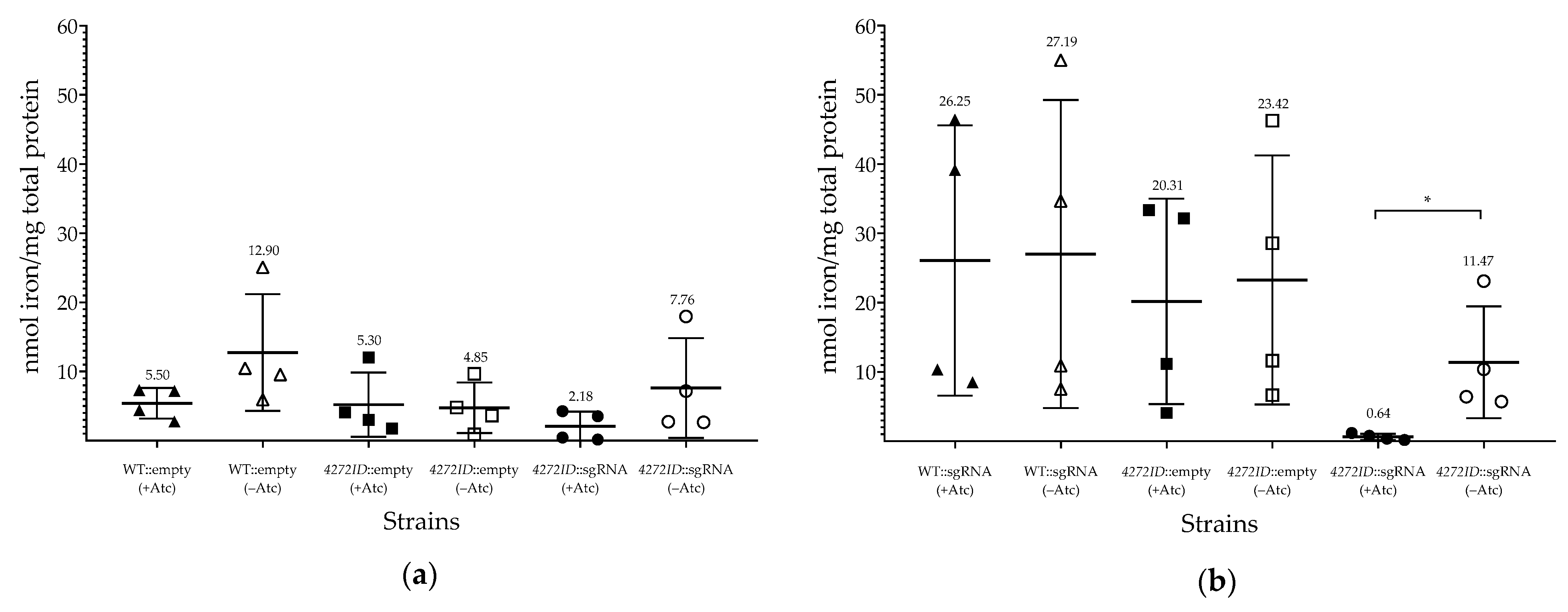
3.2. Phenotypic Impact of MSMEG_4272 Transcriptional Silencing on M. smegmatis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mandin, P.; Chareyre, S.; Barras, F. A Regulatory Circuit Composed of a Transcription Factor, IscR, and a Regulatory RNA, RyhB, Controls Fe-S Cluster Delivery. mBio 2016, 7, e00966-16. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-Sulfur Clusters: Nature’s Modular, Multipurpose Structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef]
- Rees, D.C.; Howard, J.B. The Interface Between the Biological and Inorganic Worlds: Iron-Sulfur Metalloclusters. Science 2003, 300, 929–931. [Google Scholar] [CrossRef]
- Bilder, P.W.; Ding, H.; Newcomer, M.E. Crystal Structure of the Ancient, Fe-S Scaffold IscA Reveals a Novel Protein Fold. Biochemistry 2004, 43, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Pinske, C.; Sawers, R.G. A-Type Carrier Protein ErpA Is Essential for Formation of an Active Formate-Nitrate Respiratory Pathway in Escherichia Coli K-12. J. Bacteriol. 2012, 194, 346–353. [Google Scholar] [CrossRef]
- Jacobson, M.R.; Cash, V.L.; Weiss, M.C.; Laird, N.F.; Newton, W.E.; Dean, D.R. Biochemical and Genetic Analysis of the NifUSVWZM Cluster from Azotobacter Vinelandii. Mol. Gen. Genet. 1989, 219, 49–57. [Google Scholar] [CrossRef]
- Zheng, L.; Cash, V.L.; Flint, D.H.; Dean, D.R. Assembly of Iron-Sulfur Clusters. Identification of an IscSUA-HscBA-Fdx Gene Cluster from Azotobacter Vinelandii. J. Biol. Chem. 1998, 273, 13264–13272. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Tokumoto, U. A Third Bacterial System for the Assembly of Iron-Sulfur Clusters with Homologs in Archaea and Plastids. J. Biol. Chem. 2002, 277, 28380–28383. [Google Scholar] [CrossRef] [PubMed]
- Black, K.A.; Dos Santos, P.C. Shared-Intermediates in the Biosynthesis of Thio-Cofactors: Mechanism and Functions of Cysteine Desulfurases and Sulfur Acceptors. Biochim. Biophys. Acta 2015, 1853, 1470–1480. [Google Scholar] [CrossRef]
- Vinella, D.; Brochier-Armanet, C.; Loiseau, L.; Talla, E.; Barras, F. Iron-Sulfur (Fe/S) Protein Biogenesis: Phylogenomic and Genetic Studies of A-Type Carriers. PLOS Genet. 2009, 5, e1000497. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Iannuzzi, C.; Prischi, F.; Pastore, C.; Iametti, S.; Martin, S.R.; Bonomi, F.; Pastore, A. Bacterial Frataxin CyaY Is the Gatekeeper of Iron-Sulfur Cluster Formation Catalyzed by IscS. Nat. Struct. Mol. Biol. 2009, 16, 390–396. [Google Scholar] [CrossRef]
