Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. Culture and DNA Extraction
2.3. Identification of STR Loci
2.4. Whole-Genome Sequencing
2.5. Primer Design and PCR Genotyping
2.6. Data Analysis
2.7. WGS SNP Calling
2.8. Antifungal Susceptibility Testing (AFST)
2.9. Investigation Resistance-Associated Genes
3. Results
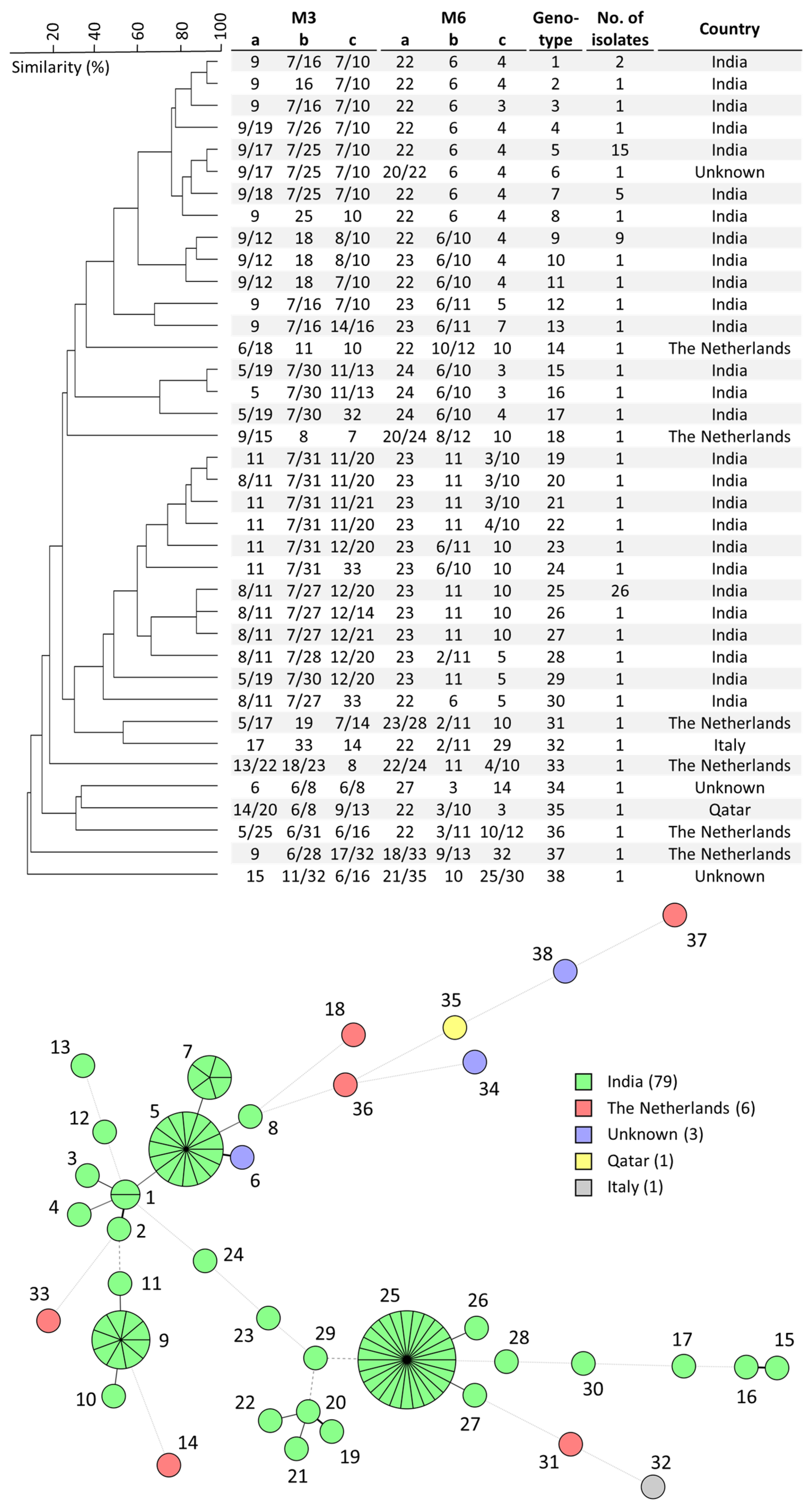
3.1. Development and Application of W. anomalus STR Genotyping
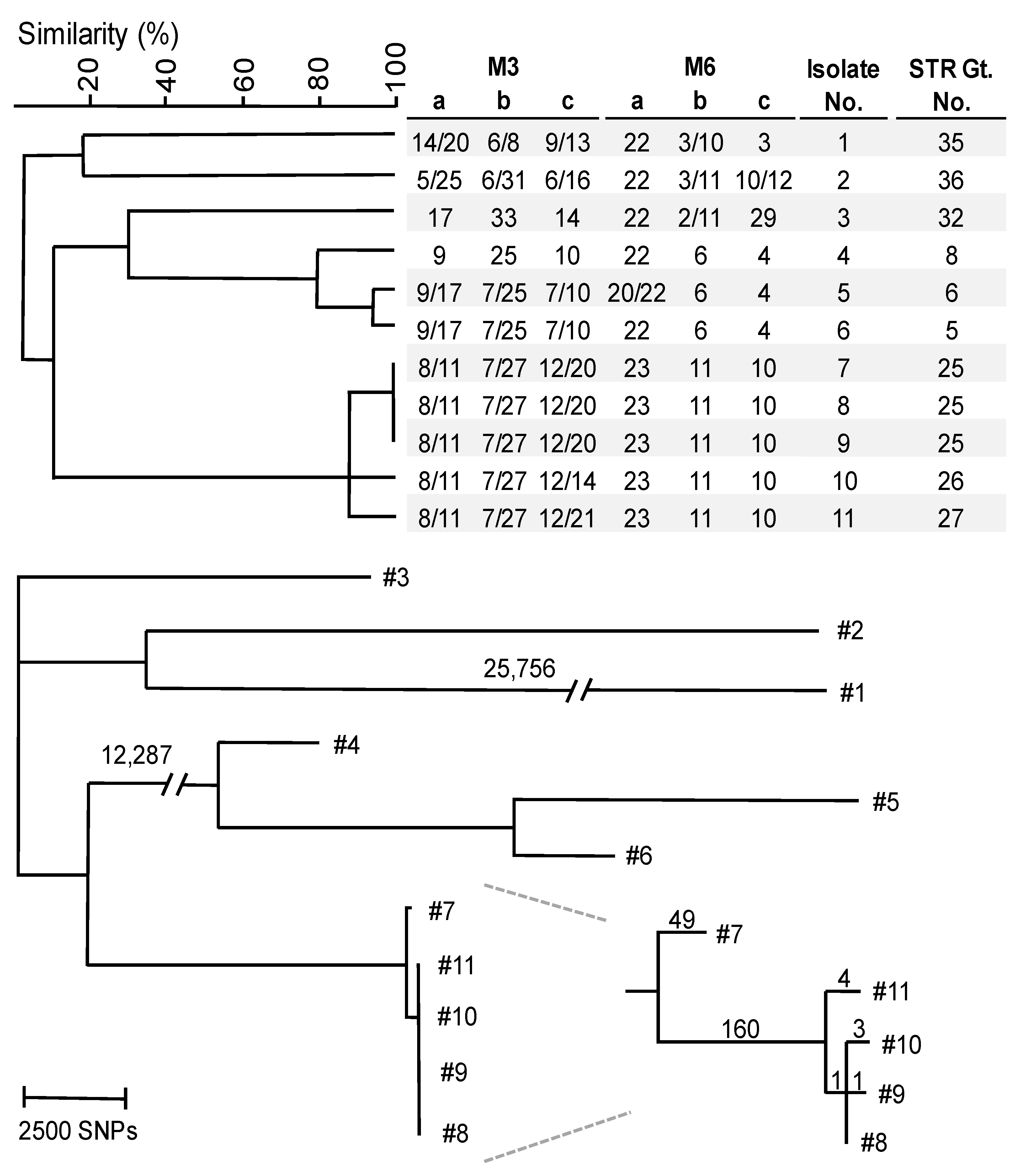
3.2. WGS SNP Analysis
3.3. AFST and Resistance-Associated Genes Investigation
4. Discussion
4.1. W. anomalus Genotyping and Loss of Heterozygosity
4.2. Simultaneous Outbreaks of Different W. anomalus Strains in Single Hospital
4.3. Antifungal Susceptibility and I469L Substitution in ERG11
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rokas, A. Evolution of the human pathogenic lifestyle in fungi. Nat. Microbiol. 2022, 7, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.; Neofytos, D.; Diekema, D.; Azie, N.; Meier-Kriesche, H.U.; Quan, S.P.; Horn, D. Epidemiology and outcomes of candidemia in 3648 patients: Data from the Prospective Antifungal Therapy (PATH Alliance(R)) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 2012, 74, 323–331. [Google Scholar] [CrossRef]
- Hoenigl, M.; Egger, M.; Salmanton-Garcia, J.; Koehler, P.; Gangneux, J.P.; Arendrup, M.C.; Bicanic, T.; Arikan-Akdagli, S.; Cornely, O.A.; ECMM Study Group. Guideline adherence and survival of patients with candidaemia in Europe: Results from the ECMM Candida III multinational European observational cohort study. Lancet Infect. Dis. 2023, 9, ofac492.038. [Google Scholar] [CrossRef]
- Lyon, G.M.; Karatela, S.; Sunay, S.; Adiri, Y.; Candida Surveillance Study Investigators. Antifungal susceptibility testing of Candida isolates from the Candida surveillance study. J. Clin. Microbiol. 2010, 48, 1270–1275. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species From 1997–2016. Open Forum. Infect. Dis. 2019, 6, S79–S94. [Google Scholar] [CrossRef]
- Sharma, M.; Chakrabarti, A. Candidiasis and Other Emerging Yeasts. Curr. Fungal Infect. Rep. 2023, 17, 15–24. [Google Scholar] [CrossRef]
