1. Introduction
Breast cancer is one of the leading causes of death in women, exhibiting a severe prognosis and high mortality rate [
1]. Advancements in the stages of breast cancer make the prevention of the disease more complex; hence, detecting breast cancer at an earlier stage is essential for a better prognosis [
2].
Breast cancer is a complex disease with multiple factors contributing to its development. For example, inherited mutations in genes such as BRCA1 and BRCA2 or exposure to toxic substances such as tobacco, smoke, and alcohol can increase a person’s risk of developing breast cancer [
3,
4,
5]. The impact of other environmental factors such as lifestyle, diet, physical activity, hormonal influences, and exposure to radiation on breast cancer development is still being studied [
4,
5]. It is important to note that the development of breast cancer is influenced by a combination of these factors and further research is needed to fully understand the mechanisms that contribute to this disease.
Carcinogenesis can be initiated by a prolonged history of inflammation, which can be triggered by pathogenic infection, leading to a cascade of inflammatory events and a defective immune response [
1,
6]. To support this hypothesis, studies show that specific bacterial infections are significant risk factors for carcinomas of different organ systems including the breast, stomach, colon, and pancreas [
7,
8,
9]. For example, a study comparing cancerous and normal ovarian samples showed that a higher percentage of samples in the ovarian cancer group tested positive for
Chlamydia trachomatis infection in comparison to the control group [
10,
11]. Additionally, infection with
Helicobacter pylori plays a crucial role in the development of gastric cancer because of the pathogen’s ability to disrupt the host’s mucosal homeostasis [
9]. Further, multiple research groups have found that the advancement of colorectal cancer (CRC) is influenced by intestinal bacteria including H. pylori and the anaerobic bacteria
Fusobacterium nucleatum [
12,
13,
14,
15]. Several studies present that
F. nucleatum facilitates colorectal carcinoma by impacting the role of tumor-infiltrating lymphocytes and suppressing the action of natural killer (NK) cells, thus producing resistance to chemotherapy [
14,
15]. It was also revealed that an increased number of
F. nucleatum in breast cancer cells enhances tumor progression and metastases by impairing the activity of T cells [
14]. Another study explored the role of microbiomes in the progression of breast cancer and metastasis [
16]. In this study, researchers identified a tumor, called the mouse mammary tumor virus-polyoma middle tumor antigen (MMTV-PyMT), that closely resembles human breast cancer in mice [
16]. Researchers observed that breast tumor cells exhibited an oxygen microenvironment that significantly reduced anaerobes while increasing facultative anaerobes [
16]. In light of this observation, it was concluded that PyMT spontaneous breast tumors contain significant quantities of live bacteria [
16]. In addition, a substantial decrease in lung metastasis was observed following intratumor bacterial reduction by antibiotic treatment administered to eliminate tumor-resident microbiota [
16]. These observations suggested that tumor-associated bacteria could assist in cancer spread and metastasis [
16]. Additionally, it was shown that microorganisms are capable of inducing DNA damage in breast cancer tissues but not in normal breasts [
17].
To further strengthen the above observation, findings from a recent microbiome study showed that several pathogenic bacteria were present in different types of breast cancer tissues [
1]. One such microorganism was
Borrelia burgdorferi, a spirochete that causes Lyme disease. The presence of
B. burgdorferi in breast cancer tissues was also found to be associated with poor prognosis [
1]. This information explains the need to further investigate the influential role of
B. burgdorferi in the development of breast cancer.
Lyme disease is a vector-borne human disease that affects various body systems, causing musculoskeletal, neurological, and cardiac conditions [
18].
B. burgdorferi manipulates the mammalian host cells after its entry by modifying its glycoproteins, which help it to invade and propagate in different body tissues [
18,
19]. It was reported that
B. burgdorferi could invade human brain cells in vitro and induce an inflammatory process [
19]. Further, it was also observed that by the mechanism of intracellular localization
B. burgdorferi could stay hidden from the host’s immune system [
19]. Our research group has recently found evidence that
B. burgdorferi can invade the triple-negative MDA-MB-231 breast cancer cells at a significantly higher rate in comparison to normal mammary epithelial cells [
20]. This study also showed that the in vitro invasion and migratory capability of breast cancer cells was significantly increased following
B. burgdorferi infection [
20]. Our current research focused on identifying the molecular changes that occur when
B. burgdorferi infects breast tissues, and whether these changes could initiate or progress tumor development. In recent years, microRNAs (miRNA or miR) have been identified as novel cancer biomarkers that can play a vital role in the regulation of molecular mechanisms leading to tumorigenesis [
21]. Therefore, the goal of this study was to identify changes in miRNA expression levels in
B. burgdorferi-infected versus uninfected breast cancer cells to better understand the potential relationship between this infection and breast cancer development.