- Adinolfi, S.; Puglisi, R.; Crack, J.C.; Iannuzzi, C.; Dal Piaz, F.; Konarev, P.V.; Svergun, D.I.; Martin, S.; Le Brun, N.E.; Pastore, A. The Molecular Bases of the Dual Regulation of Bacterial Iron Sulfur Cluster Biogenesis by CyaY and IscX. Front. Mol. Biosci. 2017, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yang, J.; Tan, G.; Ding, H. Complementary Roles of SufA and IscA in the Biogenesis of Iron–Sulfur Clusters in Escherichia Coli. Biochem. J. 2008, 409, 535. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Clark, R.J. Characterization of Iron Binding in IscA, an Ancient Iron-Sulphur Cluster Assembly Protein. Biochem. J. 2004, 379, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Mapolelo, D.T.; Zhang, B.; Naik, S.G.; Huynh, B.H.; Johnson, M.K. Spectroscopic and Functional Characterization of Iron-Bound Forms of Azotobacter Vinelandii Nif IscA. Biochemistry 2012, 51, 8056–8070. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Sendra, M.; Naik, S.G.; Chahal, H.K.; Huynh, B.H.; Outten, F.W.; Fontecave, M.; de Choudens, S.O.; Native, E. Coli SufA, Co-Expressed with SufBCDSE, Purifies as a [2Fe-2S] Protein and Acts as an Fe-S Transporter to Fe-S Target Enzymes. J. Am. Chem. Soc. 2009, 131, 6149–6153. [Google Scholar] [CrossRef] [PubMed]
- Loiseau, L.; Gerez, C.; Bekker, M.; Choudens, S.O.; Py, B.; Sanakis, Y.; Teixeira de Mattos, J.; Fontecave, M.; Barras, F. ErpA, an Iron–Sulfur (Fe–S) Protein of the A-Type Essential for Respiratory Metabolism in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2007, 104, 13626–13631. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Lu, J.; Bitoun, J.P.; Huang, H.; Ding, H. IscA/SufA Paralogs Are Required for the [4Fe-4S] Cluster Assembly in Enzymes of Multiple Physiological Pathways in Escherichia Coli under Aerobic Growth Conditions. Biochem. J. 2009, 420, 463–472. [Google Scholar] [CrossRef]
- Py, B.; Gerez, C.; Angelini, S.; Planel, R.; Vinella, D.; Loiseau, L.; Talla, E.; Brochier-Armanet, C.; Garcia Serres, R.; Latour, J.-M.; et al. Molecular Organization, Biochemical Function, Cellular Role and Evolution of NfuA, an Atypical Fe-S Carrier. Mol. Microbiol. 2012, 86, 155–171. [Google Scholar] [CrossRef]
- Ollagnier-de-Choudens, S.; Mattioli, T.; Takahashi, Y.; Fontecave, M. Iron-sulfur cluster assembly characterization of isca and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 2001, 276, 22604–22607. [Google Scholar] [CrossRef]
- Wollenberg, M.; Berndt, C.; Bill, E.; Schwenn, J.D.; Seidler, A. A Dimer of the FeS Cluster Biosynthesis Protein IscA from Cyanobacteria Binds a [2Fe2S] Cluster between Two Protomers and Transfers It to [2Fe2S] and [4Fe4S] Apo Proteins. Eur. J. Biochem. 2003, 270, 1662–1671. [Google Scholar] [CrossRef]
- Krebs, C.; Agar, J.N.; Smith, A.D.; Frazzon, J.; Dean, D.R.; Huynh, B.H.; Johnson, M.K. IscA, an Alternate Scaffold for Fe-S Cluster Biosynthesis. Biochemistry 2001, 40, 14069–14080. [Google Scholar] [CrossRef] [PubMed]
