Multi-Approach Characterization of Novel Pyrene-Degrading Mycolicibacterium austroafricanum Isolates Lacking nid Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Cultivation Conditions
2.2. Qualitative Assays of Pyrene Degradation Using the 2,6-DCPIP Assay and Double-Layer Plates
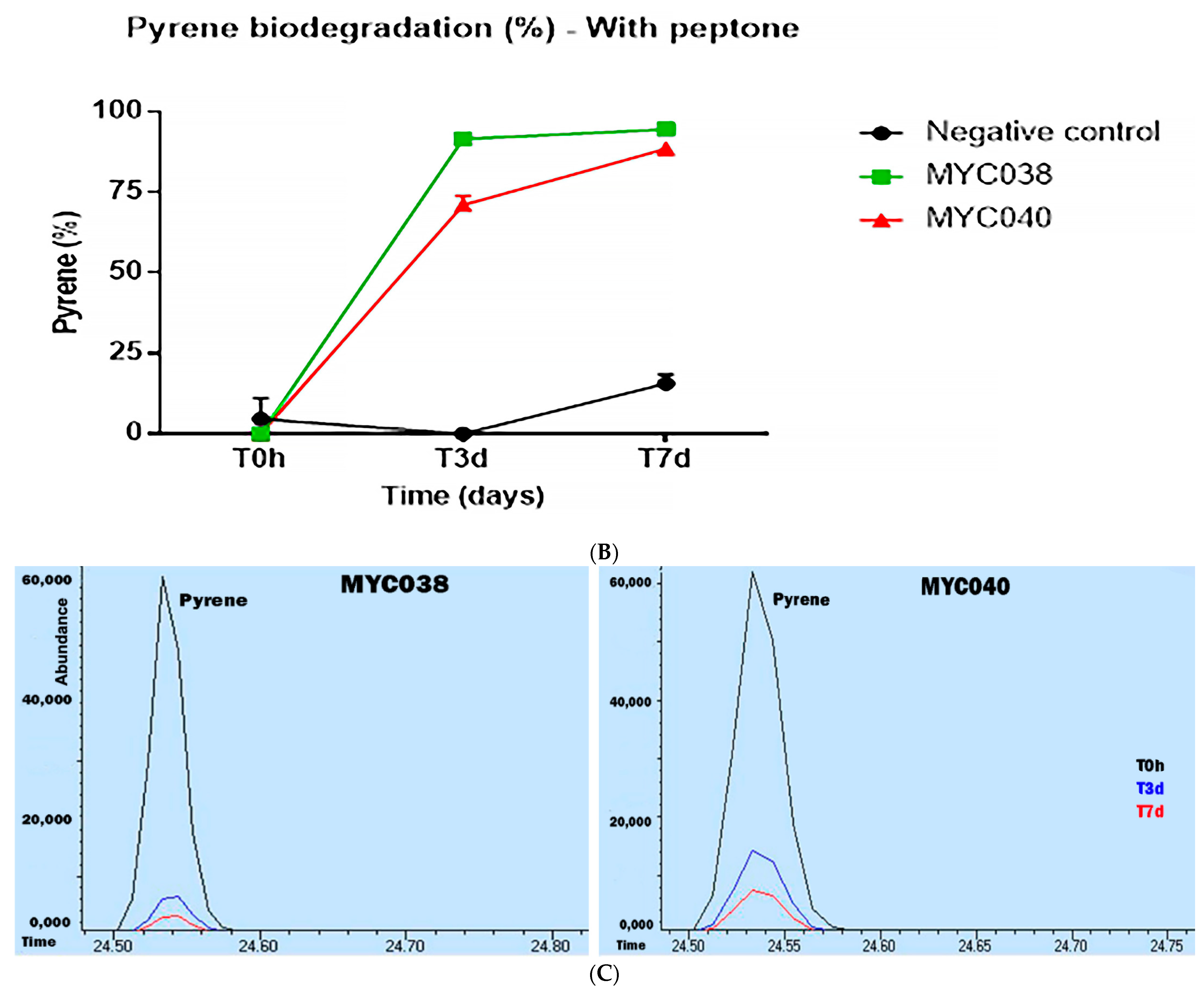
2.3. Evaluation of Pyrene Degradation via GC/MS
2.4. Amplification of Dioxygenase (nid) and Cytochrome P450 (cyp) Genes via PCR
2.5. Genome Sequencing, Assembly, and Annotation
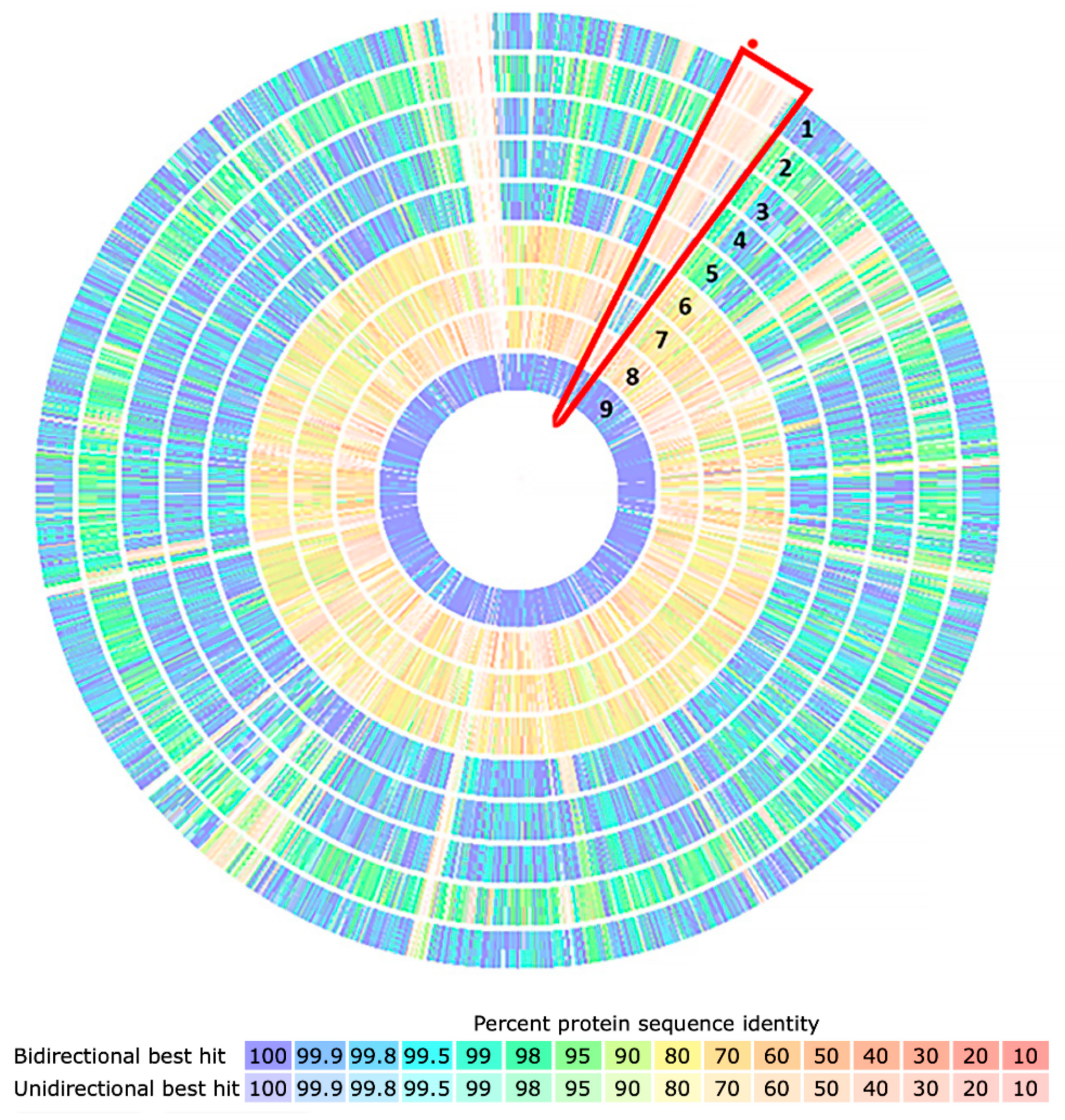
2.6. Bioinformatic Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, C.; Anderson, A.J. Multiplicity of Genes for Aromatic Ring-Hydroxylating Dioxygenases in Mycobacterium Isolate KMS and Their Regulation. Biodegradation 2012, 23, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ren, D.; Sun, W.; Li, X.; Huang, B.; Chen, R.; Lin, C.; Pan, X. Polycyclic Aromatic Hydrocarbons Associated with Total Suspended Particles and Surface Soils in Kunming, China: Distribution, Possible Sources, and Cancer Risks. Environ. Sci. Pollut. Res. 2015, 22, 6696–6712. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.I.; Mansour, M.S.M. A Review on Polycyclic Aromatic Hydrocarbons: Source, Environmental Impact, Effect on Human Health and Remediation. Egypt. J. Pet. 2016, 25, 107–123. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef]
- Haritash, A.K. A Comprehensive Review of Metabolic and Genomic Aspects of PAH-Degradation. Arch. Microbiol. 2020, 202, 2033–2058. [Google Scholar] [CrossRef]
- Patel, S.; Homaei, A.; Patil, S.; Daverey, A. Microbial Biosurfactants for Oil Spill Remediation: Pitfalls and Potentials. Appl. Microbiol. Biotechnol. 2018, 103, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Choi, I.G. Genomic and Transcriptomic Perspectives on Mycoremediation of Polycyclic Aromatic Hydrocarbons. Appl. Microbiol. Biotechnol. 2020, 104, 6919–6928. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Gadd, G.M. Fungal Bioremediation of Soil Co-Contaminated with Petroleum Hydrocarbons and Toxic Metals. Appl. Microbiol. Biotechnol. 2020, 104, 8999–9008. [Google Scholar] [CrossRef]
- Sarkar, J.; Saha, A.; Roy, A.; Bose, H.; Pal, S.; Sar, P.; Kazy, S.K. Development of Nitrate Stimulated Hydrocarbon Degrading Microbial Consortia from Refinery Sludge as Potent Bioaugmenting Agent for Enhanced Bioremediation of Petroleum Contaminated Waste. World J. Microbiol. Biotechnol. 2020, 36, 156. [Google Scholar] [CrossRef]
