Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Determination of Antibiotic Susceptibility of Bacterial Isolates
2.3. Bacteriophage Isolation
2.4. Determination of Bacteriophage Titer
2.5. Bacteriophage Purification and Propagation
2.6. Multiplicity of Infection (MOI)
2.7. Host Range Determination
2.8. Influence of Temperature and pH on Bacteriophage Stability
2.9. Adsorption Assay and One-Step Growth Curve
2.10. Concentration of Bacteriophages by Ultracentrifugation
2.11. Electron Microscopy
2.12. Phage DNA Isolation and Sequencing
2.13. Bioinformatic Analysis
2.14. Statistics and Graphic Display of Results
3. Results
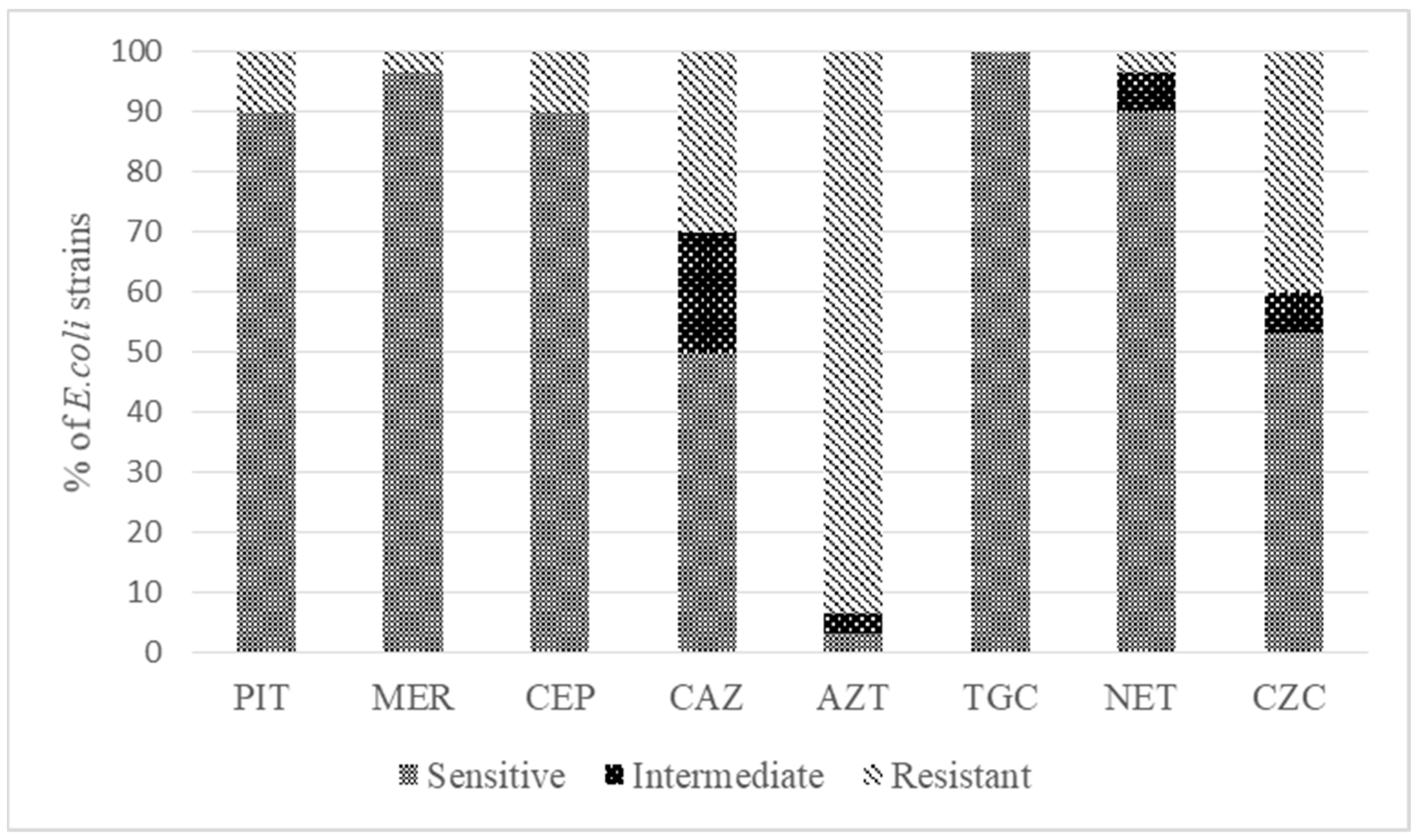
3.1. Determination of the Antibiotic Resistance of Isolates
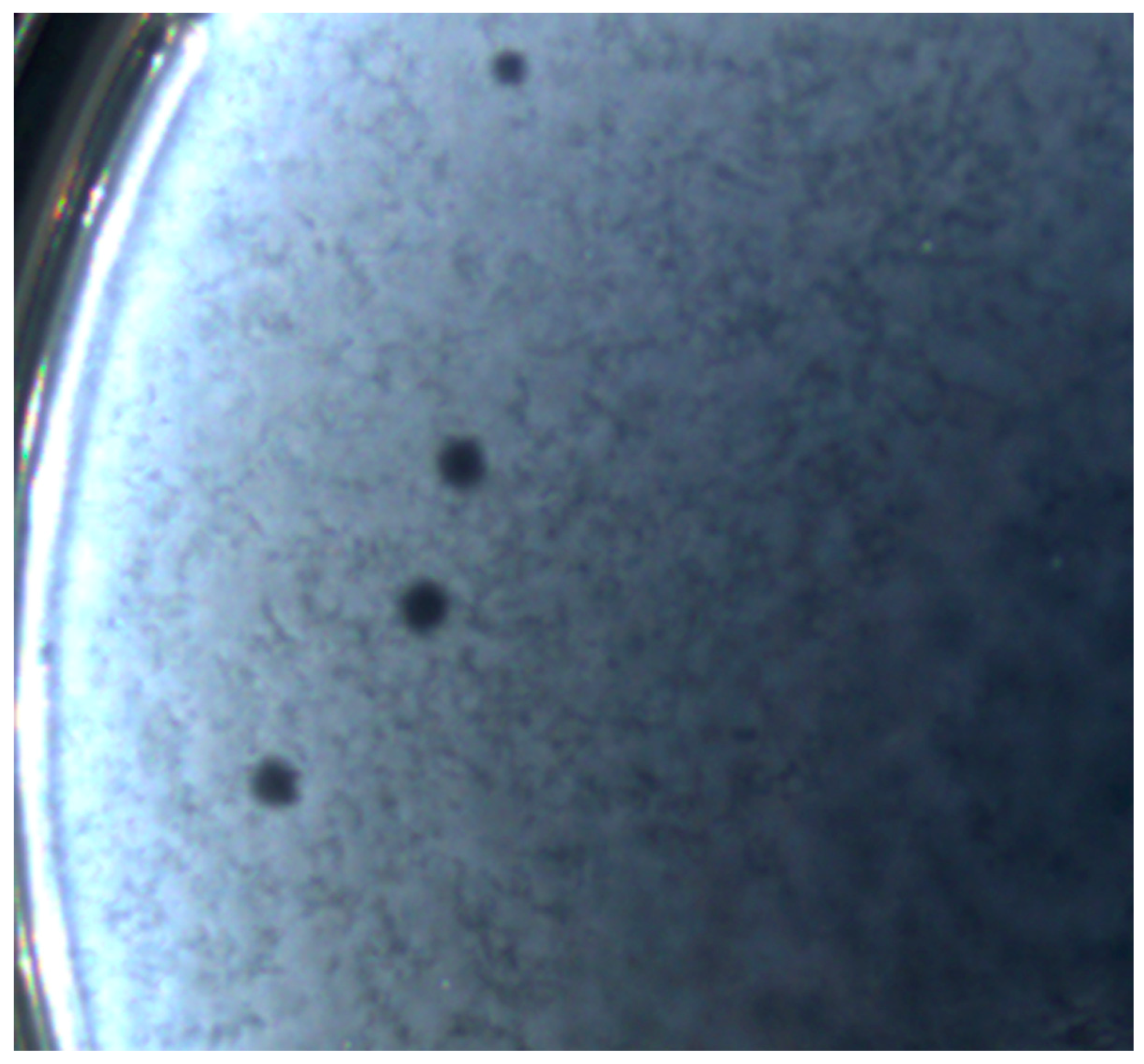
3.2. Bacteriophage Isolation and Plaque Morphology
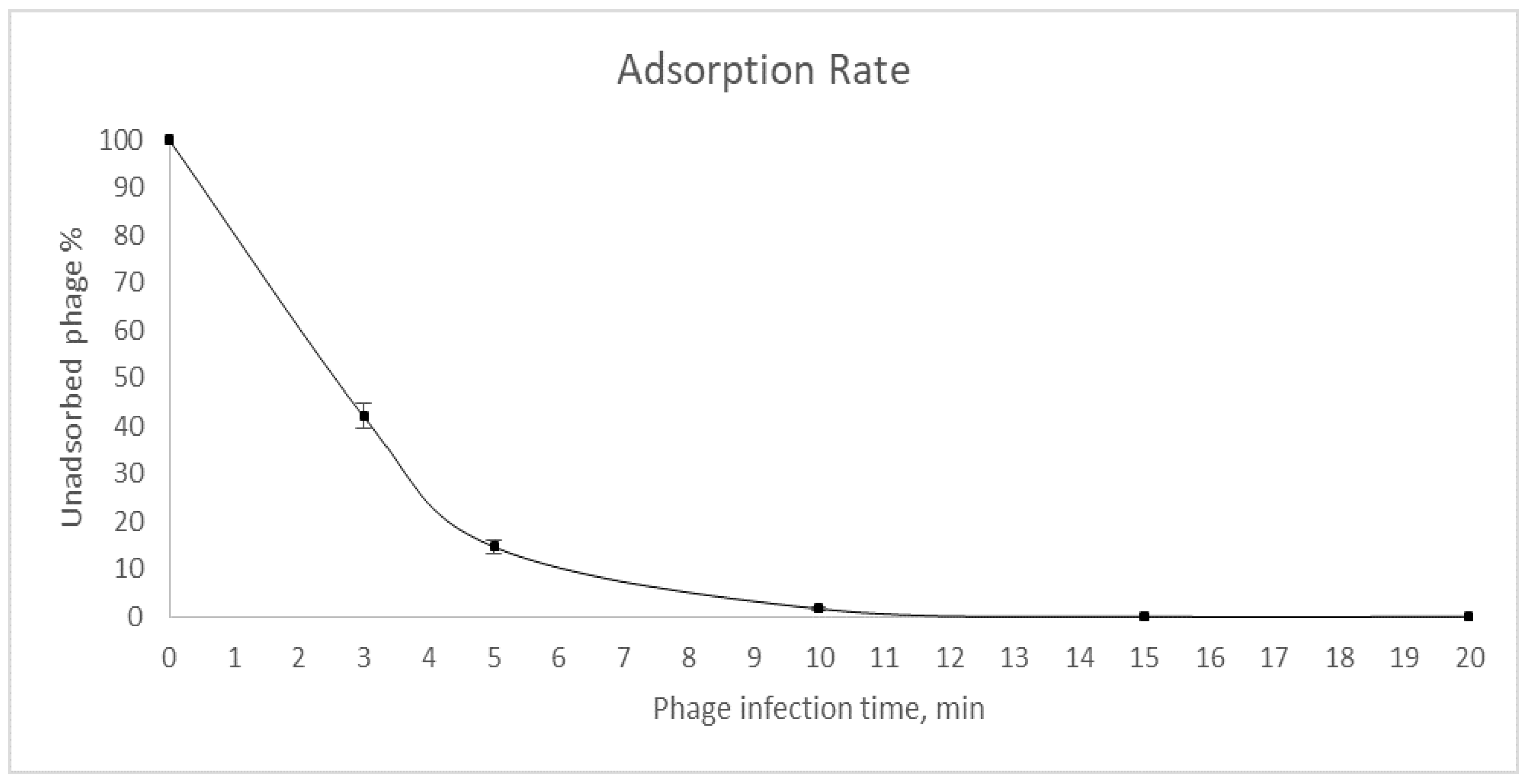
3.3. Determination of Multiplicity of Infection, Adsorption Rate and One-Step Growth Curve of the Bacteriophage
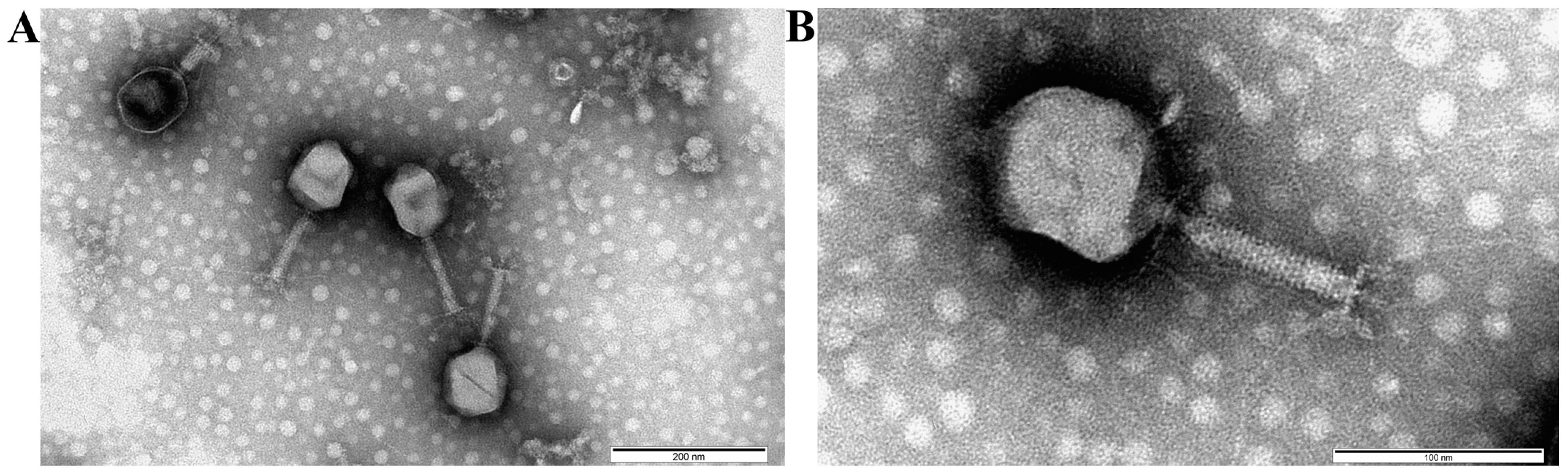
3.4. Bacteriophage Morphology
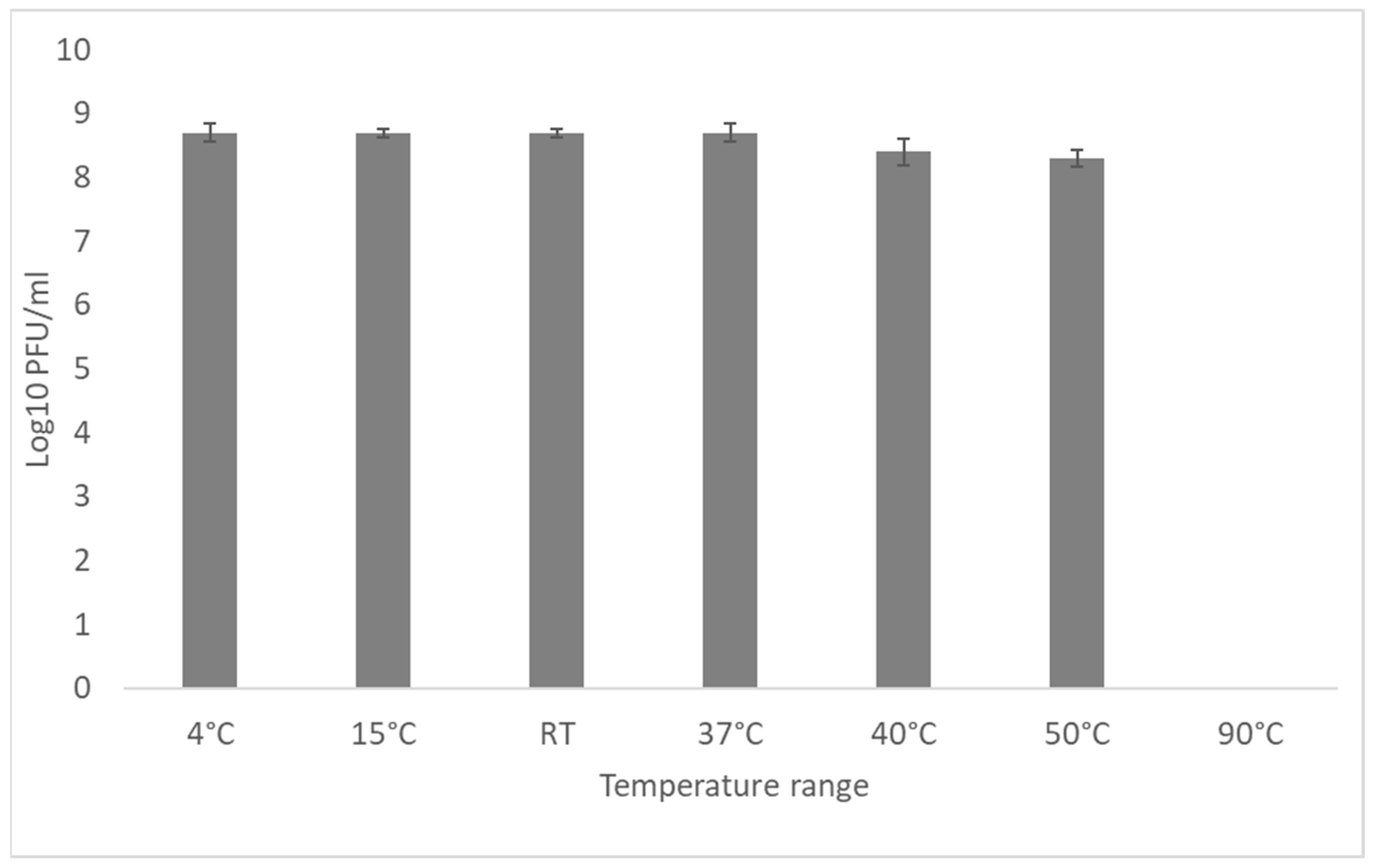
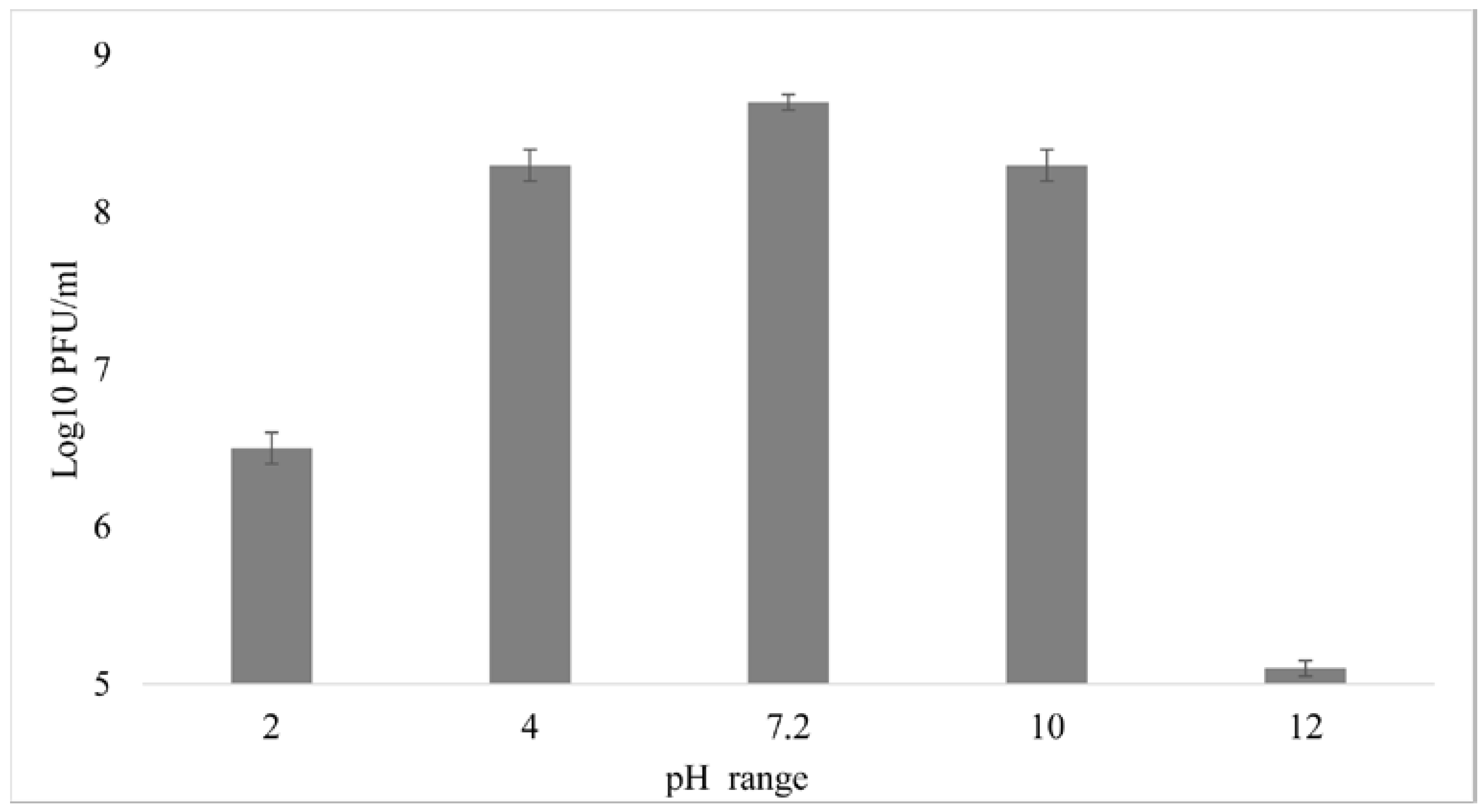
3.5. External Factors Stability Tests on the Phage
3.6. Host Range Determination
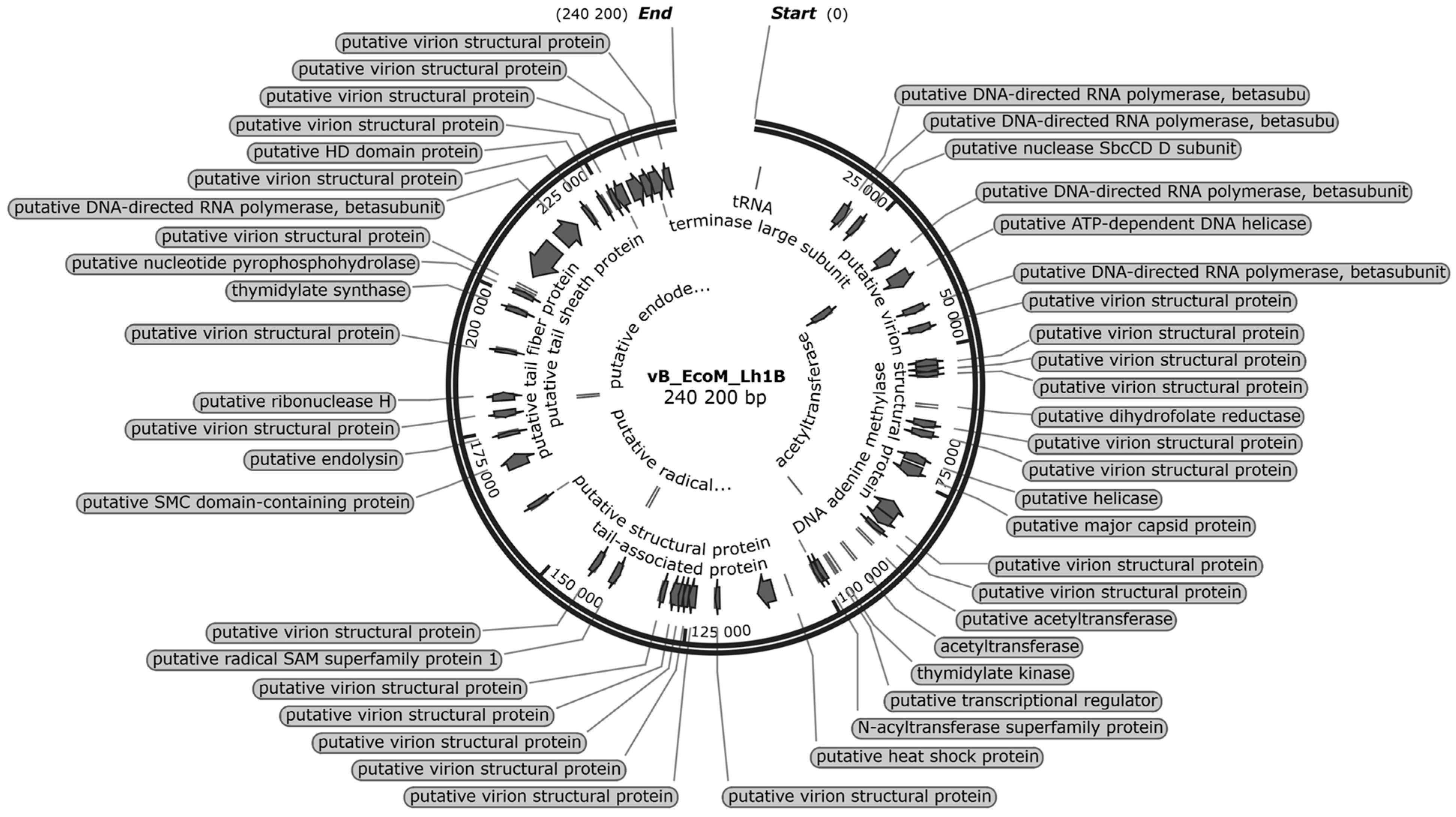
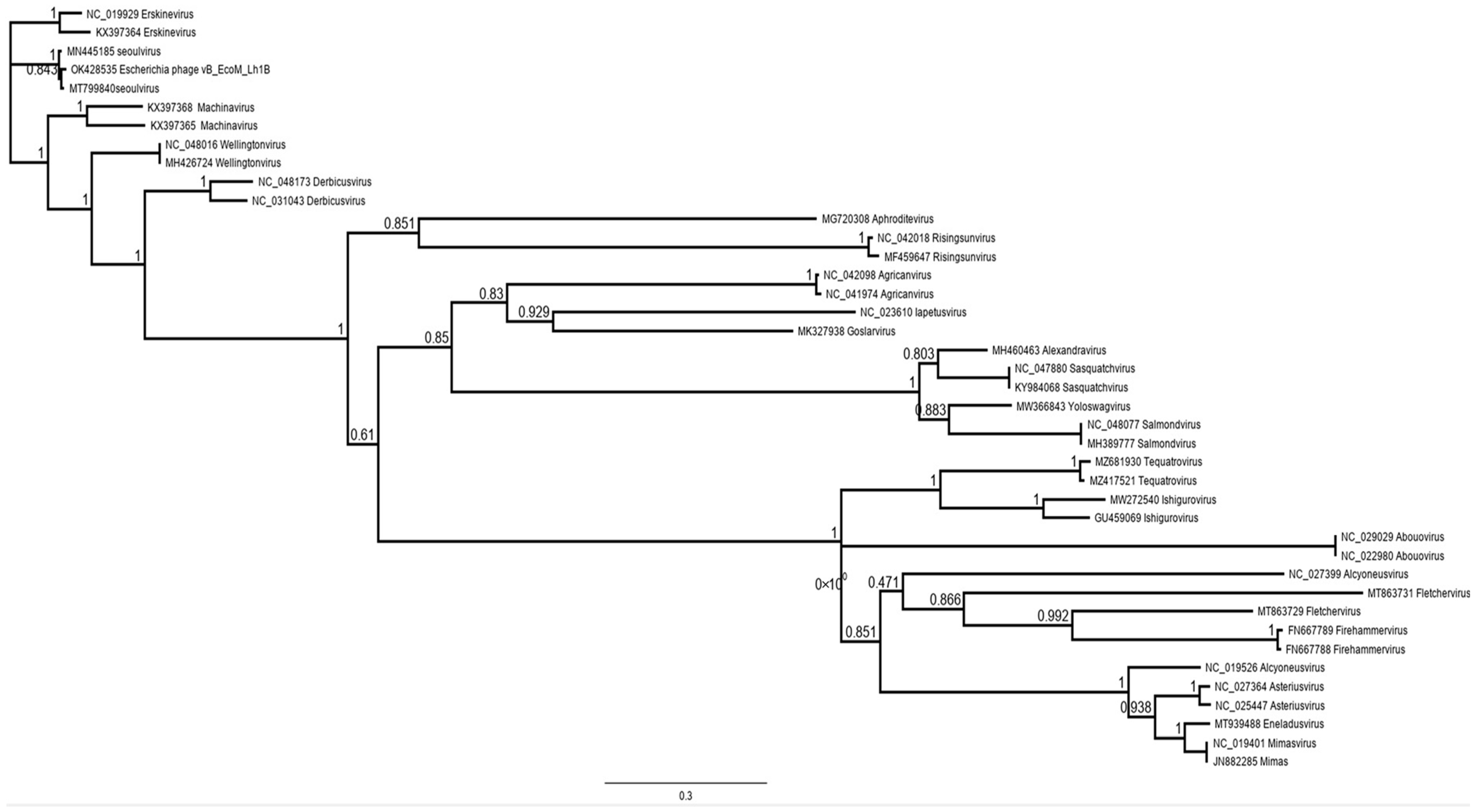
3.7. Bioinformatic Analysis of the vB_EcoM_Lh1B Bacteriophage Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, H.; Chen, S.; White, D.G.; Zhao, S.; McDermott, P.; Walker, R.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Escherichia coli Isolates from Diseased Chickens and Swine in China. J. Clin. Microbiol. 2004, 42, 3483–3489. [Google Scholar] [CrossRef] [PubMed]
