Multi-Omics Analysis Demonstrates the Critical Role of Non-Ethanolic Components of Alcoholic Beverages in the Host Microbiome and Metabolome: A Human- and Animal-Based Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects, Fecal Sample Collection, and Preservation
2.2. Study on the Effect of Alcoholic Beverages in Animal Model
2.3. DNA Extraction, PCR Amplification, and Denaturing Gradient Gel Electrophoresis (DGGE)
2.4. DGGE Band Sequencing
2.5. Next-Generation Sequencing (NGS) of Fecal 16S rDNA
2.6. Gas Chromatography–Mass Spectrometry (GC-MS)-Based Untargeted Metabolomic Profiling of Fecal and Plasma Samples
2.7. Chemical Analysis of Alcoholic Beverages
2.8. Programs for Gas Chromatography–Mass Spectrometry
2.9. Interleukin Assays
2.10. Statistical Analysis
3. Results
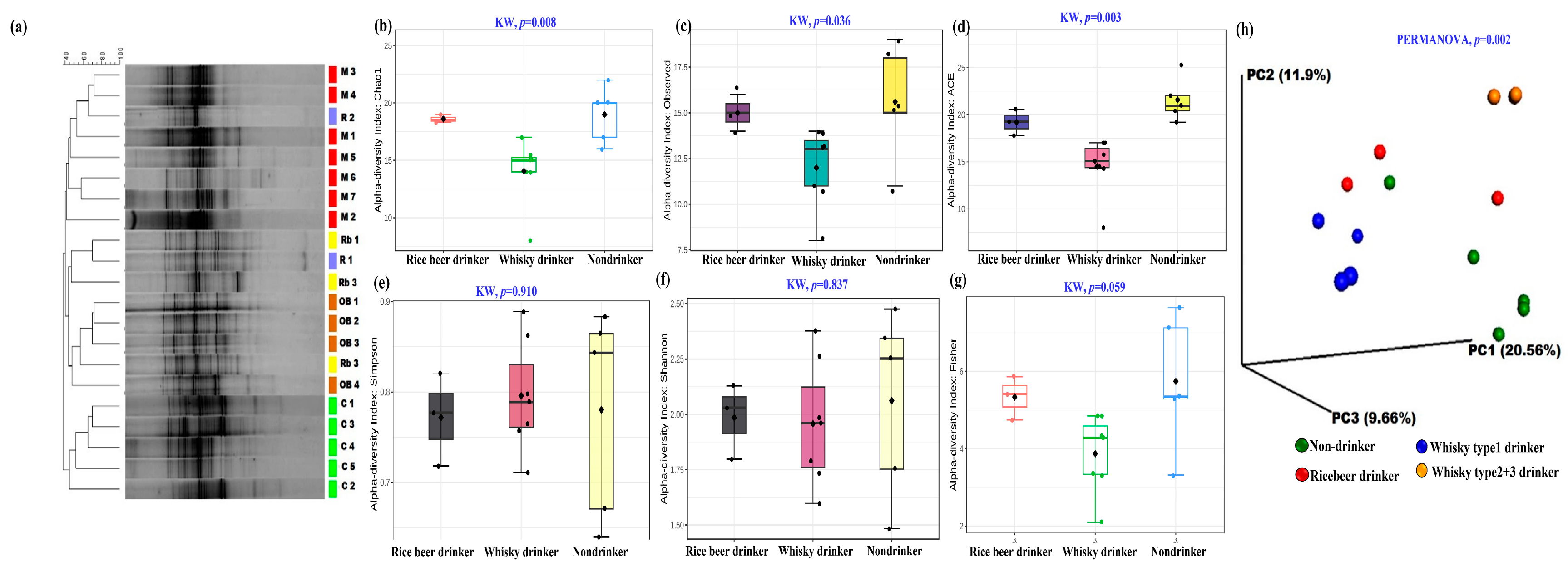
3.1. Effect of Alcoholic Beverages on the Human Gut Microbiome (Pilot Study)
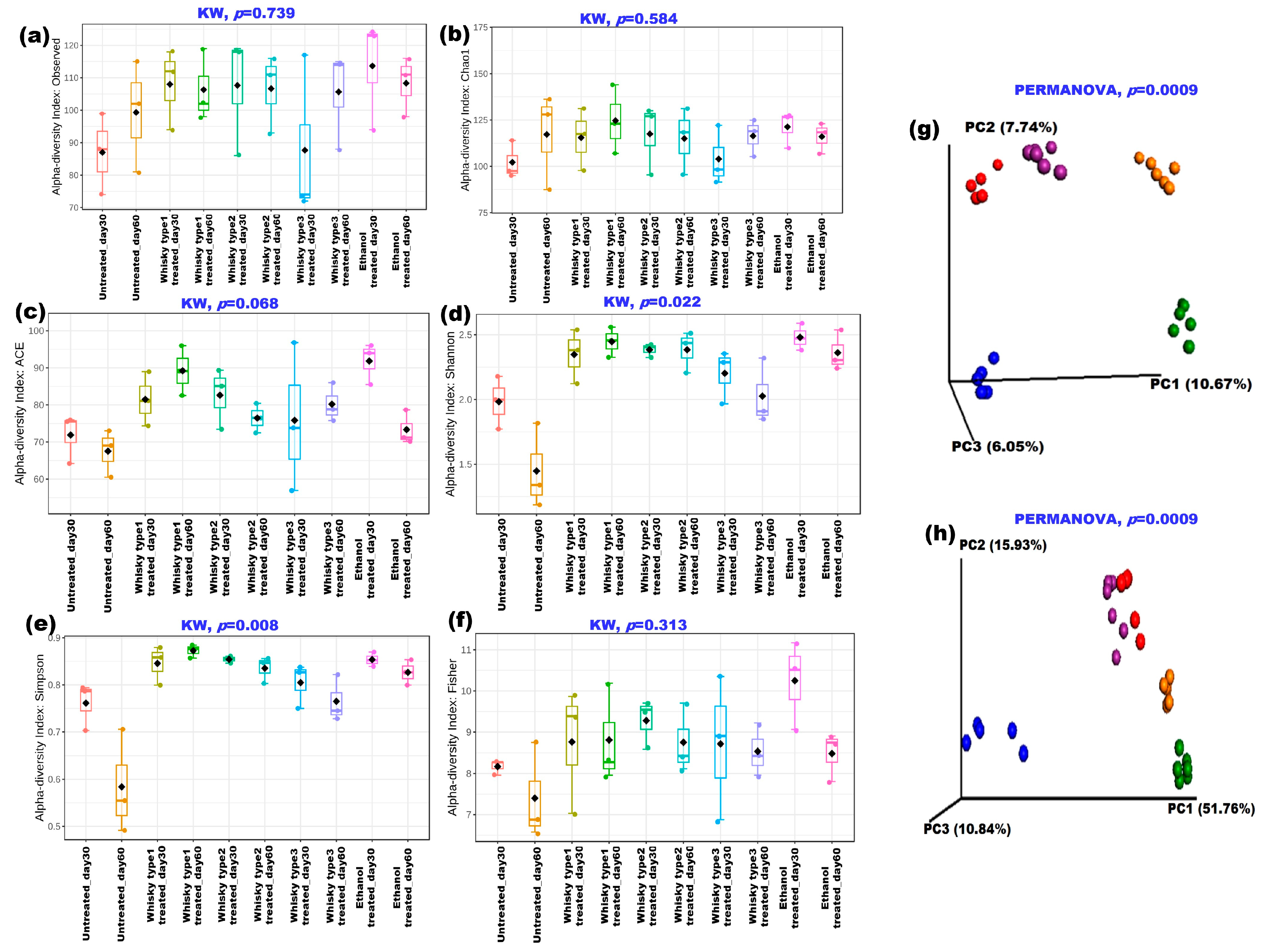
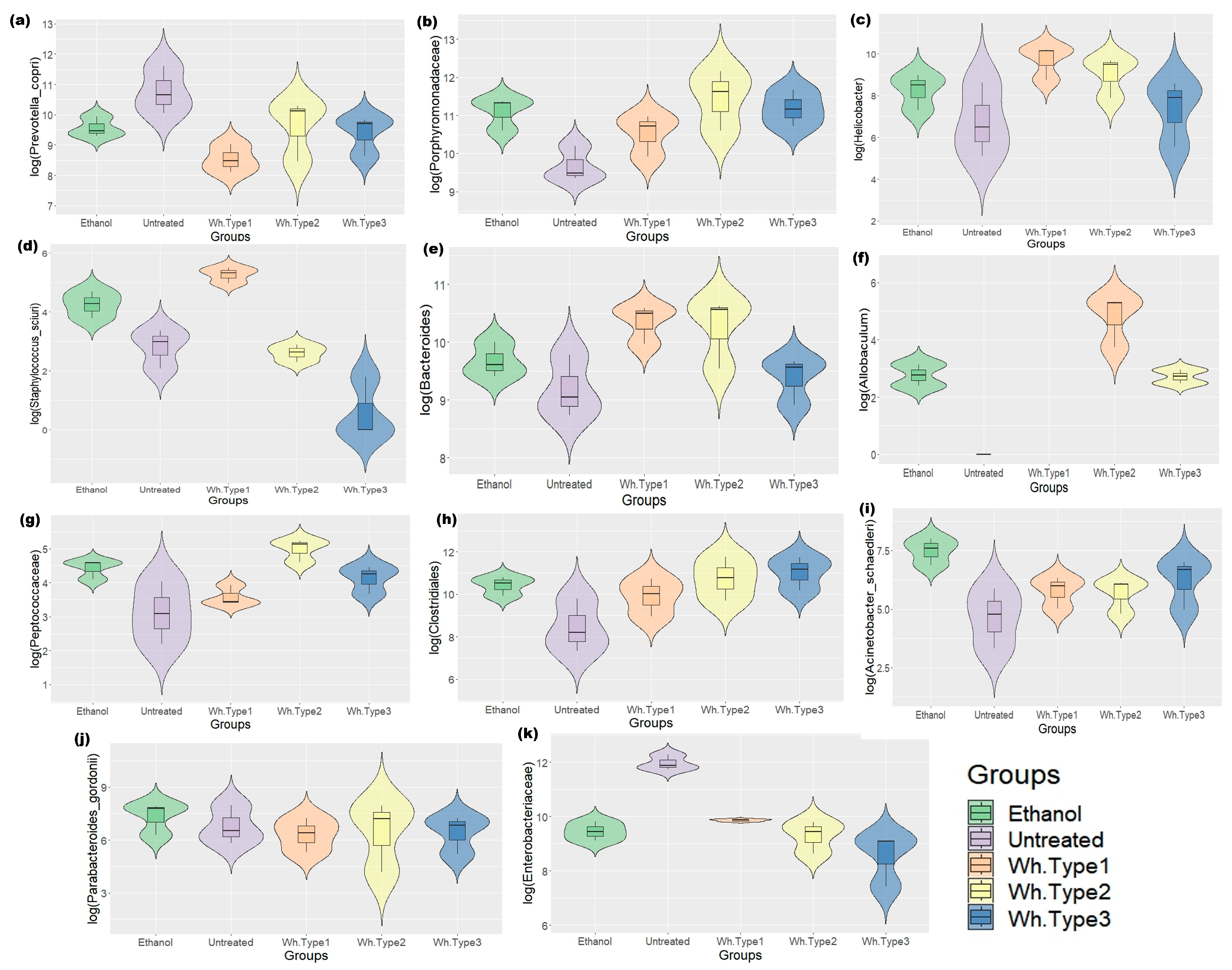
3.2. Effect of Alcoholic Beverages on Mouse Gut Microbiome (Validation Phase)
3.3. Effect of Whisky Brands on Fecal and Plasma Metabolites in Animal Model
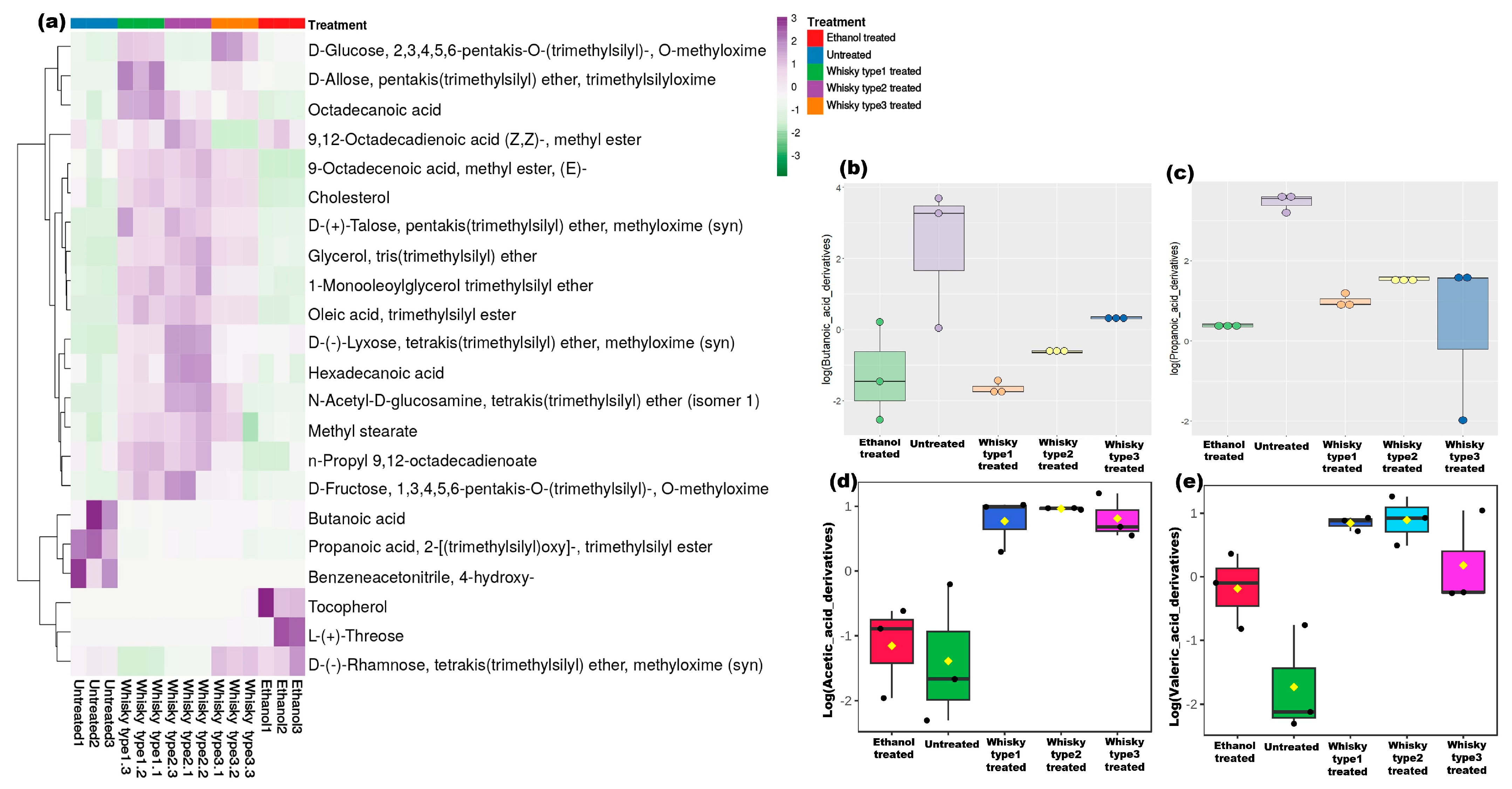
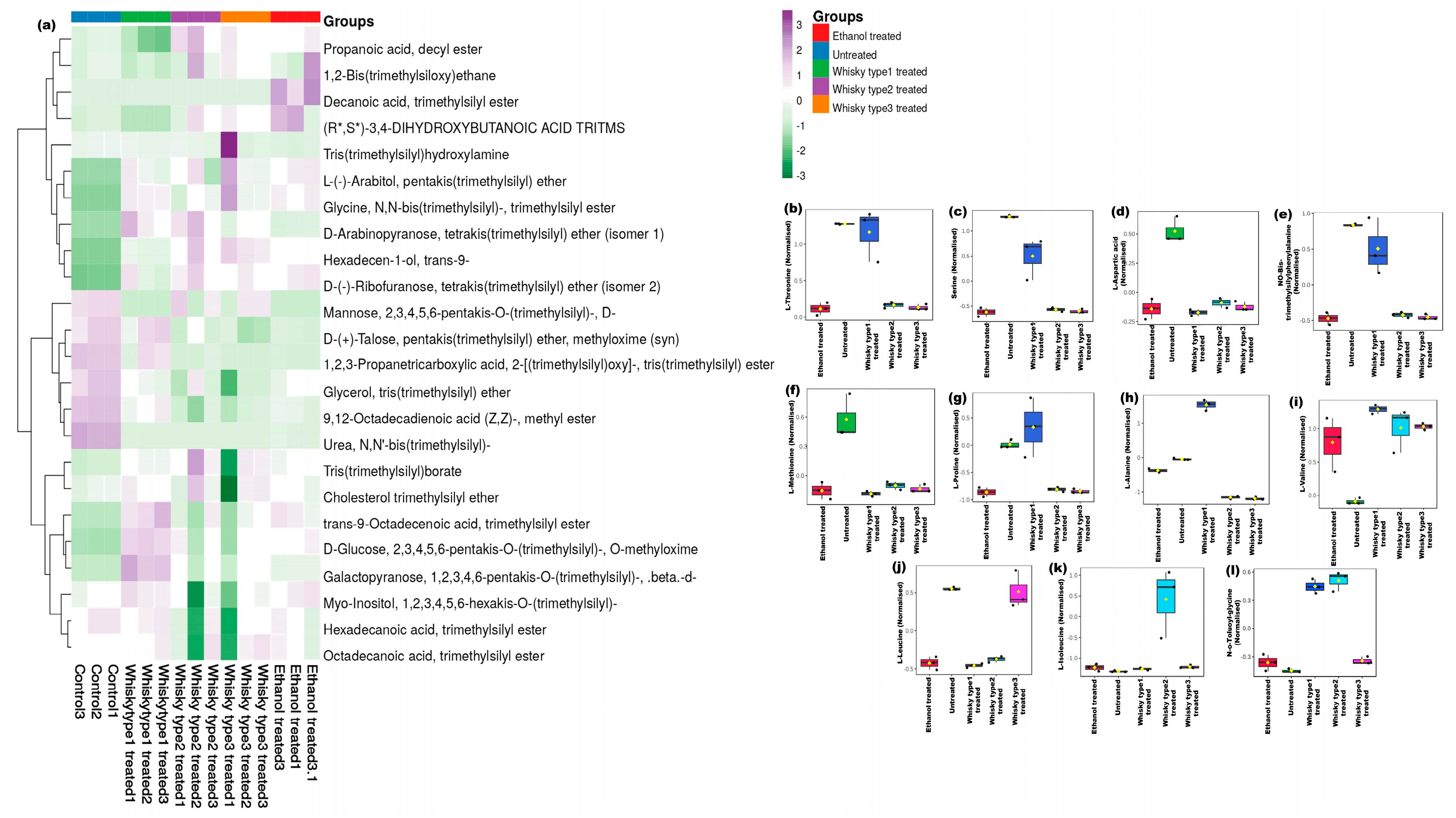
3.3.1. Fecal Metabolites
3.3.2. Plasma Metabolites
3.3.3. Crosstalk between Host Microbiome and Metabolome