- Curiel-Alegre, S.; Velasco-Arroyo, B.; Rumbo, C.; Khan, A.H.A.; Tamayo-Ramos, J.A.; Rad, C.; Gallego, J.L.R.; Barros, R. Evaluation of Biostimulation, Bioaugmentation, and Organic Amendments Application on the Bioremediation of Recalcitrant Hydrocarbons of Soil. Chemosphere 2022, 307, 135638. [Google Scholar] [CrossRef]
- Lawniczak, Ł.; Woźniak-Karczewska, M.; Loibner, A.P.; Heipieper, H.J.; Chrzanowski, Ł. Molecules Microbial Degradation of Hydrocarbons-Basic Principles for Bioremediation: A Review. Molecules 2020, 25, 856. [Google Scholar] [CrossRef] [PubMed]
- Heitkamp, M.A.; Franklin, W.; Cerniglia, C.E. Microbial Metabolism of Polycyclic Aromatic Hydrocarbons: Isolation and Characterization of a Pyrene-Degrading Bacterium. Appl. Environ. Microbiol. 1988, 54, 2549–2555. [Google Scholar] [CrossRef]
- Khan, A.A.; Kim, S.J.; Paine, D.D.; Cerniglia, C.E. Classification of a Polycyclic Aromatic Hydrocarbon-Metabolizing Bacterium, Mycobacterium sp. Strain PYR-1, as Mycobacterium vanbaalenii sp. Nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1997–2002. [Google Scholar] [CrossRef]
- Schneider, J.; Grosser, R.; Jayasimhulu, K.; Xue, W.; Warshawsky, D. Degradation of Pyrene, Benz[a]Anthracene, and Benzo[a]Pyrene by Mycobacterium sp. Strain RJGII-135, Isolated from a Former Coal Gasification Site. Appl. Environ. Microbiol. 1996, 62, 13–19. [Google Scholar] [CrossRef]
- Churchill, P.F.; Morgan, A.C.; Kitchens, E. Characterization of a Pyrene-Degrading Mycobacterium sp. Strain CH-2. J. Environ. Sci. Health 2008, 43, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Churchill, S.A.; Harper, J.P.; Churchill, P.F. Isolation and Characterization of a Mycobacterium Species Capable of Degrading Three-and Four-Ring Aromatic and Aliphatic Hydrocarbons. Appl. Environ. Microbiol. 1999, 65, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.D.; Hall, K.; Liang, Y.N.; Nieman, K.; Sorensen, D.; Issa, B.; Anderson, A.J.; Sims, R.C. Isolation and Characterization of Polycyclic Aromatic Hydrocarbon-Degrading Mycobacterium Isolates from Soil. Microb. Ecol. 2004, 48, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Lin, X.; Zhang, J.; Li, X. Isolation of Polycyclic Aromatic Hydrocarbons (PAHs)-Degrading Mycobacterium Spp. and the Degradation in Soil. J. Hazard. Mater. 