- Abreu, R.; Semedo-Lemsaddek, T.; Cunha, E.; Tavares, L.; Oliveira, M. Antimicrobial Drug Resistance in Poultry Production: Current Status and Innovative Strategies for Bacterial Control. Microorganisms 2023, 11, 953. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S.; Hofacre, C.L. Potential Impacts of Antibiotic Use in Poultry Production. Avian. Dis. 2006, 50, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Zhussupova, G.; Utepova, D.; Orazova, G.; Zhaldybayeva, S.; Skvirskaya, G.; Tossekbayev, K. Evaluation of Antibiotic Use in Kazakhstan for the Period 2017–2019 Based on WHO Access, Watch and Reserve Classification (AWaRe 2019). Antibiotics 2021, 10, 58. [Google Scholar] [CrossRef]
- Shah, M.; Zaneb, H.; Masood, S.; Khan, R.U.; Mobashar, M.; Khan, I.; Din, S.; Khan, M.S.; Rehman, H.U.; Tinelli, A. Single or Combined Applications of Zinc and Multi-Strain Probiotic on Intestinal Histomorphology of Broilers Under Cyclic Heat Stress. Probiot. Antimicrob. Proteins 2020, 12, 473–480. [Google Scholar] [CrossRef]
- Haq, I.u.; Hafeez, A.; Khan, R.U. Protective Effect of Nigella sativa and Saccharomyces cerevisiae on Zootechnical Characteristics, Fecal Escherichia coli and Hematopoietic Potential in Broiler Infected with Experimental Colibacillosis. Livest. Sci. 2020, 239, 104119. [Google Scholar] [CrossRef]