miRNAs are small non-coding RNAs that post-transcriptionally regulate gene expression [
21]. miRNAs are involved in regulating many molecular pathways, and their dysregulation can lead to disease development, including various cancers [
21]. Some miRNAs act as tumor suppressors and exhibit decreased expression in certain cancers, while other miRNAs are considered oncogenes and are expressed at higher levels in some cancer types [
22]. miRNAs are also implicated in cancer-related processes such as inflammation and tumor progression [
22,
23]. Numerous studies have reported that the development of breast cancer involves the dysregulated expression of miRNAs [
21]. Furthermore, previous research has reported that
B. burgdorferi infection can affect the expression of miRNAs in host cells [
24,
25]. It is thus essential to understand the role of miRNAs in
B. burgdorferi-infected breast carcinoma pathogenesis.
This study focused on analyzing the expression levels of several cancer-associated miRNAs in breast cancer cells following
B. burgdorferi infection. These potential target miRNAs were selected based on results from an exploratory miRNA breast cancer array and a scientific literature review. A confirmatory study was carried out using quantitative real-time reverse transcription PCR (qRT-PCR), which is considered a standard method for gene expression measurement. This study also focused on analyzing the pathways regulated by our target miRNAs and their associated genes using DIANA software [
26]. This research helps in better understanding the miRNAs that contribute to
B. burgdorferi infection-induced responses and breast cancer development, as well as their associated pathways and genes.
4. Discussion
Previous research has demonstrated that the dysregulation of miRNA expression plays a significant role in breast cancer development [
21,
28,
29,
30,
31]. Other reports have indicated that
B. burgdorferi infection can affect the expression of certain miRNAs in host cells [
25]. Therefore, we aimed to understand the impact of
B. burgdorferi infection on the expression and function of miRNAs in breast cancer. In this study, miRNAs of interest were first identified from an exploratory breast cancer array and extensive literature reviews [
21,
28,
29,
30,
31]. The expression of the target miRNAs was then further validated by qRT-PCR.
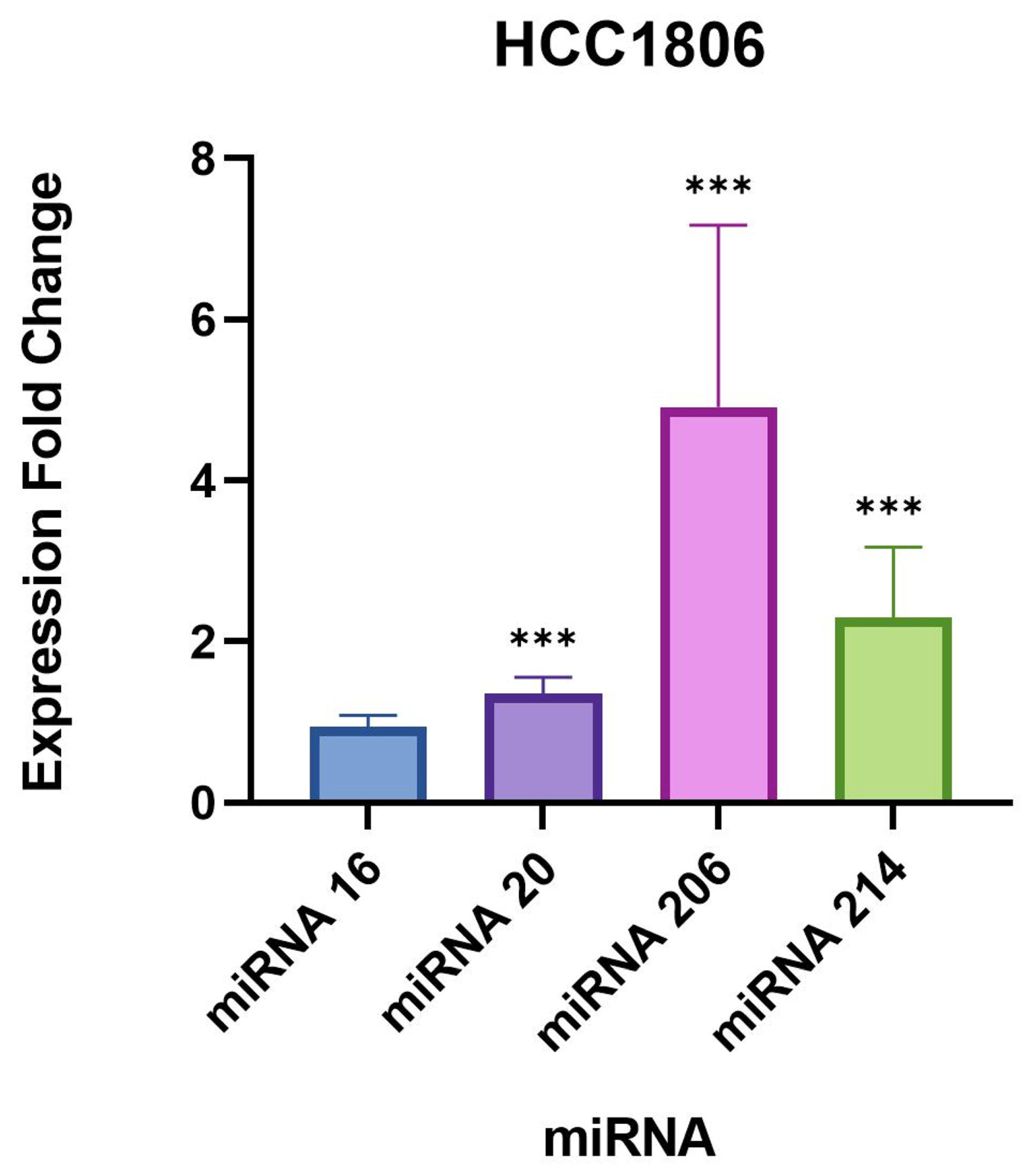
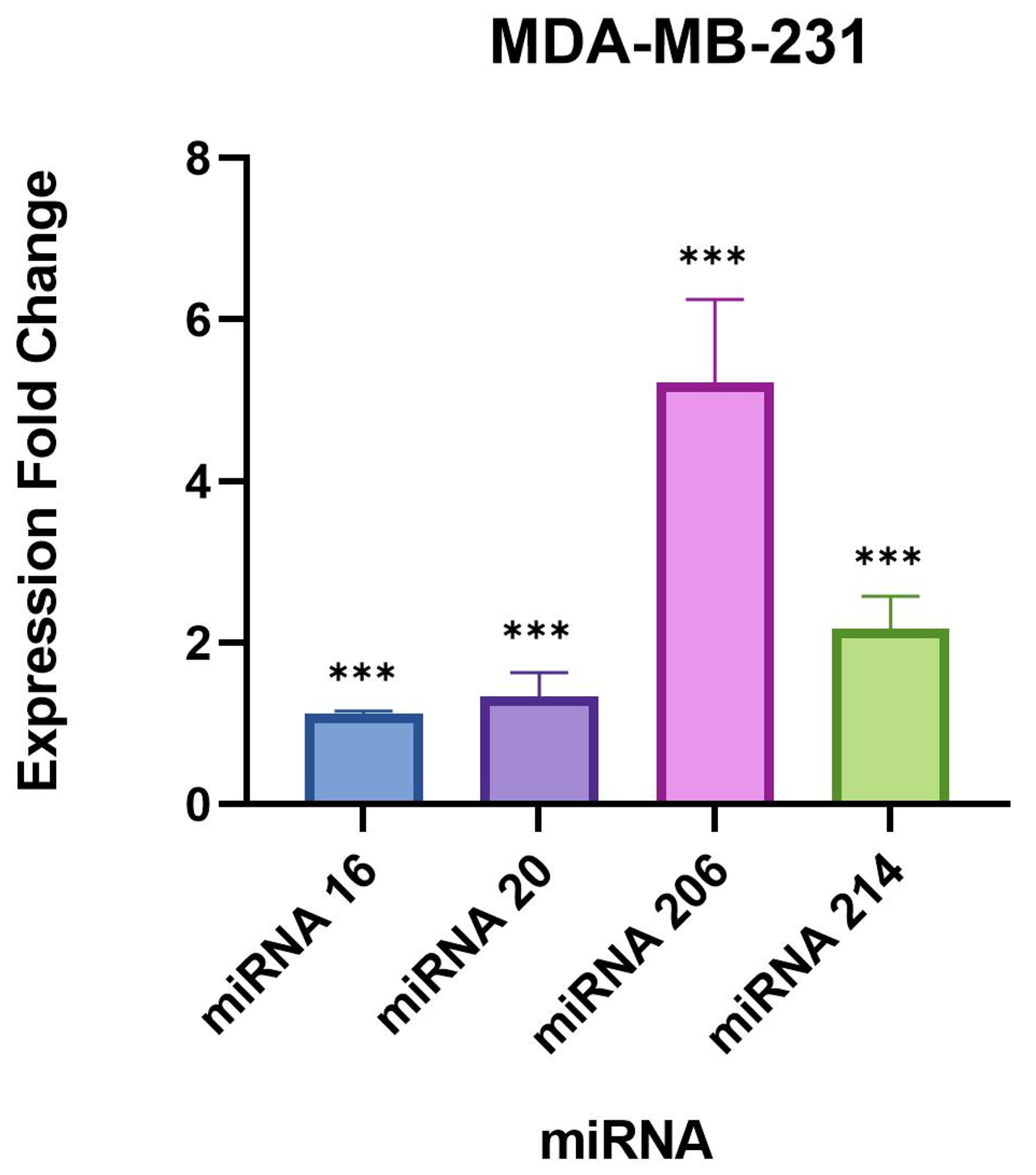
We found a very significant expression change of two miRNAs, miR-206 and 214-3p, in
B. burgdorferi-infected triple-negative breast cancer cell lines. Studies have previously demonstrated an upregulated expression of these miRNAs in breast cancer cells [
28,
41], especially miR-206 which was found to be associated with poor prognosis in breast cancer patients [
28]. Another study found that miR-206 facilitates the invasion, migration, and proliferation of breast cancer cells by suppressing the expression of the G-protein-coupled receptor neurokinin-1 (NK1R-FL) [
41]. Moreover, the proliferation of breast cancer cells was significantly reduced with the downregulation of miR-206 [