2010, 183, 718–723. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, K.; Lee, D.H.; Cha, C.J. Comparative Genomic Analysis of Pyrene-Degrading Mycobacterium Species: Genomic Islands and Ring-Hydroxylating Dioxygenases Involved in Pyrene Degradation. J. Microbiol. 2018, 56, 798–804. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Chen, Y.; Lou, J.; Wu, L.; Xu, J. Salicylate and Phthalate Pathways Contributed Differently on Phenanthrene and Pyrene Degradations in Mycobacterium sp. WY10. J. Hazard. Mater. 2019, 364, 509–518. [Google Scholar] [CrossRef]
- Golubev, S.N.; Muratova, A.Y.; Panchenko, L.V.; Shchyogolev, Y.; Turkovskaya, O.V. Mycolicibacterium sp. Strain PAM1, an Alfalfa Rhizosphere Dweller, Catabolizes PAHs and Promotes Partner-Plant Growth. Microbiol. Res. 2021, 253, 126885. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Luan, T.; Lin, L.; Liu, H.; Tam, N.F.Y. Production of Metabolites in the Biodegradation of Phenanthrene, Fluoranthene and Pyrene by the Mixed Culture of Mycobacterium sp. and Sphingomonas sp. Bioresour. Technol. 2010, 102, 2965–2972. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.B.; Wang, C.Y.; Lv, C.Y.; Lun, Z.M.; Zheng, C.G. Removal Capacities of Polycyclic Aromatic Hydrocarbons (PAHs) by a Newly Isolated Strain from Oilfield Produced Water. Int. J. Environ. Res. Public Health 2017, 14, 215. [Google Scholar] [CrossRef] [PubMed]
- Moody, J.D.; Freeman, J.P.; Doerge, D.R.; Cerniglia, C.E. Degradation of Phenanthrene and Anthracene by Cell Suspensions of Mycobacterium sp. Strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 1476–1483. [Google Scholar] [CrossRef]
- Brezna, B.; Kweon, O.; Stingley, R.L.; Freeman, J.P.; Khan, A.A.; Polek, B.; Jones, R.C.; Cerniglia, C.E. Molecular Characterization of Cytochrome P450 Genes in the Polycyclic Aromatic Hydrocarbon Degrading Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 2006, 71, 522–532. [Google Scholar] [CrossRef]
- Kim, S.J.; Kweon, O.; Jones, R.C.; Edmondson, R.D.; Cerniglia, C.E. Genomic Analysis of Polycyclic Aromatic Hydrocarbon Degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation 2008, 19, 859–881. [Google Scholar] [CrossRef]