- Khan, R.U.; Naz, S. The Applications of Probiotics in Poultry Production. World’s Poult. Sci. J. 2013, 69, 621–632. [Google Scholar] [CrossRef]
- Pearlin, B.V.; Muthuvel, S.; Govidasamy, P.; Villavan, M.; Alagawany, M.; Ragab Farag, M.; Dhama, K.; Gopi, M. Role of Acidifiers in Livestock Nutrition and Health: A Review. J. Anim. Physiol. Anim. Nutr. 2020, 104, 558–569. [Google Scholar] [CrossRef]
- Adil, S.; Banday, T.; Bhat, G.A.; Mir, M.S.; Rehman, M. Effect of Dietary Supplementation of Organic Acids on Performance, Intestinal Histomorphology, and Serum Biochemistry of Broiler Chicken. Vet. Med. Int. 2010, 2010, 479485. [Google Scholar] [CrossRef]
- Hernández, F.; García, V.; Madrid, J.; Orengo, J.; Catalá, P.; Megías, M.D. Effect of Formic Acid on Performance, Digestibility, Intestinal Histomorphology and Plasma Metabolite Levels of Broiler Chickens. Br. Poult. Sci. 2006, 47, 50–56. [Google Scholar] [CrossRef]
- Lopez-Romero, J.C.; González-Ríos, H.; Borges, A.; Simões, M. Antibacterial Effects and Mode of Action of Selected Essential Oils Components against Escherichia coli and Staphylococcus Aureus. Evid.-Based Complement. Altern. Med. 2015, 2015, 795435. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage Therapy: An Alternative to Antibiotics in the Age of Multi-Drug Resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Prangishvili, D.; Hendrix, R.W.; Bamford, D.H. Genomics of Bacterial and Archaeal Viruses: Dynamics within the Prokaryotic Virosphere. Microbiol. Mol. Biol. Rev. 2011, 75, 610–635. [Google Scholar] [CrossRef]
