Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Growth and Alkane Degradation Assays
2.3. Analytical Methods
2.4. Reverse Transcription and Real-Time Quantitative PCR (RT-qPCR)
2.5. Construction of alkB Gene Knockout Mutants
2.6. Heterologous Expression of alkB Genes
3. Results and Discussion
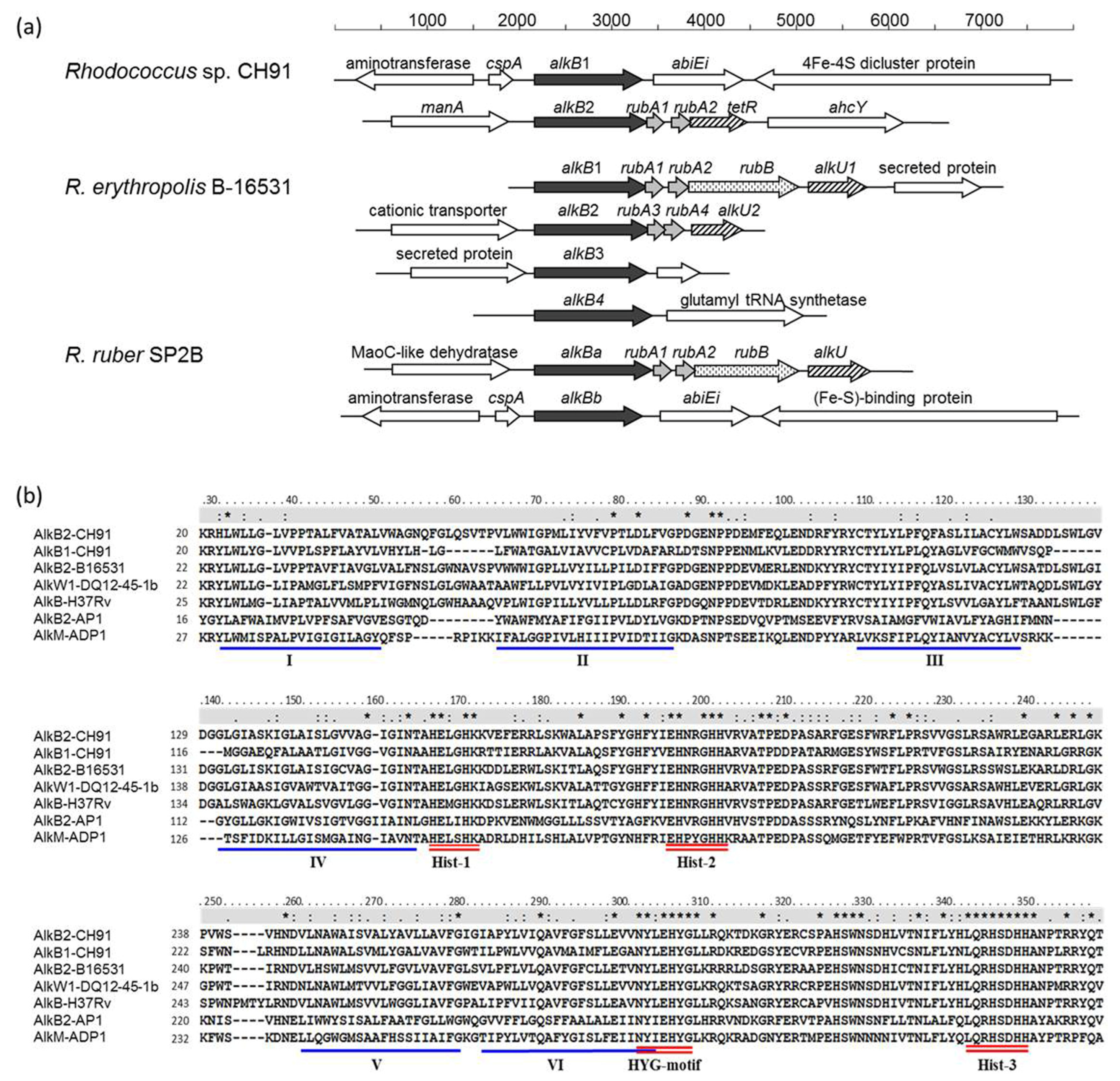
3.1. Genetic Characteristics of alkB Genes in Rhodococcus sp. CH91
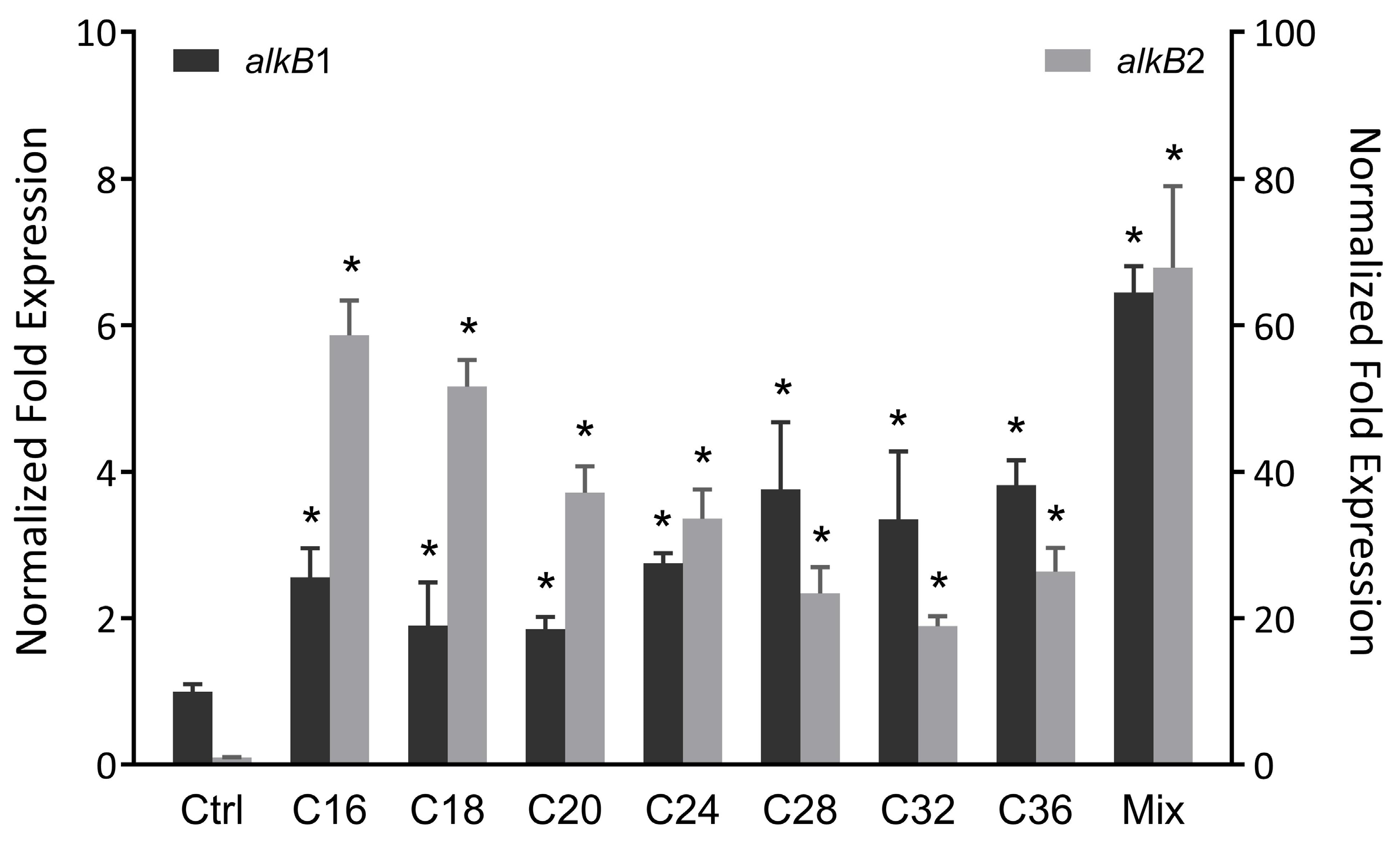
3.2. Transcriptional Expression of n-Alkane Hydroxylase Genes in Rhodococcus sp. Strain CH91
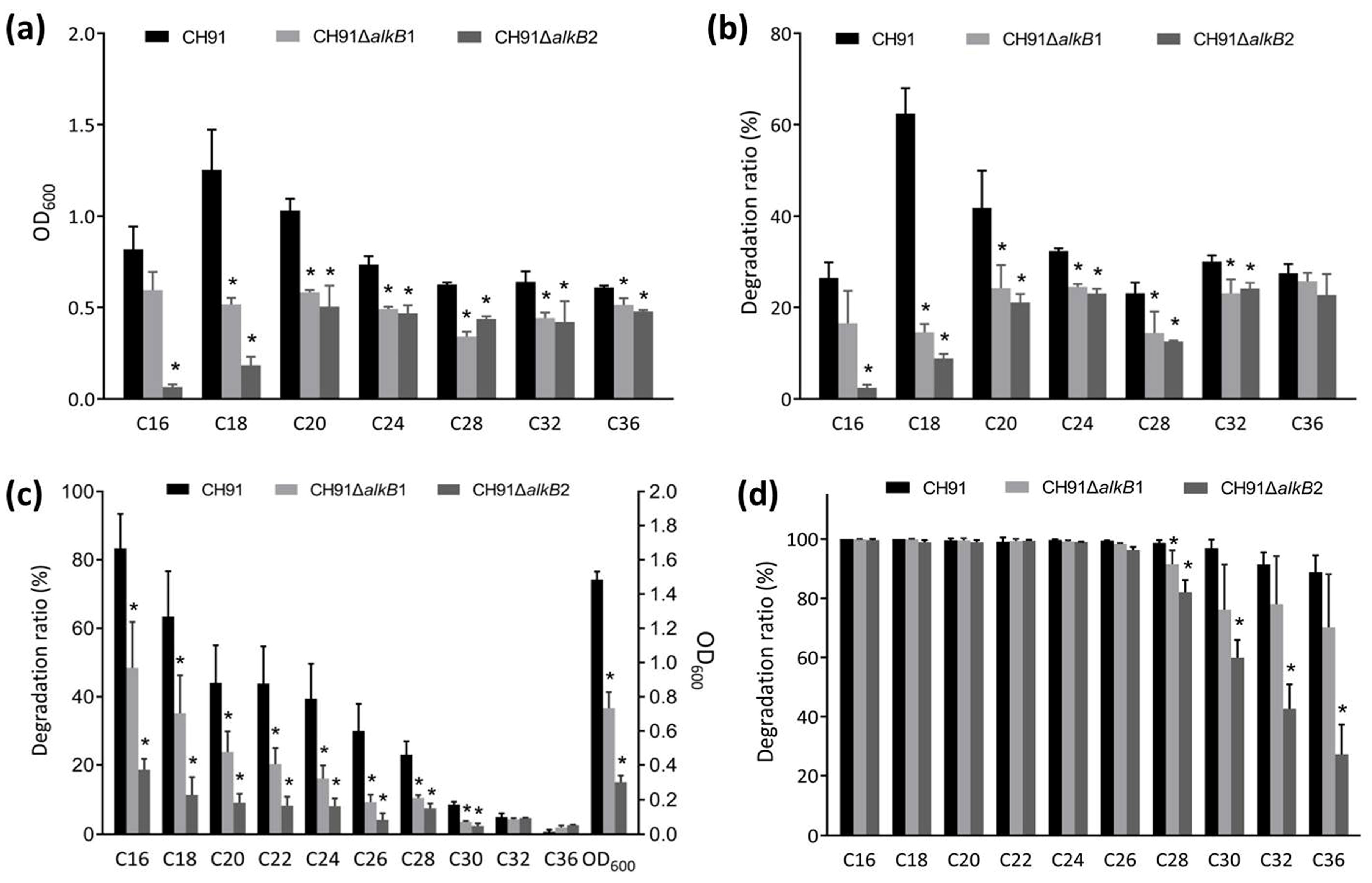
3.3. Utilization of n-Alkanes by Wild-Type Strain CH91 and Its Mutant Derivatives
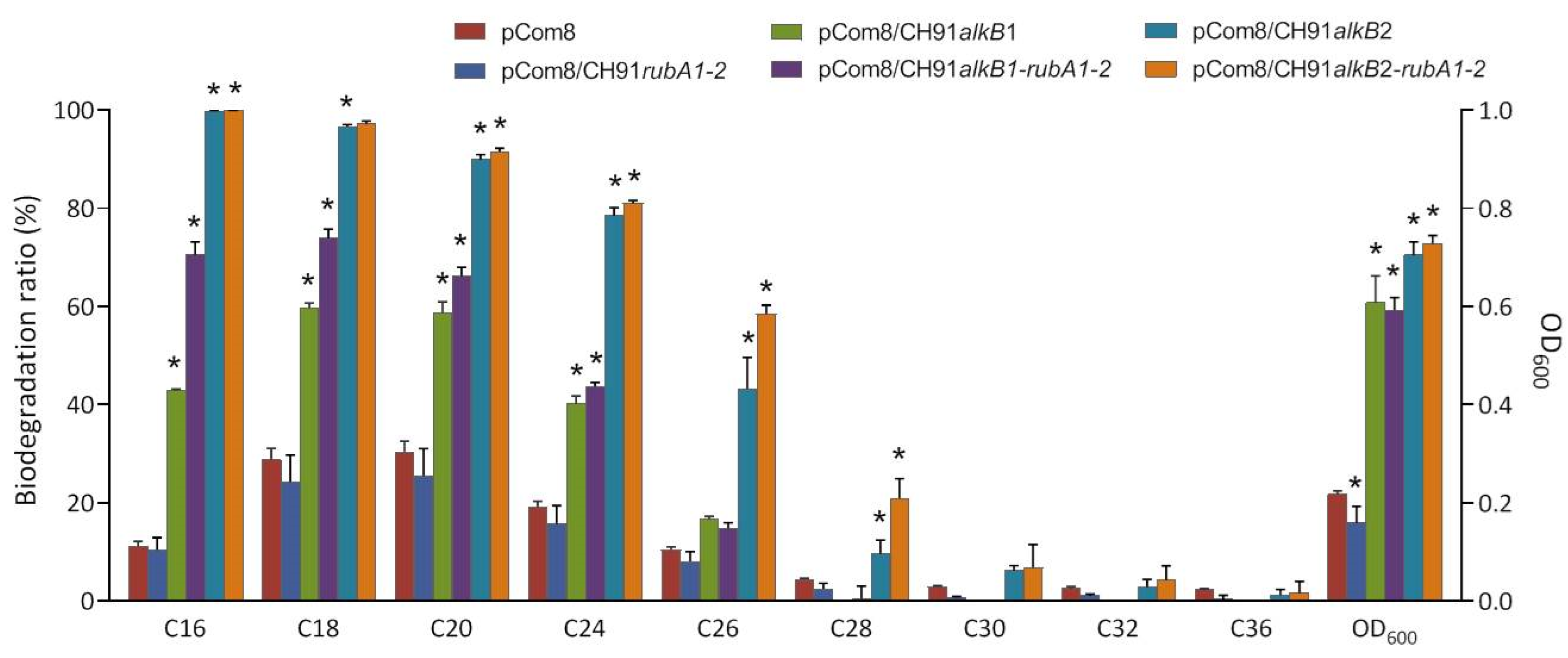
3.4. Functional Complementation of Strain CH91 alkB Genes in P. fluorescens KOB2Δ1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.J.C.; de Carvalho, C.C.C.R. Phenotypic adaptations help Rhodococcus erythropolis cells during the degradation of paraffin wax. Biotechnol. J. 2019, 14, e1800598. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhao, C.; Gao, X.; Wang, L.; Tian, Q.; Liu, Y.; Xue, S.; Han, Z.; Chen, F.; Wang, S. Characterization and transcriptome analysis of a long-chain n-alkane-degrading strain Acinetobacter pittii SW-1. Int. J. Environ. Res. Public Health 2021, 18, 6365. [Google Scholar] [CrossRef] [PubMed]