41]. These previous studies indicate that miR-206 likely plays a role in inducing a tumorigenic response.
Another important effect of miR-206 is the regulation of matrix metalloproteinase 9 (MMP9) expression, a protein that acts in tissue reassembly by activating cytokines and chemokines that degenerate the extracellular matrix [
32]. In a mycobacterial infection study, miR-206 was found to regulate the expression of MMP9 by influencing tissue inhibitors of metalloproteinases such as TIMP3 [
33]. Another study identified an upregulated expression of miR-206 in the failing hearts of mice and cardiac fibroblast cells and was found to be associated with reduced TIMP 3 activity [
42]. Previously, our research group detected a significant increase in the expression of MMP9 in
B. burgdorferi-infected MDA-MB-231 cells compared to the uninfected control group [
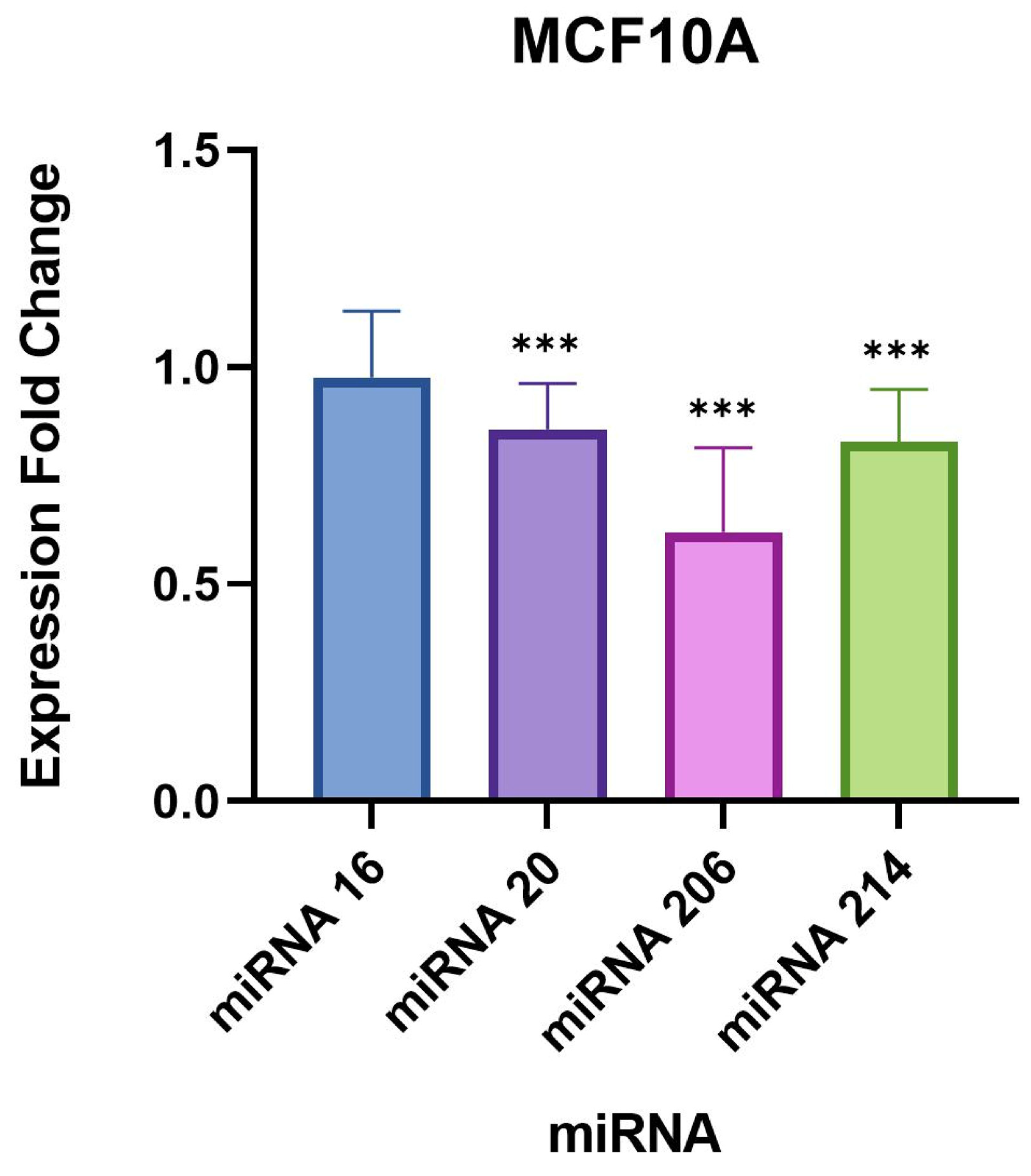
20]. In contrast, the normal mammary epithelial cell line, MCF10A, did not show any change in MMP9 expression after infection with
B. burgdorferi [
20]. This finding demonstrated that the migratory and invasive characteristics of breast cancer cells were influenced by the level of tissue remodeling markers following infection with
B. burgdorferi [
20].
miR-214-3p also exhibited significant upregulation in our study which is supported by previous studies that have found miR-214-3p upregulation to be associated with the progression of triple-negative breast cancer [
29,