- Gildea, L.; Ayariga, J.A.; Robertson, B.K. Bacteriophages as Biocontrol Agents in Livestock Food Production. Microorganisms 2022, 10, 2126. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Wernicki, A.; Urban-Chmiel, R. Efficacy of Experimental Phage Therapies in Livestock. Anim. Health Res. Rev. 2020, 21, 69–83. [Google Scholar] [CrossRef]
- Alomari, M.M.M.; Dec, M.; Urban-Chmiel, R. Bacteriophages as an Alternative Method for Control of Zoonotic and Foodborne Pathogens. Viruses 2021, 13, 2348. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowska, J.; Casey, E.; Lugli, G.A.; Ventura, M.; Clarke, D.J.; van Sinderen, D.; Mahony, J. In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions. Microorganisms 2021, 9, 1869. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Żbikowska, K.; Michalczuk, M.; Dolka, B. The Use of Bacteriophages in the Poultry Industry. Animals 2020, 10, 872. [Google Scholar] [CrossRef]
- Barrow, P.; Lovell, M.; Berchieri, A. Use of Lytic Bacteriophage for Control of Experimental Escherichia coli Septicemia and Meningitis in Chickens and Calves. Clin. Diagn. Lab. Immunol. 1998, 5, 294–298. [Google Scholar] [CrossRef]
- Xie, H.; Zhuang, X.; Kong, J.; Ma, G.; Zhang, H. Bacteriophage Esc-A Is an Efficient Therapy for Escherichia coli 3-1 Caused Diarrhea in Chickens. J. Gen. Appl. Microbiol. 2005, 51, 159–163. [Google Scholar] [CrossRef]