- Romanova, V.; Markelova, M.; Boulygina, E.; Siniagina, M.; Müller, R.; Grigoryeva, T.; Laikov, A. Significance of both alkB and P450 alkane-degrading systems in Tsukamurella tyrosinosolvens: Proteomic evidence. Appl. Microbiol. Biotechnol. 2022, 106, 3153–3171. [Google Scholar] [CrossRef]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef]
- SadrAzodi, S.M.; Shavandi, M.; Amoozegar, M.A.; Mehrnia, M.R. Biodegradation of long chain alkanes in halophilic conditions by Alcanivorax sp. strain Est-02 isolated from saline soil. 3 Biotech. 2019, 9, 141. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Wubbolts, M.G.; Witholt, B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 1994, 5, 161–174. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Panke, S.; Lucchini, S.; Franchini, A.G.; Röthlisberger, M.; Witholt, B. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: Evolution and regulation of the alk genes. Microbiology 2001, 147 Pt 6, 1621–1630. [Google Scholar] [CrossRef]
- Williams, S.C.; Austin, R.N. An overview of the electron-transfer proteins that activate alkane monooxygenase (AlkB). Front. Microbiol. 2022, 13, 845551. [Google Scholar] [CrossRef]
- Geissdörfer, W.; Kok, R.G.; Ratajczak, A.; Hellingwerf, K.J.; Hillen, W. The genes rubA and rubB for alkane degradation in Acinetobacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 1999, 181, 4292–4298. [Google Scholar] [CrossRef]