3.4. Effect of Whisky on Inflammatory Cytokines
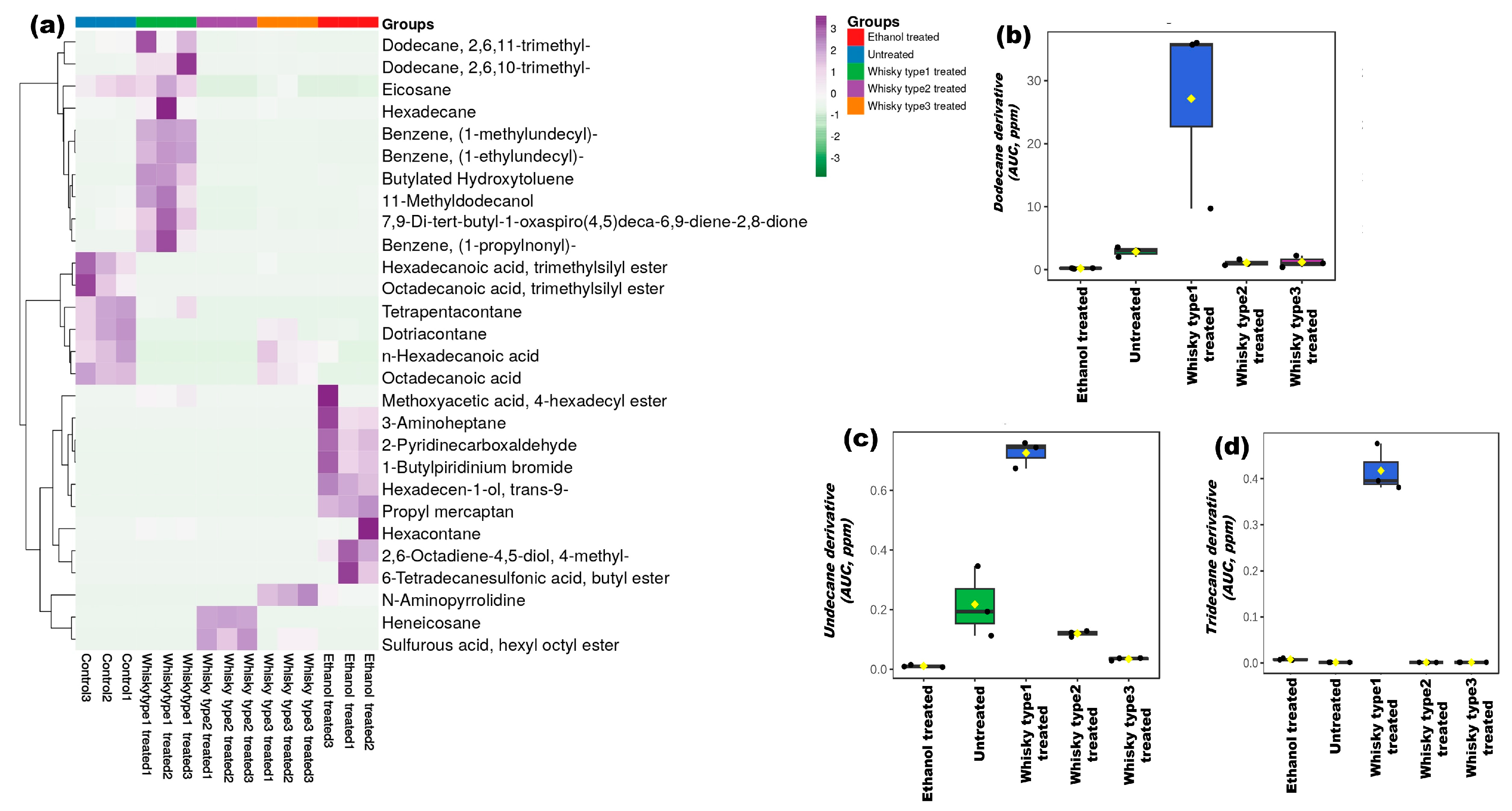
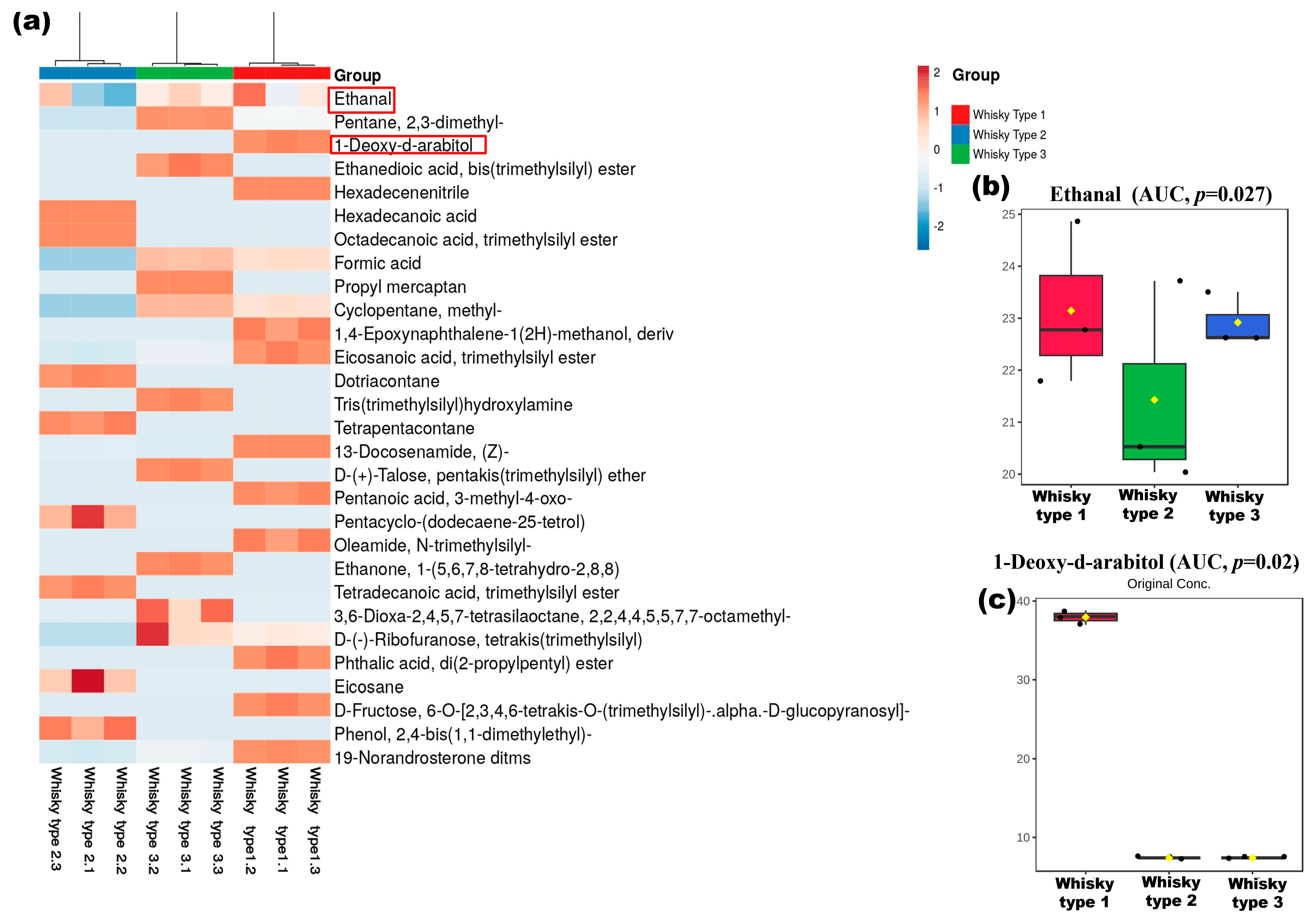
3.5. Chemical Profiles of the Whisky Brands
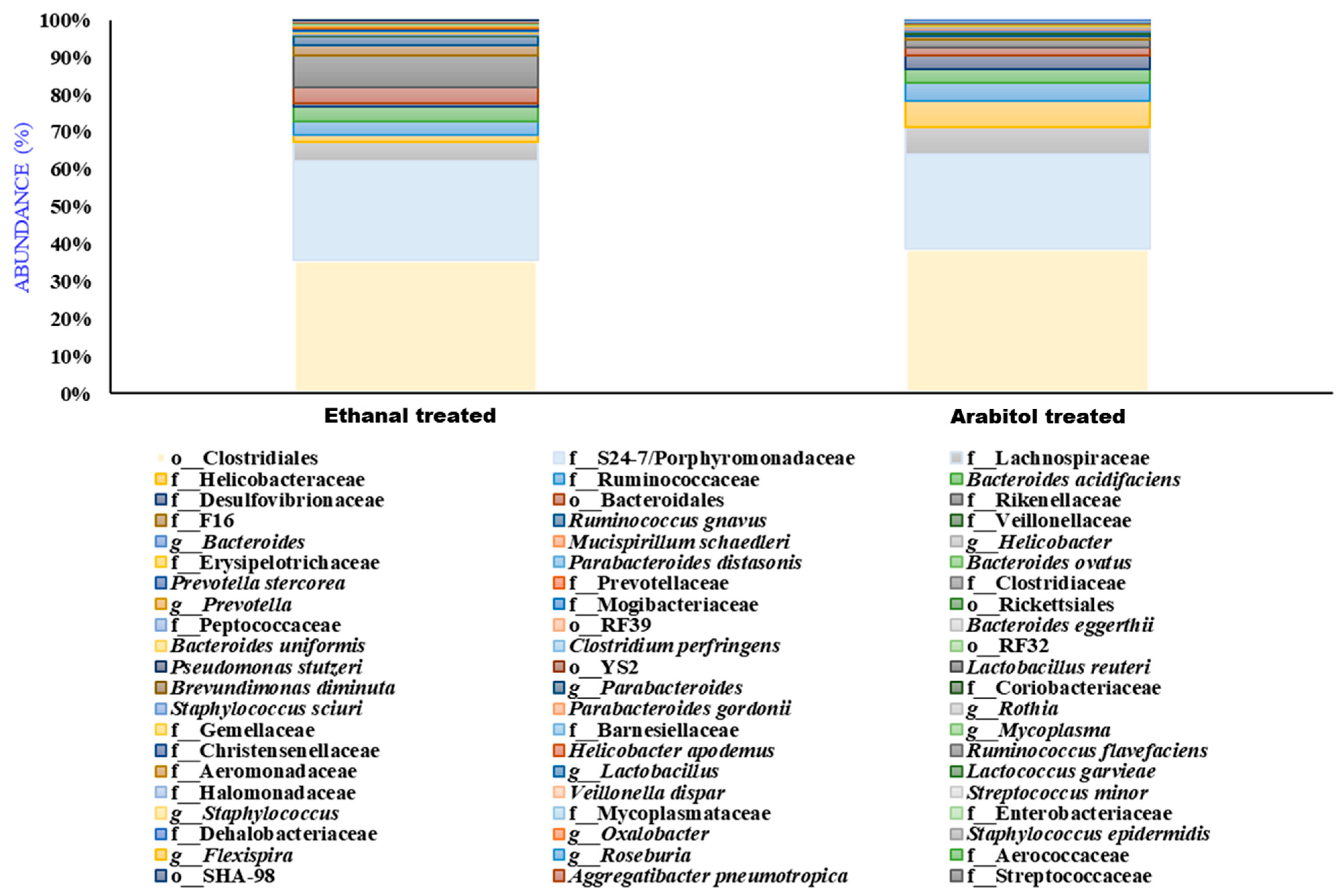
3.6. Effect of Arabitol and Ethanal on Gut Microbiome on the Mouse Gut Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaut, M.; Clavel, T. Metabolic diversity of the intestinal microbiota: Implications for health and disease. J. Nutr. 2007, 137, 751S–755S. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; DeCoffe, D.; Molcan, E.; Gibson, D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [PubMed]
- Tuomisto, S.; Pessi, T.; Collin, P.; Vuento, R.; Aittoniemi, J.; Karhunen, P. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014, 14, 1–8. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Mutlu, E.A.; Gillevet, P.M.; Rangwala, H.; Sikaroodi, M.; Naqvi, A.; Engen, P.A.; Kwasny, M.; Lau, C.K.; Keshavarzian, A.; Physiology, L. Colonic microbiome is altered in alcoholism. Am. J. Physiol. Liver Physiol. 2012, 302, G966–G978. [Google Scholar] [CrossRef]
- Martínez-Montoro, J.I.; Quesada-Molina, M.; Gutiérrez-Repiso, C.; Ruiz-Limón, P.; Subiri-Verdugo, A.; Tinahones, F.J.; Moreno-Indias, I. Effect of moderate consumption of different phenolic-content beers on the human gut microbiota composition: A randomized crossover trial. Antioxidants 2022, 11, 696. [Google Scholar] [CrossRef]