- Kim, S.J.; Kweon, O.; Jones, R.C.; Freeman, J.P.; Edmondson, R.D.; Cerniglia, C.E. Complete and Integrated Pyrene Degradation Pathway in Mycobacterium vanbaalenii PYR-1 Based on Systems Biology. J. Bacteriol. 2007, 189, 464–472. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.J.; Blom, J.; Kim, S.K.; Kim, B.S.; Baek, D.H.; Park, S.I.; Sutherland, J.B.; Cerniglia, C.E. Comparative Functional Pan-Genome Analyses to Build Connections between Genomic Dynamics and Phenotypic Evolution in Polycyclic Aromatic Hydrocarbon Metabolism in the Genus Mycobacterium. BMC Evol. Biol. 2015, 15, 21. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kweon, O.; Cerniglia, C.E. Degradation of Polycyclic Aromatic Hydrocarbons by Mycobacterium Strains. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1865–1879. [Google Scholar] [CrossRef]
- Zada, S.; Zhou, H.; Xie, J.; Hu, Z.; Ali, S.; Sajjad, W.; Wang, H. Bacterial Degradation of Pyrene: Biochemical Reactions and Mechanisms. Int. Biodeterior. Biodegrad. 2021, 162, 105233. [Google Scholar] [CrossRef]
- Wu, F.; Guo, C.; Liu, S.; Liang, X.; Lu, G.; Dang, Z. Pyrene Degradation by Mycobacterium gilvum: Metabolites and Proteins Involved. Water Air Soil Pollut. 2019, 230, 67. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Li, L.; Lin, C.; Dong, W.; Shen, W.; Yong, X.; Jia, H.; Wu, X.; Zhou, J. Anaerobic Biodegradation of Pyrene by Klebsiella sp. LZ6 and Its Proposed Metabolic Pathway. Environ. Technol. 2020, 41, 2130–2139. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, C.L.; Pellegrino KC, M.; Silva, N.M.; Brianesi, U.A.; Leão, S.C.; Rabello, M.C.D.S.; Viana-Niero, C. Diversity of Mycobacteriaceae from Aquatic Environment at the São Paulo Zoological Park Foundation in Brazil. PLoS ONE 2020, 15, e0227759. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Daisuke, A.E.; Ae, K.; Matsumiya, Y.; Seon-Yong, A.E.; Ae, C.; Kubo, M. Phylogenetic Analysis of Long-Chain Hydrocarbon-Degrading Bacteria and Evaluation of Their Hydrocarbon-Degradation by the 2,6-DCPIP Assay. Biodegradation 2008, 19, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.M.; De Oliveira, A.M.S.A.; Pegorin, S.; Giusti, C.E.; Ferrari, V.B.; Barbosa, D.; Martins, L.F.; Morais, C.; Setubal, J.C.; Vasconcellos, S.P.; et al. Characterization of Novel Hydrocarbon-Degrading Gordonia Paraffinivorans and Gordonia Sihwensis Strains Isolated from Composting. PLoS ONE 2019, 14, e0215396. [Google Scholar] [CrossRef]