- Martini, C.; Torelli, R.; de Groot, T.; De Carolis, E.; Morandotti, G.A.; De Angelis, G.; Posteraro, B.; Meis, J.F.; Sanguinetti, M. Prevalence and Clonal Distribution of Azole-Resistant Candida parapsilosis Isolates Causing Bloodstream Infections in a Large Italian Hospital. Front. Cell. Infect. Microbiol. 2020, 10, 232. [Google Scholar] [CrossRef]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017, 13, e1006290. [Google Scholar] [CrossRef]
- Souza, A.C.; Fuchs, B.B.; Pinhati, H.M.S.; Siqueira, R.A.; Hagen, F.; Meis, J.F.; Mylonakis, E.; Colombo, A.L. Candida parapsilosis Resistance to Fluconazole: Molecular Mechanisms and In Vivo Impact in Infected Galleria mellonella Larvae. Antimicrob. Agents Chemother. 2015, 59, 6581–6587. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Baqueiro, C.C.S.Z.; Colombo, A.L.; Meijer, E.F.J.; de Almeida, J.N., Jr.; Berrio, I.; Fernández, N.B.; Chaves, G.M.; Meis, J.F.; de Groot, T.; et al. Short Tandem Repeat Genotyping and Antifungal Susceptibility Testing of Latin American Candida tropicalis Isolates. J. Fungi 2023, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Kalkanci, A.; Dizbay, M.; Turan, O.; Fidan, I.; Yalçin, B.; Hirfanoğlu, I.; Kuştimur, S.; Aktaş, F.; Sugita, T. Nosocomial transmission of Candida pelliculosa fungemia in a pediatric intensive care unit and review of the literature. Turk. J. Pediatr. 2010, 52, 42–49. [Google Scholar] [PubMed]
- Yang, Y.; Wu, W.; Ding, L.; Yang, L.; Su, J.; Wu, B. Two different clones of Candida pelliculosa bloodstream infection in a tertiary neonatal intensive care unit. J. Infect. Dev. Ctries. 2021, 15, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Song, K.Y.; Park, C.; Byun, J.H.; Chun, H.S.; Choi, J.H.; Han, E.H.; Lee, S.O.; Jeong, Y.; Kim, Y.J.; Kim, S.H. Fungal arthritis with adjacent osteomyelitis caused by Candida pelliculosa: A case report. BMC Infect. Dis. 2020, 20, 438. [Google Scholar] [CrossRef] [PubMed]
- Mehta, V.; Mohanty, A.; Meena, S.; Rahul, J.S.; Kumar, N.U.; Chattopadhyay, D.; Bakliwal, A.; Choudhary, R.; Gupta, P. Wickerhamomyces anomalous: A Rare Cause of Fungemia Causing Febrile Neutropenia in Acute Lymphoblastic Leukemia. Case Rep. Infect. Dis. 2020, 2020, 8847853. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.W.; Cartwright, E.J.; Reddy, S.C.; Kraft, C.S.; Wang, Y.F. Pichia anomala (Candida pelliculosa) fungemia in a patient with sickle cell disease. Mycopathologia 2013, 176, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Singh, K.; Narang, A.; Singhi, S.; Batra, R.; Rao, K.L.N.; Ray, P.; Gopalan, S.; Das, S.; Gupta, A.K.; et al. Outbreak of Pichia anomala infection in the pediatric service of a tertiary-care center in Northern India. J. Clin. Microbiol. 