- Sarkar, P.; Kalita, M.C.; Khan, M.R. Fecal Metabolite Profiles of Whisky Drinkers in India-A Pilot Study. Trends Biosci. 2017, 10, 9445. [Google Scholar]
- Dehingia, M.; Thangjam Devi, K.; Talukdar, N.C.; Talukdar, R.; Reddy, N.; Mande, S.S.; Deka, M.; Khan, M. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 2015, 5, 18563. [Google Scholar] [CrossRef]
- Abbaszadegan, M.R.; Tavasoli, A.; Velayati, A.; Sima, H.R.; Vosooghinia, H.; Farzadnia, M.; Asadzedeh, H.; Gholamin, M.; Dadkhah, E.; Aarabi, A. Stool-based DNA testing, a new noninvasive method for colorectal cancer screening, the first report from Iran. World J. Gastroenterol. World J. Gastroenterol. 2007, 13, 1528. [Google Scholar] [CrossRef] [PubMed]
- Gafan, G.P.; Lucas, V.S.; Roberts, G.J.; Petrie, A.; Wilson, M.; Spratt, D. Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J. Clin. Microbiol. 2005, 43, 3971–3978. [Google Scholar] [CrossRef] [PubMed]
- Tapia, E.; Donoso-Bravo, A.; Cabrol, L.; Alves, M.; Pereira, A.; Rapaport, A.; Ruiz-Filippi, G. A methodology for a quantitative interpretation of DGGE with the help of mathematical modelling: Application in biohydrogen production. Water Sci. Technol. 2014, 69, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Xu, X.; Gao, Y.; Cao, X.; Wang, X.; Song, G.; Zhao, J.; Hu, Y. Derivatization followed by gas chromatography-mass spectrometry for quantification of ethyl carbamate in alcoholic beverages. J. Sep. Sci. 2012, 35, 804–810. [Google Scholar] [CrossRef]