- Ratajczak, A.; Geissdörfer, W.; Hillen, W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl. Environ. Microbiol. 1998, 64, 1175–1179. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Markussen, S.; Winnberg, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: The role of two AlkB-type alkane hydroxylases. Appl. Microbiol. Biotechnol. 2006, 72, 353–360. [Google Scholar] [CrossRef]
- Whyte, L.G.; Smits, T.H.; Labbé, D.; Witholt, B.; Greer, C.W.; van Beilen, J.B. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 2002, 68, 5933–5942. [Google Scholar] [CrossRef]
- Smits, T.H.; Balada, S.B.; Witholt, B.; van Beilen, J.B. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 2002, 184, 1733–1742. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Li, Y.P.; Pan, J.C.; Ma, Y.L. Elucidation of multiple alkane hydroxylase systems in biodegradation of crude oil n-alkane pollution by Pseudomonas aeruginosa DN1. J. Appl. Microbiol. 2020, 128, 151–160. [Google Scholar] [CrossRef]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef]
- Lo Piccolo, L.; De Pasquale, C.; Fodale, R.; Puglia, A.M.; Quatrini, P. Involvement of an alkane hydroxylase system of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Appl. Environ. Microbiol. 2011, 77, 1204–1213. [Google Scholar] [CrossRef]