- Zhou, H.W.; Luan, T.G.; Zou, F.; Tam, N.F.Y. Different Bacterial Groups for Biodegradation of Three- and Four-Ring PAHs Isolated from a Hong Kong Mangrove Sediment. J. Hazard. Mater. 2008, 152, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Al-Thukair, A.A.; Malik, K.; Nzila, A. Biodegradation of Selected Hydrocarbons by Novel Bacterial Strains Isolated from Contaminated Arabian Gulf Sediment. Sci. Rep. 2020, 10, 21846. [Google Scholar] [CrossRef]
- Yuan, K.; Xie, X.; Wang, X.; Lin, L.; Yang, L.; Luan, T.; Chen, B. Transcriptional Response of Mycobacterium sp. Strain A1-PYR to Multiple Polycyclic Aromatic Hydrocarbon Contaminations. Environ. Pollut. 2018, 243, 824–832. [Google Scholar] [CrossRef]
- Kim, S.J.; Song, J.; Kweon, O.; Holland, R.D.; Kim, D.W.; Kim, J.; Yu, L.R.; Cerniglia, C.E. Functional Robustness of a Polycyclic Aromatic Hydrocarbon Metabolic Network Examined in a NidA Aromatic Ring-Hydroxylating Oxygenase Mutant of Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2012, 78, 3715–3723. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.J.; Freeman, J.P.; Song, J.; Baek, S.; Cerniglia, C.E. Substrate Specificity and Structural Characteristics of the Novel Rieske Nonheme Iron Aromatic Ring-Hydroxylating Oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 2010, 1, e00135-10. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef]
- Galardini, M.; Biondi, E.G.; Bazzicalupo, M.; Mengoni, A. CONTIGuator: A Bacterial Genomes Assembler for Structural Insights on Draft Genomes. Source Code Biol. Med. 2011, 6, 11. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Delineating Bacterial Subspecies Additional File 3 to: “Complete Genome Sequence of DSM 30083 T, the Type Strain (U5/41 T) of Escherichia coli, and a Proposal for Delineating Subspecies in Microbial Taxonomy”. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lo, B.; Son, J. Corrigendum: Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2019, 67, 10. [Google Scholar] [CrossRef]
- Santiago, C.R.N.; Digiampietre, L.A.; Moreira, L.M. Porto Alegre: Sociedade Brasileira de Computação. 2020, pp. 23–30. Available online: https://sol.sbc.org.br/index.php/sbcas_estendido/article/view/11553 (accessed on 10 December 2022).