- Ng, J.S.Y.; Ryan, U.; Trengove, R.D.; Maker, G.L. Development of an untargeted metabolomics method for the analysis of human fecal samples using Cryptosporidium-infected samples. Mol. Biochem. Parasitol. 2012, 185, 145–150. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [PubMed]
- Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Murri, M.; Gomez-Zumaquero, J.M.; Clemente-Postigo, M.; Estruch, R.; Cardona Diaz, F.; Andres-Lacueva, C.; Tinahones, F.J. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012, 95, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Litwinowicz, K.; Gamian, A. Microbiome Alterations in Alcohol Use Disorder and Alcoholic Liver Disease. Int. J. Mol. Sci. 2023, 24, 2461. [Google Scholar] [CrossRef] [PubMed]
- Avivi-Green, C.; Polak-Charcon, S.; Madar, Z.; Schwartz, B.; Therapeutics, C.C. Apoptosis cascade proteins are regulated in vivo by high intracolonic butyrate concentration: Correlation with colon cancer inhibition. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2001, 12, 83–95. [Google Scholar] [CrossRef]
- Greer, J.B.; O’Keefe, S.J. Microbial induction of immunity, inflammation, and cancer. Front. Physiol. 2011, 1, 168. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Martin, P.R.; Singleton, C.K.; Hiller-Sturmhöfel, S. The role of thiamine deficiency in alcoholic brain disease. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2003, 27, 134. [Google Scholar]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Fischer, A.; Siegmund, B.; Kupz, A.; Niebergall, J.; Fuchs, D.; Jahn, H.-K.; Freudenberg, M.; Loddenkemper, C.; Batra, A. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS ONE 2007, 2, e662. [Google Scholar] [CrossRef]
- Happel, K.I.; Nelson, S. Alcohol, Immunosuppression, and the Lung. In Proceedings of the American Thoracic Society, San Diego, CA, USA, 20–25 May 2005; Volume 2, pp. 428–432. [Google Scholar]
- Huang, W.K.; Chang, J.C.; See, L.C.; Tu, H.T.; Chen, J.S.; Liaw, C.C.; Lin, Y.C.; Yang, T.S. Higher rate of colorectal cancer among patients with pyogenic liver abscess with Klebsiella pneumoniae than those without: An 11-year follow-up study. Color. Dis. 2012, 14, e794–e801. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, R.A.; McCubbin, T.; Navone, L.; Stowers, C.; Nielsen, L.K.; Marcellin, E. Microbial propionic acid production. Fermentation 2017, 3, 21. [Google Scholar] [CrossRef]
- Ishibashi, N.; Yaeshima, T.; Hayasawa, H. Bifidobacteria: Their significance in human intestinal health. Malays. J. Nutr. 1997, 3, 149–159. [Google Scholar]