- Park, C.; Shin, B.; Jung, J.; Lee, Y.; Park, W. Metabolic and stress responses of Acinetobacter oleivorans DR1 during long-chain alkane degradation. Microb. Biotechnol. 2017, 10, 1809–1823. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Liang, Y.; Hong, S.; Wang, G.; You, J.; Xue, Y.; Ma, Y. Degradation of long-chain n-alkanes by a novel thermal-tolerant Rhodococcus strain. Arch. Microbiol. 2022, 204, 259. [Google Scholar] [CrossRef] [PubMed]
- Lageveen, R.G.; Huisman, G.W.; Preusting, H.; Ketelaar, P.; Eggink, G.; Witholt, B. Formation of polyesters by Pseudomonas oleovorans: Effect of Substrates on Formation and Composition of Poly-(R)-3-Hydroxyalkanoates and Poly-(R)-3-Hydroxyalkenoates. Appl. Environ. Microbiol. 1988, 54, 2924–2932. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.; Higa, A. Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 1970, 53, 159–162. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Liang, Y.; Jiao, S.; Wang, M.; Yu, H.; Shen, Z. A CRISPR/Cas9-based genome editing system for Rhodococcus ruber TH. Metab. Eng. 2020, 57, 13–22. [Google Scholar] [CrossRef]
- Smits, T.H.; Seeger, M.A.; Witholt, B.; van Beilen, J.B. New alkane-responsive expression vectors for Escherichia coli and Pseudomonas. Plasmid 2001, 46, 16–24. [Google Scholar] [CrossRef]