- Huff, W.E.; Huff, G.R.; Rath, N.C.; Balog, J.M.; Xie, H.; Moore, P.A.; Donoghue, A.M. Prevention of Escherichia coli Respiratory Infection in Broiler Chickens with Bacteriophage (SPR02). Poult. Sci. 2002, 81, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Wernicki, A.; Nowaczek, A.; Urban-Chmiel, R. Bacteriophage Therapy to Combat Bacterial Infections in Poultry. Virol. J. 2017, 14, 179. [Google Scholar] [CrossRef]
- Tiwari, R.; Dhama, K.; Wani, M.Y.; Verma, V.; Vaid, R.; Chauhan, R. Bacteriophage Therapy: A Novel Tool for Combating Bacterial Diseases of Poultry—A Review. J. Immunol. Immunopathol. 2011, 13, 55–66. [Google Scholar]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Donelli, G. Isolation of a Bacteriophage of Exceptional Dimensions Active in R. Megatherium. Atti Accad. Naz. Lincei Rend.-Cl. Sci. Fis.-Mat. Nat. 1968, 44, 95. [Google Scholar]
- Nazir, A.; Ali, A.; Qing, H.; Tong, Y. Emerging Aspects of Jumbo Bacteriophages. Infect. Drug Resist. 2021, 14, 5041–5055. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, M. Jumbo Bacteriophages: An Overview. Front. Microbiol. 2017, 8, 403. [Google Scholar] [CrossRef]
- Wagemans, J.; Tsonos, J.; Holtappels, D.; Fortuna, K.; Hernalsteens, J.-P.; De Greve, H.; Estrozi, L.F.; Bacia-Verloop, M.; Moriscot, C.; Noben, J.-P.; et al. Structural Analysis of Jumbo Coliphage PhAPEC6. Int. J. Mol. Sci. 2020, 21, 3119. [Google Scholar] [CrossRef]
- Serwer, P.; Hayes, S.J.; Thomas, J.A.; Hardies, S.C. Propagating the Missing Bacteriophages: A Large Bacteriophage in a New Class. Virol. J. 2007, 4, 21. [Google Scholar] [CrossRef]