- Payne, A.; Chassard, C.; Lacroix, C. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: Implications for host–microbe interactions contributing to obesity. Obes. Rev. 2012, 13, 799–809. [Google Scholar] [CrossRef]
- Tamura, M.; Hoshi, C.; Hori, S. Xylitol affects the intestinal microbiota and metabolism of daidzein in adult male mice. Int. J. Mol. Sci. 2013, 14, 23993–24007. [Google Scholar] [CrossRef]
- Spruss, A.; Kanuri, G.; Wagnerberger, S.; Haub, S.; Bischoff, S.C.; Bergheim, I. Toll-like receptor 4 is involved in the development of fructose-induced hepatic steatosis in mice. Hepatology 2009, 50, 1094–1104. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef]
- Candela, M.; Turroni, S.; Biagi, E.; Carbonero, F.; Rampelli, S.; Fiorentini, C.; Brigidi, P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. WJG 2014, 20, 908. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, V.; Betrapally, N.S.; Hylemon, P.B.; White, M.B.; Gillevet, P.M.; Unser, A.B.; Fagan, A.; Daita, K.; Heuman, D.M.; Zhou, H. Impaired gut-liver-brain axis in patients with cirrhosis. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Yan, A.W.; Fouts, D.E.; Brandl, J.; Stärkel, P.; Torralba, M.; Schott, E.; Tsukamoto, H.; Nelson, K.E.; Brenner, D.A.; Schnabl, B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 2011, 53, 96–105. [Google Scholar] [CrossRef]