43]. One study observed that miR-214 promotes breast cancer progression by impairing the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR) signal transduction (PI3K/Akt/mTOR) pathway [
29]. This pathway is one of the most important cell signaling pathways as it plays an essential role in regulating cell proliferation, apoptosis, and differentiation [
29]. Another study indicated that miR-214 was significantly upregulated in sporadic breast cancer patients compared to the hereditary form [
43]. Furthermore, triple-negative breast cancer patients with a higher expression of miR-214 exhibited a poorer prognosis than patients with a lower expression of miR-214 [
43]. In yet another study, miR-214 was among the miRNAs found to be differentially expressed in primary astrocytes after infection with
B. burgdorferi [
35]. Moreover, a research study observed that triple-negative breast cancer cells’ viability, migration, and invasion capacity were higher with miR-214-3p upregulation and that inhibiting this miRNA suppressed these effects [
44]. Furthermore, miR-214-3p was found to target ST6GAL1 by negative feedback regulation [
44]. ST6GAL1 is a sialyltransferase whose transformation relates to the growth, invasion, and metastasis of the tumor [
44,
45]. It was identified that knocking down ST6GAL1 repressed triple-negative breast cancer cells’ viability, migration, and invasion abilities, which were restored by lowering the expression of miR-214-3p, indicating the association of this miRNA in breast cancer development [
44].