- Wragg, P.; Randall, L.; Whatmore, A.M. Comparison of Biolog GEN III MicroStation Semi-Automated Bacterial Identification System with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and 16S Ribosomal RNA Gene Sequencing for the Identification of Bacteria of Veterinary Interest. J. Microbiol. Methods 2014, 105, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of Minimal Predictors of Avian Pathogenic Escherichia coli Virulence for Use as a Rapid Diagnostic Tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [PubMed]
- Gratia, A. The Numerical Relation between Lysogenic Bacteria and the Phage Particles which they carry. Ann. Inst. Pasteur. 1936, 57, 652–676. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 69–76. ISBN 978-1-60327-164-6. [Google Scholar]
- Baer, A.; Kehn-Hall, K. Viral Concentration Determination Through Plaque Assays: Using Traditional and Novel Overlay Systems. J. Vis. Exp. 2014, 93, 52065. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Therapy Dosing: The Problem(s) with Multiplicity of Infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef] [PubMed]
- Alexyuk, P.; Bogoyavlenskiy, A.; Alexyuk, M.; Akanova, K.; Moldakhanov, Y.; Berezin, V. Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail. Viruses 2022, 14, 2381. [Google Scholar] [CrossRef]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of Bacteriophages: Morphological and Biological Properties of a Large Group of Phages Isolated from Urban Sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef]