- Kalmokoff, M.; Franklin, J.; Petronella, N.; Green, J.; Brooks, S.P. Phylum level change in the cecal and fecal gut communities of rats fed diets containing different fermentable substrates supports a role for nitrogen as a factor contributing to community structure. Nutrients 2015, 7, 3279–3299. [Google Scholar] [CrossRef]
- Murakami, Y.; Hanazawa, S.; Nishida, K.; Iwasaka, H.; Kitano, S. N-acetyl-D-galactosamine inhibits TNF-α gene expression induced in mouse peritoneal macrophages by fimbriae of Porphyromonas (Bacteroides) gingivalis, an oral anaerobe. Biochem. Biophys. Res. Commun. 1993, 192, 826–832. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Villegas, A.; Sánchez-Tapia, M.; Avila-Nava, A.; Ramírez, V.; Tovar, A.R.; Torres, N. Differential effect of sucrose and fructose in combination with a high fat diet on intestinal microbiota and kidney oxidative stress. Nutrients 2017, 9, 393. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Zheng, H.; Joglekar, P.; Higginbottom, S.K.; Firbank, S.J.; Bolam, D.N.; Sonnenburg, J.L. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 2010, 141, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://mousecyc.jax.org:1555/META/NEWIMAGE?type=ORGANISM&object=TAX-210&detail-level=3%27A (accessed on 7 September 2017).