2001, 39, 1702–1706. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Sood, P.; Rudramurthy, S.M.; Chen, S.; Jillwin, J.; Iyer, R.; Sharma, A.; Harish, B.N.; Roy, I.; Kindo, A.J.; et al. Characteristics, outcome and risk factors for mortality of paediatric patients with ICU-acquired candidemia in India: A multicentre prospective study. Mycoses 2020, 63, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Passoth, V.; Olstorpe, M.; Schnurer, J. Past, present and future research directions with Pichia anomala. Antonie Van Leeuwenhoek 2011, 99, 121–125. [Google Scholar] [CrossRef]
- Walker, G.M. Pichia anomala: Cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek 2011, 99, 25–34. [Google Scholar] [CrossRef]
- Steyn, A.; Roets, F.; Botha, A. Yeasts Associated with Culex pipiens and Culex theileri Mosquito Larvae and the Effect of Selected Yeast Strains on the Ontogeny of Culex pipiens. Microb. Ecol. 2016, 71, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cao, Y.; Li, Y.; Chen, X.; Ding, C.; Liu, Y. Risk factors and biofilm formation analyses of hospital-acquired infection of Candida pelliculosa in a neonatal intensive care unit. BMC Infect. Dis. 2021, 21, 620. [Google Scholar] [CrossRef] [PubMed]
- Kidd, S.E.; Abdolrasouli, A.; Hagen, F. Fungal Nomenclature: Managing Change is the Name of the Game. Open Forum. Infect. Dis. 2023, 10, ofac559. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek 2011, 99, 13–23. [Google Scholar] [CrossRef]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Duong Vu Cardinali, G.; Arthur, I.; Norman, A.C.; Giraldo, A. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Arendrup, M.C. Update on antifungal resistance in Aspergillus and Candida. Clin. Microbiol. Infect. 2014, 20, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Lupetti, A.; Danesi, R.; Campa, M.; Del Tacca, M.; Kelly, S. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 2002, 8, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.J.; Liu, J.Y.; Ni, P.H.; Wang, S.; Shi, C.; Wei, B.; Ni, Y.X.; Ge, H.L. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013, 13, 386–393. [Google Scholar] [CrossRef]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2016, 7, 2173. [Google Scholar] [CrossRef]