- Boonmak, C.; Takahashi, Y.; Morikawa, M. Cloning and expression of three ladA-type alkane monooxygenase genes from an extremely thermophilic alkane-degrading bacterium Geobacillus thermoleovorans B23. Extremophiles 2014, 18, 515–523. [Google Scholar] [CrossRef]
- Casini, A.; MacDonald, J.T.; De Jonghe, J.; Christodoulou, G.; Freemont, P.S.; Baldwin, G.S.; Ellis, T. One-pot DNA construction for synthetic biology: The Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 2014, 42, e7. [Google Scholar] [CrossRef]
- Højberg, O.; Schnider, U.; Winteler, H.V.; Sørensen, J.; Haas, D. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. Appl. Environ. Microbiol. 1999, 65, 4085–4093. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Shanklin, J.; Whittle, E.; Fox, B.G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 1994, 33, 12787–12794. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Penninga, D.; Witholt, B. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 1992, 267, 9194–9201. [Google Scholar] [CrossRef]
- Tani, A.; Ishige, T.; Sakai, Y.; Kato, N. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 2001, 183, 1819–1823. [Google Scholar] [CrossRef]
- Takei, D.; Washio, K.; Morikawa, M. Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol. Lett. 2008, 30, 1447–1452. [Google Scholar] [CrossRef]
- Laczi, K.; Kis, Á.; Horváth, B.; Maróti, G.; Hegedüs, B.; Perei, K.; Rákhely, G. Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons. Appl. Microbiol. Biotechnol. 2015, 99, 9745–9759. [Google Scholar] [CrossRef]
- Marín, M.M.; Yuste, L.; Rojo, F. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 3232–3237. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Marín, M.M.; Smits, T.H.; Röthlisberger, M.; Franchini, A.G.; Witholt, B.; Rojo, F. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 2004, 6, 264–273. [Google Scholar] [CrossRef]
- Nie, Y.; Liang, J.; Fang, H.; Tang, Y.Q.; Wu, X.L. Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl. Environ. Microbiol. 2011, 77, 7279–7288. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, W.; Hong, S.; Xue, Y.; Ma, Y. Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91. Microorganisms 2023, 11, 1537. https://doi.org/10.3390/microorganisms11061537
Xiang W, Hong S, Xue Y, Ma Y. Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91. Microorganisms. 2023; 11(6):1537. https://doi.org/10.3390/microorganisms11061537
Chicago/Turabian StyleXiang, Wei, Shan Hong, Yanfen Xue, and Yanhe Ma. 2023. "Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91" Microorganisms 11, no. 6: 1537. https://doi.org/10.3390/microorganisms11061537
APA StyleXiang, W., Hong, S., Xue, Y., & Ma, Y. (2023). Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91. Microorganisms, 11(6), 1537. https://doi.org/10.3390/microorganisms11061537