- Ollagnier-de Choudens, S.; Nachin, L.; Sanakis, Y.; Loiseau, L.; Barras, F.; Fontecave, M. SufA from Erwinia Chrysanthemi. Characterization of a Scaffold Protein Required for Iron-Sulfur Cluster Assembly. J. Biol. Chem. 2003, 278, 17993–18001. [Google Scholar] [CrossRef] [PubMed]
- Pala, Z.R.; Saxena, V.; Saggu, G.S.; Yadav, S.K.; Pareek, R.P.; Kochar, S.K.; Kochar, D.K.; Garg, S. Structural and Functional Characterization of an Iron–Sulfur Cluster Assembly Scaffold Protein-SufA from Plasmodium Vivax. Gene 2016, 585, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Naik, S.G.; O’Carroll, I.P.; Huynh, B.-H.; Dean, D.R.; Johnson, M.K.; Dos Santos, P.C. A Proposed Role for the Azotobacter Vinelandii NfuA Protein as an Intermediate Iron-Sulfur Cluster Carrier. J. Biol. Chem. 2008, 283, 14092–14099. [Google Scholar] [CrossRef] [PubMed]
- Angelini, S.; Gerez, C.; Choudens, S.O.; Sanakis, Y.; Fontecave, M.; Barras, F.; Py, B. NfuA, a New Factor Required for Maturing Fe/S Proteins in Escherichia Coli under Oxidative Stress and Iron Starvation Conditions. J. Biol. Chem. 2008, 283, 14084–14091. [Google Scholar] [CrossRef] [PubMed]
- Py, B.; Gerez, C.; Huguenot, A.; Vidaud, C.; Fontecave, M.; Ollagnier de Choudens, S.; Barras, F. The ErpA/NfuA Complex Builds an Oxidation-Resistant Fe-S Cluster Delivery Pathway. J. Biol. Chem. 2018, 293, 7689–7702. [Google Scholar] [CrossRef] [PubMed]
- Huet, G.; Daffé, M.; Saves, I. Identification of the Mycobacterium Tuberculosis SUF Machinery as the Exclusive Mycobacterial System of [Fe-S] Cluster Assembly: Evidence for Its Implication in the Pathogen’s Survival. J. Bacteriol. 2005, 187, 6137–6146. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the Biology of Mycobacterium Tuberculosis from the Complete Genome Sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes Required for Mycobacterial Growth Defined by High Density Mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Rengarajan, J.; Bloom, B.R.; Rubin, E.J. Genome-Wide Requirements for Mycobacterium Tuberculosis Adaptation and Survival in Macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 8327–8332. [Google Scholar] [CrossRef] [PubMed]
- Frazzon, J.; Fick, J.R.; Dean, D.R. Biosynthesis of Iron-Sulphur Clusters Is a Complex and Highly Conserved Process. Biochem. Soc. Trans. 2002, 30, 680–685. [Google Scholar] [CrossRef]
- Lu, H.-M.; Li, J.-D.; Zhang, Y.-D.; Lu, X.-L.; Xu, C.; Huang, Y.; Gribskov, M. The Evolution History of Fe–S Cluster A-Type Assembly Protein Reveals Multiple Gene Duplication Events and Essential Protein Motifs. Genome Biol. Evol. 2020, 12, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Gold, B.; Rodriguez, G.M.; Marras, S.A.E.; Pentecost, M.; Smith, I. The Mycobacterium Tuberculosis IdeR Is a Dual Functional Regulator That Controls Transcription of Genes Involved in Iron Acquisition, Iron Storage and Survival in Macrophages. Mol. Microbiol. 2001, 42, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Rodriguez, G.M. IdeR Is Required for Iron Homeostasis and Virulence in Mycobacterium Tuberculosis. Mol. Microbiol. 2014, 91, 98–109. [Google Scholar] [CrossRef]