- Barchiesi, F.; Tortorano, A.M.; Flaconi Di Francesco, L.; Rigoni, A.; Giacometti, A.; Spreghini, E.; Scalise, G.; Viviani, M.A. Genotypic variation and antifungal susceptibilities of Candida pelliculosa clinical isolates. J. Med. Microbiol. 2005, 54, 279–285. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, M.; Arastehfar, A.; Ilkit, M.; Zou, J.; Deng Yuchen Xu, Y.; Liao, W.; Zhao, J.; Fang, W. Investigation of the Emerging Nosocomial Wickerhamomyces anomalus Infections at a Chinese Tertiary Teaching Hospital and a Systemic Review: Clinical Manifestations, Risk Factors, Treatment, Outcomes, and Anti-fungal Susceptibility. Front. Microbiol. 2021, 12, 744502. [Google Scholar] [CrossRef]
- Lin, H.C.; Lin, H.Y.; Su, B.H.; Ho, M.W.; Ho, C.M.; Lee, C.Y.; Lin, M.H.; Hsieh, H.Y.; Lin, H.C.; Li, T.C.; et al. Reporting an outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. J. Microbiol. Immunol. Infect. 2013, 46, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Moon, Y.S.; Yoo, J.A.; Lim, J.H.; Jeong, J.; Jun, J.B. Investigation of a nosocomial outbreak of fungemia caused by Candida pelliculosa (Pichia anomala) in a Korean tertiary care center. J. Microbiol. Immunol. Infect. 2018, 51, 794–801. [Google Scholar] [CrossRef]
- Yilmaz-Semerci, S.; Demirel, G.; Tastekin, A. Wickerhamomyces anomalus blood stream infection in a term newborn with pneumonia. Turk. J. Pediatr. 2017, 59, 349–351. [Google Scholar] [CrossRef]
- Suhr, M.J.; Gomes-Neto, J.C.; Banjara, N.; Florescu, D.F.; Mercer, D.F.; Iwen, P.C.; Hallen-Adams, H.E. Epidemiological investigation of Candida species causing bloodstream infection in paediatric small bowel transplant recipients. Mycoses 2017, 60, 366–374. [Google Scholar] [CrossRef]
- de Groot, T.; Spruijtenburg, B.; Parnell, L.A.; Chow, N.A.; Meis, J.F. Optimization and Validation of Candida auris Short Tandem Repeat Analysis. Microbiol. Spectr. 2022, 10, e0264522. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Bombassaro, A.; Meijer, E.F.J.; Rodrigues, A.M.; Grisolia, M.E.; Vicente, V.A.; de Queiroz-Telles, F.; Meis, J.F.; de Groot, T. Sporothrix brasiliensis genotyping reveals numerous independent zoonotic introductions in Brazil. J. Infect. 2023, 86, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Spruijtenburg, B.; van Haren, M.H.I.; Chowdhary, A.; Meis, J.F.; de Groot, T. Development and Application of a Short Tandem Repeat Multiplex Typing Assay for Candida tropicalis. Microbiol. Spectr. 2023, 11, e0461822. [Google Scholar] [CrossRef]