In our study, the remaining targets, miR-16-5p and 20b-5p, did not demonstrate drastic expression changes compared to miR-206 and 214-3p, but were still statistically significant in the evaluated breast cell lines. A recent study published in 2021 concluded that the overexpression of miR-16-5p in breast cancer tissues resulted in cell cycle arrest and inhibited breast cancer cell proliferation [
30]. An upregulation of ANLN (Anillin actin-binding protein) was identified in breast carcinoma and was a target of miR-16-5p in breast cancer cells [
30]. ANLN is involved in cell division and is upregulated in various cancers such as pancreatic and cervical cancer [
46,
47]. Furthermore, it was discovered that miR-16-5p suppressed ANLN expression contributing to the inhibition of breast cancer cells, similar to the overexpression of miR-16-5p [
30]. Additionally, it was reported that infection with
Helicobacter pylori (
H. pylori) induces an inflammatory process by the persistent colonization of the gastric mucosa, which could ultimately progress to more severe diseases such as gastric carcinoma [
36,
48]. Research also demonstrated that infection of the gastric epithelial cells resulted in an upregulation of miR-16, suggesting the role of bacterial infection in the dysregulation of this miRNA [
48]. miR-20b-5p, evaluated in our study, is also implicated in playing a critical role in the development of breast cancer [
31,
37]. One study observed that miR-20b-5p significantly promoted breast cancer cell migration and proliferation capacity [
31]. Researchers also demonstrated that upregulation of miR-20b-5p in breast cancer stem cells (BCSCs) inhibited their apoptosis and facilitated proliferation [
31]. Further, an in vivo experiment involving xenograft tumors of breast cancer cells indicated that miR-20b-5p overexpression led to their significant enlargement [
31]. Another study revealed the overexpression of miR-20b in breast cancer cells inhibited a critical tumor suppressor gene, PTEN, by binding to its 3′-UTR [
37]. Additionally, the upregulation of miR-20b significantly promoted the proliferation and colony formation of breast cancer cells, whereas its downregulation reduced the growth of breast cancer cells [
37]. PTEN downregulation activates the critical oncogenic PI3K/Akt signaling pathway, hence regulating cell proliferation, migration, cell cycle, and apoptosis during the development of cancer [
37,
49].
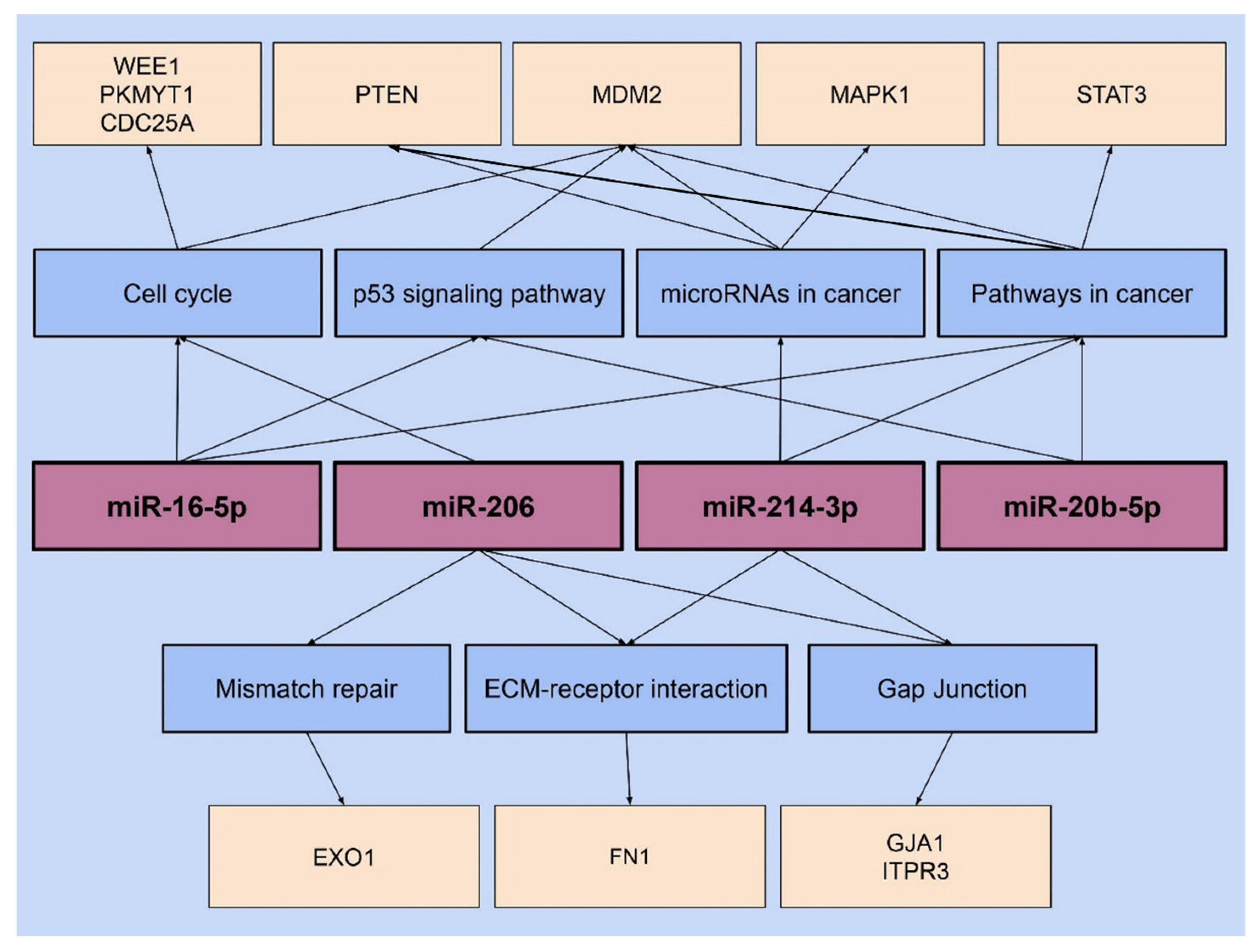