- Hong, Y.; Pan, Y.; Ebner, P.D. Meat Science and Muscle Biology Symposium: Development of Bacteriophage Treatments to Reduce Escherichia coli O157:H7 Contamination of Beef Products and Produce. J. Anim. Sci. 2014, 92, 1366–1377. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Jeong, D.; Park, S.C. Isolation and Characterisation of PVa-21, a Giant Bacteriophage with Anti-Biofilm Potential against Vibrio Alginolyticus. Sci. Rep. 2019, 9, 6284. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, S.B.; Giri, S.S.; Kim, H.J.; Kim, S.W.; Kwon, J.; Park, J.; Roh, E.; Park, S.C. Characterization of Novel Erwinia Amylovora Jumbo Bacteriophages from Eneladusvirus Genus. Viruses 2020, 12, 1373. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Ackermann, H.-W. Basic Phage Electron Microscopy. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 113–126. ISBN 978-1-60327-164-6. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.-M.; et al. Genome Detective: An Automated System for Virus Identification from High-Throughput Sequencing Data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Furuno, M.; Kasukawa, T.; Saito, R.; Adachi, J.; Suzuki, H.; Baldarelli, R.; Hayashizaki, Y.; Okazaki, Y. CDS Annotation in Full-Length CDNA Sequence. Genome Res. 2003, 13, 1478–1487. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The TRNAscan-SE, Snoscan and SnoGPS Web Servers for the Detection of TRNAs and SnoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One Health—Its Importance in Helping to Better Control Antimicrobial Resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-151552-8.