- van Haren, M.H.I.; de Groot, T.; Spruijtenburg, B.; Jain, K.; Chowdhary, A.; Meis, J.F. Development of a Multiplex PCR Short Tandem Repeat Typing Scheme for Candida krusei. J. Clin. Microbiol. 2022, 60, e0203221. [Google Scholar] [CrossRef] [PubMed]
- De Carolis, E.; Vella, A.; Vaccaro, L.; Torelli, R.; Posteraro, P.; Ricciardi, W.; Sanguinetti, M.; Posteraro, B. Development and validation of an in-house database for matrix-assisted laser desorption ionization-time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J. Clin. Microbiol. 2014, 52, 1453–1458. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Skinner, M.E.; Uzilov, A.V.; Stein, L.D.; Mungall, C.J.; Holmes, I.H. JBrowse: A next-generation genome browser. Genome Res. 2009, 19, 1630–1638. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Welsh, R.M.; Misas, E.; Forsberg, K.; Lyman, M.; Chow, N.A. Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines. J. Fungi 2021, 7, 214. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Badali, H.; Abastabar, M.; Mirhendi, H.; Khodavaisy, S.; Sharifisooraki, J.; Armaki, M.T.; de Groot, T.; Meis, J.F. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes. Infect. 2022, 11, 2405–2411. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Ramasamy, U.; Chen, D. VCF2PopTree: A client-side software to construct population phylogeny from genome-wide SNPs. PeerJ 2019, 7, e8213. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th ed.; Approved Standard M27; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Clinical and Laboratory Standards Institute. Epidemiological Cutoff Values for Antifungal Susceptiblity Testing, M57S, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics Systems, Version 1.2r3pre, Schrödinger LLC. Available online: https://pymol.org/2/ (accessed on 22 August 2022).
- Guinea, J.; Mezquita, S.; Gómez, A.; Padilla, B.; Zamora, E.; Sánchez-Luna, M.; Sánchez-Carrillo, C.; Muñoz, P.; Escribano, P. Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med. Mycol. 2021, 60, myab068. [Google Scholar] [CrossRef]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2011, 2, e00129-11. [Google Scholar] [CrossRef]