Our study further analyzed the pathways and genes associated with each target miRNA using the mirPath v.3 DIANA software. Multiple pathways were found to be associated with each miRNA studied, and each of these pathways involved multiple genes regulated by the corresponding miRNA. For example, miR-206 and 214-5p were found to influence two pathways, ECM–receptor interaction and Gap junction. The ECM–receptor interaction pathway involves communication between cells and the extracellular matrix, a process that assists in the development of cell–matrix interactions and ECM remodeling [
50]. Mutation in this pathway can result in the development of a tumor, augment the migration of tumor cells, and initiate metastatic progression by redesigning the ECM in distant organs [
50]. Relevantly, one research study has demonstrated a direct association between ECM remodeling and breast cancer progression [
51]. The study identified advanced tumor growth caused by increased collagen-I deposition in mouse mammary epithelial cells, along with aggressive tumor progression and invasive lung metastasis [
51]. In both miR-206 and 214-5p, the genes FN1 and GJA1 were found to be associated with the ECM–receptor interaction and Gap junction pathway, respectively [
26].
The Gap junction pathway, regulated by miR-206 and 214-5p, consists of intercellular channels that lead to the direct transmission of ions and small molecules between the cytosolic compartments of adjacent cells [
52]. The upregulated expression of the FN1 gene has been correlated with the development of breast cancer and poor prognosis [
53]. It has also been demonstrated that, when upregulated, miR-206 targets and leads to the irregular expression of the FN1 gene in Hirschsprung disease patients [
54]. Moreover,
B. burgdorferi targets the FN1 protein by expressing BBK32, a fibronectin-binding protein [
55]. An upregulated expression of the GJA1 gene has been demonstrated to play a significant role in the vesicle-mediated transport of the transcellular location of
Borrelia bavariensis and resulting in infection of the human brain microvascular endothelial cells [
56]. This report, therefore, contributes to a better understanding of the resulting molecular changes caused by the dysregulation of miR-206 and 214-5p related to pathways and the associated genes [
53,
55,
56]. Further, our data support the hypothesis that changes in miRNA expression can be a contributing factor to the development of breast cancer after