- Wei, J.-R.; Krishnamoorthy, V.; Murphy, K.; Kim, J.-H.; Schnappinger, D.; Alber, T.; Sassetti, C.M.; Rhee, K.Y.; Rubin, E.J. Depletion of Antibiotic Targets Has Widely Varying Effects on Growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4176–4181. [Google Scholar] [CrossRef]
- Parish, T.; Stoker, N.G. Use of a Flexible Cassette Method to Generate a Double Unmarked Mycobacterium Tuberculosis TlyA PlcABC Mutant by Gene Replacement. Microbiology 2000, 146, 1969–1975. [Google Scholar] [CrossRef]
- de Wet, T.J.; Gobe, I.; Mhlanga, M.M.; Warner, D.F. CRISPRi-Seq for the Identification and Characterisation of Essential Mycobacterial Genes and Transcriptional Units. bioRxiv 2018, 358275. [Google Scholar] [CrossRef]
- Riemer, J.; Hoepken, H.H.; Czerwinska, H.; Robinson, S.R.; Dringen, R. Colorimetric Ferrozine-Based Assay for the Quantitation of Iron in Cultured Cells. Anal. Biochem. 2004, 331, 370–375. [Google Scholar] [CrossRef]
- Willemse, D.; Weber, B.; Masino, L.; Warren, R.M.; Adinolfi, S.; Pastore, A.; Williams, M.J. Rv1460, a SufR Homologue, Is a Repressor of the Suf Operon in Mycobacterium Tuberculosis. PLoS ONE 2018, 13, e0200145. [Google Scholar] [CrossRef] [PubMed]
- Leite, C.Q.; Beretta, A.L.; Anno, I.S.; Telles, M.A. Standardization of Broth Microdilution Method for Mycobacterium Tuberculosis. Mem. Inst. Oswaldo Cruz 2000, 95, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Alam, M.M.; Choudhury, M.E.; Kobayashi, N.; Ahmed, M.U. Determination of Minimum Inhibitory Concentration (MIC) of Cloxacilin for Selected Isolates of Methicilin-Resistant Staphylococcus Aureus (MRSA) with Their Antibiogram. Bangladesh J. Vet. Med. 2008, 6, 121–126. [Google Scholar] [CrossRef]
- Tian, J.; Bryk, R.; Itoh, M.; Suematsu, M.; Nathan, C. Variant Tricarboxylic Acid Cycle in Mycobacterium Tuberculosis: Identification of α-Ketoglutarate Decarboxylase. Proc. Natl. Acad. Sci. USA 2005, 102, 10670–10675. [Google Scholar] [CrossRef] [PubMed]
- Tamuhla, T.; Joubert, L.; Willemse, D.; Williams, M.J.Y. SufT Is Required for Growth of Mycobacterium Smegmatis under Iron Limiting Conditions. Microbiology 2020, 166, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.M.; Hopkins, F.F.; Chavez, A.; Diallo, M.; Chase, M.R.; Gerrick, E.R.; Pritchard, J.R.; Church, G.M.; Rubin, E.J.; Sassetti, C.M.; et al. Programmable Transcriptional Repression in Mycobacteria Using an Orthogonal CRISPR Interference Platform. Nat. Microbiol. 2017, 2, 16274. [Google Scholar] [CrossRef]