- Diogo, D.; Bouchier, C.; d’Enfert, C.; Bougnoux, M.E. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet Biol. 2009, 46, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Coste, A.; Turner, V.; Ischer, F.; Morschhäuser, J.; Forche, A.; Selmecki, A.; Berman, J.; Bille, J.; Sanglard, D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 2006, 172, 2139–2156. [Google Scholar] [CrossRef]
- Dunkel, N.; Blass, J.; Rogers, P.D.; Morschhäuser, J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 2008, 69, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Niimi, K.; Monk, B.C.; Hirai, A.; Hatakenaka, K.; Umeyama, T.; Lamping, E.; Maki, K.; Tanabe, K.; Kamimura, T.; Ikeda, F.; et al. Clinically significant micafungin resistance in Candida albicans involves modification of a glucan synthase catalytic subunit GSC1 (FKS1) allele followed by loss of heterozygosity. J. Antimicrob. Chemother. 2010, 65, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Lo, H.S.; Wu, C.C.; Ko, H.C.; CHang, T.P.; Yang, Y.L. Loss of heterozygosity of FCY2 leading to the development of flucytosine resistance in Candida tropicalis. Antimicrob. Agents Chemother. 2011, 55, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.M.; Moons, M.C.; Huret, S.; Vrancken, G.; De Vuyst, L. Wickerhamomyces anomalus in the sourdough microbial ecosystem. Antonie Van Leeuwenhoek 2011, 99, 63–73. [Google Scholar] [CrossRef]
- Muccilli, S.; Wemhoff, S.; Restuccia, C.; Meinhardt, F. Exoglucanase-encoding genes from three Wickerhamomyces anomalus killer strains isolated from olive brine. Yeast 2013, 30, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Marichal, P.; Koymans, L.; Willemsens, S.; Bellens, D.; Verhasselt, P.; Luyten, W.; Borgers, M.; Ramaekers, F.C.S.; Odds, F.C.; Vanden Bossche, H. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 1999, 145, 2701–2713. [Google Scholar] [CrossRef] [PubMed]

| PCR Panel and Primer Name | Primer Sequence (5′–3′) | Conc b (µM) | No. of Bases of Primer-Flanking Sequence | Repeat Unit | No. of Repeats c | No. of Alleles | Intragenic/ Locus Protein Coding Gene d | ||
|---|---|---|---|---|---|---|---|---|---|
| Forward Primer a | Reverse Primer | Min | Max | ||||||
| M3 | |||||||||
| M3-a | FAM-TCTTGCAAATCGTCAGACATC | ACCATCCTTGTTCCCTTACAAC | 5 | 295 | TTG | 5 | 25 | 16 | WICANDRAFT_30999 |