- Available online: http://www.genome.jp/dbget-bin/www_bget?hca00040 (accessed on 20 March 2023).
- Nagata, Y.; Sato, T.; Enomoto, N.; Ishii, Y.; Sasaki, K.; Yamada, T. High concentrations of D-amino acids in human gastric juice. Amino Acids 2007, 32, 137–140. [Google Scholar] [CrossRef]
- Fouad, F.; Mamer, O.; Sauriol, F.; Khayyal, M.; Lesimple, A.; Ruhenstroth-Bauer, G. Cardiac heart disease in the era of sucrose polyester, Helicobacter pylori and Chlamydia pneumoniae. Med. Hypotheses 2004, 62, 257–267. [Google Scholar] [CrossRef]
- Huang, F.-Y.; Chan, A.O.-O.; Lo, R.C.-L.; Rashid, A.; Wong, D.K.-H.; Cho, C.-H.; Lai, C.-L.; Yuen, M.-F. Characterization of interleukin-1β in Helicobacter pylori-induced gastric inflammation and DNA methylation in interleukin-1 receptor type 1 knockout (IL-1R1−/−) mice. Eur. J. Cancer 2013, 49, 2760–2770. [Google Scholar] [CrossRef]
- Sepulveda, A.R. Helicobacter, inflammation, and gastric cancer. Curr. Pathobiol. Rep. 2013, 1, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Maniam, J.; Antoniadis, C.P.; Youngson, N.A.; Sinha, J.K.; Morris, M.J. Sugar consumption produces effects similar to early life stress exposure on hippocampal markers of neurogenesis and stress response. Front. Mol. Neurosci. 2016, 8, 86. [Google Scholar] [CrossRef]
- An, D.; Na, C.; Bielawski, J.; Hannun, Y.A.; Kasper, D.L. Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc. Natl. Acad. Sci. USA 2011, 108, 4666–4671. [Google Scholar] [CrossRef] [PubMed]
- Couch, R.D.; Dailey, A.; Zaidi, F.; Navarro, K.; Forsyth, C.B.; Mutlu, E.; Engen, P.A.; Keshavarzian, A. Alcohol induced alterations to the human fecal VOC metabolome. PLoS ONE 2015, 10, e0119362. [Google Scholar] [CrossRef] [PubMed]
- Kochhar, S.; Martin, F.-P. Metabonomics and Gut Microbiota in Nutrition and Disease; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De Los Reyes-gavilán, C.G.; Salazar, N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Boulangé, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.-E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarkar, P.; Kandimalla, R.; Bhattacharya, A.; Wahengbam, R.; Dehingia, M.; Kalita, M.C.; Talukdar, N.C.; Talukdar, R.; Khan, M.R. Multi-Omics Analysis Demonstrates the Critical Role of Non-Ethanolic Components of Alcoholic Beverages in the Host Microbiome and Metabolome: A Human- and Animal-Based Study. Microorganisms 2023, 11, 1501. https://doi.org/10.3390/microorganisms11061501
Sarkar P, Kandimalla R, Bhattacharya A, Wahengbam R, Dehingia M, Kalita MC, Talukdar NC, Talukdar R, Khan MR. Multi-Omics Analysis Demonstrates the Critical Role of Non-Ethanolic Components of Alcoholic Beverages in the Host Microbiome and Metabolome: A Human- and Animal-Based Study. Microorganisms. 2023; 11(6):1501. https://doi.org/10.3390/microorganisms11061501
Chicago/Turabian StyleSarkar, Priyanka, Raghuram Kandimalla, Anupam Bhattacharya, Romi Wahengbam, Madhusmita Dehingia, Mohan Chandra Kalita, Narayan Chandra Talukdar, Rupjyoti Talukdar, and Mojibur R. Khan. 2023. "Multi-Omics Analysis Demonstrates the Critical Role of Non-Ethanolic Components of Alcoholic Beverages in the Host Microbiome and Metabolome: A Human- and Animal-Based Study" Microorganisms 11, no. 6: 1501. https://doi.org/10.3390/microorganisms11061501
APA StyleSarkar, P., Kandimalla, R., Bhattacharya, A., Wahengbam, R., Dehingia, M., Kalita, M. C., Talukdar, N. C., Talukdar, R., & Khan, M. R. (2023). Multi-Omics Analysis Demonstrates the Critical Role of Non-Ethanolic Components of Alcoholic Beverages in the Host Microbiome and Metabolome: A Human- and Animal-Based Study. Microorganisms, 11(6), 1501. https://doi.org/10.3390/microorganisms11061501