- Ding, H.; Yang, J.; Coleman, L.C.; Yeung, S. Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly isca and the bacterial frataxin ortholog cyay under physiological and oxidative stress conditions. J. Biol. Chem. 2007, 282, 7997–8004. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Shen, G.; Bryant, D.A.; Golbeck, J.H. Regulatory Roles for IscA and SufA in Iron Homeostasis and Redox Stress Responses in the Cyanobacterium Synechococcus Sp. Strain PCC 7002. J. Bacteriol. 2006, 188, 3182–3191. [Google Scholar] [CrossRef]
- Lucarelli, D.; Vasil, M.L.; Meyer-Klaucke, W.; Pohl, E. The Metal-Dependent Regulators FurA and FurB from Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2008, 9, 1548–1560. [Google Scholar] [CrossRef]
- Baloni, P.; Padiadpu, J.; Singh, A.; Gupta, K.R.; Chandra, N. Identifying Feasible Metabolic Routes in Mycobacterium Smegmatis and Possible Alterations under Diverse Nutrient Conditions. BMC Microbiol. 2014, 14, 276. [Google Scholar] [CrossRef]
- Tripathi, A.; Anand, K.; Das, M.; O’Niel, R.A.; Sabarinath, P.S.; Thakur, C.; Raghunatha, R.R.L.; Rajmani, R.S.; Chandra, N.; Laxman, S.; et al. Mycobacterium Tuberculosis Requires SufT for Fe-S Cluster Maturation, Metabolism, and Survival in Vivo. PLoS Pathog. 2022, 18, e1010475. [Google Scholar] [CrossRef] [PubMed]
- Amon, J.; Titgemeyer, F.; Burkovski, A. A Genomic View on Nitrogen Metabolism and Nitrogen Control in Mycobacteria. J. Mol. Microbiol. Biotechnol. 2009, 17, 20–29. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niemand Wolhuter, N.; Ngakane, L.; de Wet, T.J.; Warren, R.M.; Williams, M.J. The Mycobacterium smegmatis HesB Protein, MSMEG_4272, Is Required for In Vitro Growth and Iron Homeostasis. Microorganisms 2023, 11, 1573. https://doi.org/10.3390/microorganisms11061573
Niemand Wolhuter N, Ngakane L, de Wet TJ, Warren RM, Williams MJ. The Mycobacterium smegmatis HesB Protein, MSMEG_4272, Is Required for In Vitro Growth and Iron Homeostasis. Microorganisms. 2023; 11(6):1573. https://doi.org/10.3390/microorganisms11061573
Chicago/Turabian StyleNiemand Wolhuter, Nandi, Lerato Ngakane, Timothy J. de Wet, Robin M. Warren, and Monique J. Williams. 2023. "The Mycobacterium smegmatis HesB Protein, MSMEG_4272, Is Required for In Vitro Growth and Iron Homeostasis" Microorganisms 11, no. 6: 1573. https://doi.org/10.3390/microorganisms11061573
APA StyleNiemand Wolhuter, N., Ngakane, L., de Wet, T. J., Warren, R. M., & Williams, M. J. (2023). The Mycobacterium smegmatis HesB Protein, MSMEG_4272, Is Required for In Vitro Growth and Iron Homeostasis. Microorganisms, 11(6), 1573. https://doi.org/10.3390/microorganisms11061573