| M3-b | JOE-AGCTTGTAATTGTTGGGCTTG | AGCTCCAGAAACTGAACCAAC | 5 | 128 | TGT | 6 | 33 | 18 | WICANDRAFT_34460 |
| M3-c | TAMRA-AGCTCAATTCCAAGCTGAAC | AATCAATCTCTGAGGGTGAAGTC | 5 | 242 | ACA | 6 | 33 | 15 | WICANDRAFT_91930 |
| M6 | |||||||||
| M6-a | FAM-GTTCGTTTGCAGTTTCTTTCC | ATCACCAACAAACTCGCTACC | 2 | 123 | TTCACT | 18 | 38 | 12 | WICANDRAFT_78907 |
| M6-b | JOE-TGCCTTATATGAAGGATGAAGG | AGAGTCACCTCTTGGGCTATG | 2 | 169 | GAGAGT | 2 | 19 | 12 | WICANDRAFT_24702 |
| M6-c | TAMRA-CGGAAGGAATAAGAAGCAAAG | GATGTTGGGTATTGTTGTCACTG | 10 | 114 | AGCAAC | 3 | 32 | 15 | WICANDRAFT_97047 |
| Antifungal | Range (mg/L) | GM (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | n Non-Wildtype (%) |
|---|---|---|---|---|---|
| AMB (n = 87) | 0.016–2 | 0.24 | 0.25 | 0.5 | 6 (6.9) |
| FLU (n = 87) | 0.5–16 | 1.71 | 2 | 4 | 2 (2.3) |
| ITC (n = 87) | 0.016–1 | 0.21 | 0.25 | 0.5 | 1 (1.1) |
| VOR (n = 87) | 0.03–0.5 | 0.09 | 0.125 | 0.125 | 3 (3.4) |
| ISA (n = 49) | 0.031–0.125 | 0.06 | 0.063 | 0.063 | N.A. |
| AFG (n = 48) | ≤0.008–0.12 | 0.04 | 0.031 | 0.12 | N.A. |
| MFG (n = 50) | ≤0.008–0.032 | 0.01 | 0.016 | 0.031 | 0 (0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spruijtenburg, B.; Rudramurthy, S.M.; Meijer, E.F.J.; van Haren, M.H.I.; Kaur, H.; Chakrabarti, A.; Meis, J.F.; de Groot, T. Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital. Microorganisms 2023, 11, 1525. https://doi.org/10.3390/microorganisms11061525
Spruijtenburg B, Rudramurthy SM, Meijer EFJ, van Haren MHI, Kaur H, Chakrabarti A, Meis JF, de Groot T. Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital. Microorganisms. 2023; 11(6):1525. https://doi.org/10.3390/microorganisms11061525
Chicago/Turabian StyleSpruijtenburg, Bram, Shivaprakash M. Rudramurthy, Eelco F. J. Meijer, Merlijn H. I. van Haren, Harsimran Kaur, Arunaloke Chakrabarti, Jacques F. Meis, and Theun de Groot. 2023. "Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital" Microorganisms 11, no. 6: 1525. https://doi.org/10.3390/microorganisms11061525
APA StyleSpruijtenburg, B., Rudramurthy, S. M., Meijer, E. F. J., van Haren, M. H. I., Kaur, H., Chakrabarti, A., Meis, J. F., & de Groot, T. (2023). Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital. Microorganisms, 11(6), 1525. https://doi.org/10.3390/microorganisms11061525