B. burgdorferi infection although more research should investigate this connection further.
In addition, miR-206 is associated with two other pathways, cell cycle, and mismatch repair, suggesting it may play a role in maintaining cellular homeostasis [
26]. According to the DIANA analysis, the cell cycle is influenced by genes WEE1 and PKMYT1 which play a vital role in recognizing and repairing DNA damage [
26,
57]. Both these genes exhibit an upregulated expression in breast cancer cells and act as oncogenes, assisting in further growth and carcinoma progression [
57]. It has been reported that as malignant transformation is induced, WEE1 upregulation in cancer cells might promote tumorigenesis by maintaining the levels of genomic instability [
57]. The DIANA analysis further shows that the mismatch repair pathway, regulated by miR-206, involves the EXO1 gene, which regulates the mechanisms of DNA damage checkpoints and repair [
58]. The EXO1 gene was investigated in a study using MDA-MB-231 breast cancer cells where an increase in gene expression was observed compared to the normal mammary epithelial cell line, MCF10A [
58]. This report suggests that the elevated expression of this EXO1 gene is associated with breast cancer development and poor prognosis by affecting the mismatch repair pathway of the cells [
58]. The role of miR-206 may be a contributing factor in carcinogenesis, impaired cellular function, and DNA damage through its relation to the cell cycle and mismatch repair pathway [
26].
DIANA analysis of miR-214-3p shows that this miRNA can influence two other pathways, microRNAs in cancer and pathways in cancer. Both these pathways involve the PTEN and MAPK1 genes [
26]. PTEN mostly acts as a tumor suppressor, and it has been observed that loss of PTEN expression is associated with breast cancer progression [
59]. Further, a research study has shown that in breast cancer cells, the upregulated expression of the MAPK pathway is associated with reduced PTEN expression [
59]. Through DIANA analysis, miR-214-3p appears to influence another gene, the MDM2, which is involved in cancer pathways [
26]. There is evidence that the members of the MDM family play a crucial role in regulating the fate of cell development [
60]. This function of MDM is achieved by the negative control of an essential tumor suppressor gene, p53 [
60]. In addition, research has concluded that the overexpression of the MDM2 gene is related to the augmentation of breast cancer [
60]. Hence, the role of miR-214-3p in these pathways obtained by DIANA analysis describes its potential effect on breast cancer development [
26]. In this study, a higher expression of miR-206 and 214-3p in the
B. burgdorferi-infected triple-negative breast cancer cells was confirmed which provided strong evidence that these miRNAs may play a crucial role in the spirochetal infection of mammalian epithelial cells.
The DIANA analysis for miR-16-5p showed that it affects three pathways, one of which is the cell cycle pathway associated with the genes MDM2 and CDC25A [
26]. The CDC25A gene regulates the G1/S and G2/M checkpoints of the cell cycle, and its dysregulation has been implemented in the development of triple-negative breast cancer [
61]. Thus, miR-16-5p might be regulating genes involved in cell cycle pathways. Further DIANA analysis identified that miR-16-5p and 20b-5p share two other pathways. Both miRNAs were found to be associated with pathways in cancer and the p53 signaling pathway coded by the STAT3 and MDM2 genes [
26]. STAT3 regulates the progress of breast cancer and the proliferation of cells by targeting oncogenes [
62]. As mentioned earlier, miR-16-5p and 20b-5p were associated with the p53 signaling pathway, which involves the p53 tumor suppressor gene, whose dysregulation led to the growth of more aggressive breast cancer and poor prognosis [
63].
In our DIANA analysis, it was further seen that out of the four target miRNAs, the MDM2 gene was associated with three miRNAs, miR-214-3p, 16-5p, and 20b-5p [
26]. MDM2 is found to act as an oncoprotein mainly by degrading the tumor suppressor gene p53 via E3 ubiquitin ligase action [
60,
64]. The association of our target miRNAs with MDM2 has been a significant observation as this gene is found to be dysregulated in several cancers [
60,
64]. In addition, the pathway in cancer, as seen in miR-16-5p and 20b-5p, was also observed in miR-214-3p, making it the only pathway that is observed to be influenced by three miRNAs [
26]. Hence, by analyzing the miRNA pathways using the DIANA, it was established that most of these target miRNAs influence cellular mechanism pathways associated with oncogenes, suggesting a possible role in the development and progression of breast cancer cells and
B. burgdorferi infection.