- Scott, A.M.; Beller, E.; Glasziou, P.; Clark, J.; Ranakusuma, R.W.; Byambasuren, O.; Bakhit, M.; Page, S.W.; Trott, D.; Mar, C.D. Is Antimicrobial Administration to Food Animals a Direct Threat to Human Health? A Rapid Systematic Review. Int. J. Antimicrob. Agents 2018, 52, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic Resistance Is the Quintessential One Health Issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.E. Multidrug-Resistant Pathogens in the Food Supply. Foodborne Pathog. Dis. 2015, 12, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial Resistance in Bacterial Poultry Pathogens: A Review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The Influence of External Factors on Bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Rezk, N.; Fayez, M.S.; Abdelmoteleb, M.; Atteya, R.; Elhadidy, M.; El-Shibiny, A. Isolation, Characterization, and Genomic Analysis of Three Novel E. coli Bacteriophages That Effectively Infect E. coli O18. Microorganisms 2022, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Kazibwe, G.; Katami, P.; Alinaitwe, R.; Alafi, S.; Nanteza, A.; Nakavuma, J.L. Bacteriophage Activity against and Characterisation of Avian Pathogenic Escherichia coli Isolated from Colibacillosis Cases in Uganda. PLoS ONE 2020, 15, e0239107. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F.; Fleming, H.P.; Altermann, E.; Klaenhammer, T.R. Isolation and Characterization of a Lactobacillus Plantarum Bacteriophage, PhiJL-1, from a Cucumber Fermentation. Int. J. Food Microbiol. 2003, 84, 225–235. [Google Scholar] [CrossRef]
- Yu, Y.-P.; Gong, T.; Jost, G.; Liu, W.-H.; Ye, D.-Z.; Luo, Z.-H. Isolation and Characterization of Five Lytic Bacteriophages Infecting a Vibrio Strain Closely Related to Vibrio Owensii. FEMS Microbiol. Lett. 2013, 348, 112–119. [Google Scholar] [CrossRef]
- Shende, R.K.; Hirpurkar, S.D.; Sannat, C.; Rawat, N.; Pandey, V. Isolation and Characterization of Bacteriophages with Lytic Activity against Common Bacterial Pathogens. Vet. World 2017, 10, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A.; Shahin, K. Isolation, Characterization and Genomic Analysis of a Novel Bacteriophage VB_EcoS-Golestan Infecting Multidrug-Resistant Escherichia coli Isolated from Urinary Tract Infection. Sci. Rep. 2020, 10, 7690. [Google Scholar] [CrossRef] [PubMed]
- Amankwah, S.; Adisu, M.; Gorems, K.; Abdella, K.; Kassa, T. Assessment of Phage-Mediated Inhibition and Removal of Multidrug-Resistant Pseudomonas Aeruginosa Biofilm on Medical Implants. Infect. Drug Resist. 2022, 15, 2797–2811. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Hussain, T.; Das, C.R.; Andleeb, S. Isolation and Characterization of a Myoviridae MJ1 Bacteriophage against Multi-Drug Resistant Escherichia coli 3. Jundishapur. J. Microbiol. 2015, 8, e25917. [Google Scholar] [CrossRef]
- Huff, W.E.; Huff, G.R.; Rath, N.C.; Donoghue, A.M. Evaluation of the Influence of Bacteriophage Titer on the Treatment of Colibacillosis in Broiler Chickens. Poult. Sci. 2006, 85, 1373–1377. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The Factors Affecting Effectiveness of Treatment in Phages Therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Nachimuthu, R. Therapeutic Characterization and Efficacy of Bacteriophage Cocktails Infecting Escherichia coli, Klebsiella Pneumoniae, and Enterobacter Species. Front. Microbiol. 2019, 10, 574. [Google Scholar] [CrossRef]
- Dalmasso, M.; Strain, R.; Neve, H.; Franz, C.M.A.P.; Cousin, F.J.; Ross, R.P.; Hill, C. Three New Escherichia coli Phages from the Human Gut Show Promising Potential for Phage Therapy. PLoS ONE 2016, 11, e0156773. [Google Scholar] [CrossRef]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.-L.; Brüssow, H. Phage-Host Interaction: An Ecological Perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; van Raaij, M.J.; Leiman, P.G. Contractile Injection Systems of Bacteriophages and Related Systems. Mol. Microbiol. 2018, 108, 6–15. [Google Scholar] [CrossRef]


| APEC Isolate ID | The Level of Lytic Activity of the vB_EcoM_Lh1B Phage * | APEC Isolate ID | The Level of Lytic Activity of the vB_EcoM_Lh1B Phage |
|---|---|---|---|
| E. coli F/18 | 100% +++ | Int WP #4 | 30% + |
| Sor. lv. bc2 | 100% +++ | Br. m/L 10.06 | 28% + |
| Yun 3d pas | 100% +++ | Sor. st. Sc1 | 25% + |
| Sor. lb. sc2 | 100% +++ | Alag 1 st. | 25% + |
| Int. m/L 04.06 | 85% ++ | Sor. st. bc1 | - |
| Sor. br. sc2 | 75% ++ | Int WR #4 | - |
| Sor. int. bc2 | 64% ++ | Yun#2 | - |
| Kd. m/L 04.06 | 61% ++ | St. m/L 04.06 | - |
| Sor. st. sc2 | 58% ++ | Lv. m/L 04.06 | - |
| Faec. H.bl. Uz. | 55% ++ | St. m/L 10.06 | - |
| Sor. br.bc1 | 51% ++ | F.ch. m/L Uz. | - |
| Sor. kd. sc2 | 44% + | Sor. lv. Bc1 | - |
| Sor. lv. sc2 | 43% + | Sor. lv. Sc1 | - |
| Sor. br. sc1 | 42% + | Sor. kd. bc1 | - |
| F.H. pr. Uz. | 34% + | Sor. br.bc2 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexyuk, P.; Bogoyavlenskiy, A.; Alexyuk, M.; Akanova, K.; Moldakhanov, Y.; Berezin, V. Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis. Microorganisms 2023, 11, 1524. https://doi.org/10.3390/microorganisms11061524
Alexyuk P, Bogoyavlenskiy A, Alexyuk M, Akanova K, Moldakhanov Y, Berezin V. Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis. Microorganisms. 2023; 11(6):1524. https://doi.org/10.3390/microorganisms11061524
Chicago/Turabian StyleAlexyuk, Pavel, Andrey Bogoyavlenskiy, Madina Alexyuk, Kuralay Akanova, Yergali Moldakhanov, and Vladimir Berezin. 2023. "Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis" Microorganisms 11, no. 6: 1524. https://doi.org/10.3390/microorganisms11061524
APA StyleAlexyuk, P., Bogoyavlenskiy, A., Alexyuk, M., Akanova, K., Moldakhanov, Y., & Berezin, V. (2023). Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis. Microorganisms, 11(6), 1524. https://doi.org/10.3390/microorganisms11061524