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Anderson, A.J. Polycyclic Aromatic Hydrocarbon Degrading Gene Islands in Five Pyrene-Degrading Mycobacterium Isolates from Different Geographic Locations. Can. J. Microbiol. 2012, 58, 102–111. [Google Scholar] [CrossRef]
- Van Hamme, J.D.; Singh, A.; Ward, O.P. Recent Advances in Petroleum Microbiology. Microbiol. Mol. Biol. Rev. 2003, 67, 503–549. [Google Scholar] [CrossRef]
- Hanson, K.G.; Desai, J.D.; Desai, A.J. A Rapid and Simple Screening Technique for Potential Crude Oil Degrading Microorganisms. Biotechnol. Tech. 1993, 7, 745–748. [Google Scholar] [CrossRef]
- Roy, S.; Hens, D.; Biswas, D.; Biswas, D.; Kumar, R. Survey of Petroleum-Degrading Bacteria in Coastal Waters of Sunderban Biosphere Reserve. World J. Microbiol. Biotechnol. 2002, 18, 575–581. [Google Scholar] [CrossRef]
- Vieira, G.A.L.; Magrini, M.J.; Bonugli-Santos, R.C.; Rodrigues, M.V.N.; Sette, L.D. Environmental Microbiology Polycyclic Aromatic Hydrocarbons Degradation by Marine-Derived Basidiomycetes: Optimization of the Degradation Process. Braz. J. Microbiol. 2018, 49, 749–756. [Google Scholar] [CrossRef]
- Rein, A.; Adam, I.K.U.; Miltner, A.; Brumme, K.; Kästner, M.; Trapp, S. Impact of Bacterial Activity on Turnover of Insoluble Hydrophobic Substrates (Phenanthrene and Pyrene)—Model Simulations for Prediction of Bioremediation Success. J. Hazard. Mater. 2016, 306, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Elyamine, A.M.; Kan, J.; Meng, S.; Tao, P.; Wang, H.; Hu, Z.; Leitão, J.H. Molecular Sciences Aerobic and Anaerobic Bacterial and Fungal Degradation of Pyrene: Mechanism Pathway Including Biochemical Reaction and Catabolic Genes. Int. J. Mol. Sci. 2021, 22, 8202. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kweon, O.; Freeman, J.P.; Jones, R.C.; Adjei, M.D.; Jhoo, J.W.; Edmondson, R.D.; Cerniglia, C.E. Molecular Cloning and Expression of Genes Encoding a Novel Dioxygenase Involved in Low- and High-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation in Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2006, 72, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gu, Y.; Chen, Z.; Song, Y.; Sun, F.; Liu, J.; Waigi, M.G. Colonization and Performance of a Pyrene-Degrading Bacterium Mycolicibacterium sp. Pyr9 on Root Surfaces of White Clover. Chemosphere 2021, 263, 127918. [Google Scholar] [CrossRef]
- Fida, T.T.; Moreno-Forero, S.K.; Breugelmans, P.; Heipieper, H.J.; Röling, W.F.M.; Springael, D. Physiological and Transcriptome Response of the Polycyclic Aromatic Hydrocarbon Degrading Novosphingobium sp. LH128 after Inoculation in Soil. Environ. Sci. Technol. 2017, 51, 1570–1579. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Xu, L.; Zhang, R.; Zhou, Y.; Xiao, Y.; Su, X.; Shen, C.; Sun, F.; Hashmi, M.Z.; Lin, H.; et al. Viable but Nonculturable State of Yeast Candida sp. Strain LN1 Induced by High Phenol Concentrations. Appl. Environ. Microbiol. 2021, 87, e01110-21. [Google Scholar] [CrossRef]
- Vila, J.; Tauler, M.; Grifoll, M. Bacterial PAH Degradation in Marine and Terrestrial Habitats. Curr. Opin. Biotechnol. 2015, 33, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.C.; Choi, C.W.; Badejo, A.O.; Shin, K.H.; Hyun, J.H.; Lee, Y.G.; Kim, S.I.; Park, K.S.; Kim, S.H.; Jung, K.H.; et al. A Global Proteome Study of Mycobacterium gilvum PYR-GCK Grown on Pyrene and Glucose Reveals the Activation of Glyoxylate, Shikimate and Gluconeogenetic Pathways through the Central Carbon Metabolism Highway. Biodegradation 2013, 24, 741–752. [Google Scholar] [CrossRef]
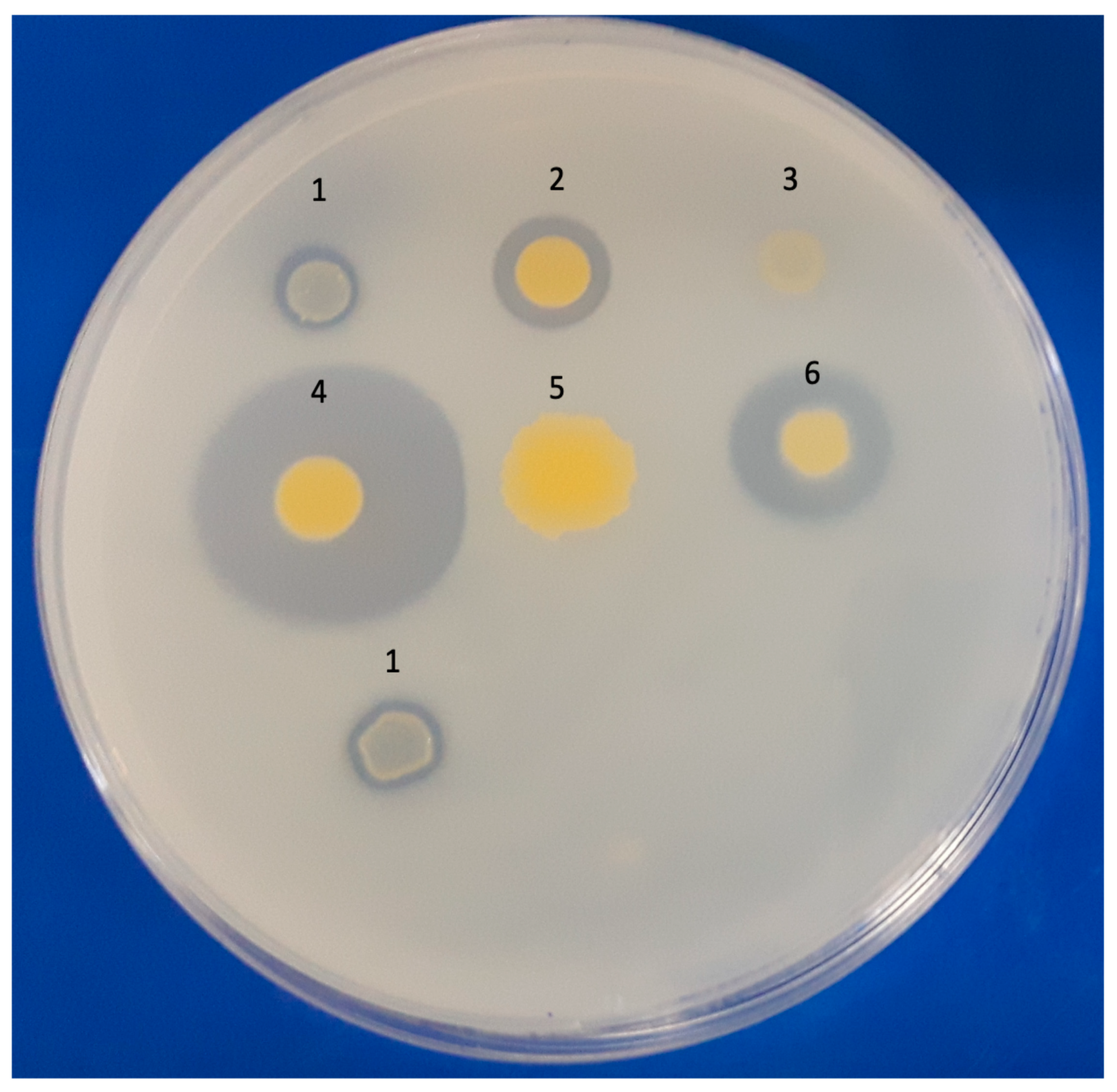


| Isolates | 2,6-DCPIP | PCR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| nidA | GP | nidB | nidB2 | nidA3 | nidB3 | cyp150 | cyp151 | ||
| M. vanbaalenii PYR-1 | + | + | + | + | + | + | + | + | + |
| MYC038 | + | − | − | − | − | − | − | + | − |
| MYC040 | + | − | − | − | − | − | − | + | − |
| MYC211 | + | − | − | − | − | − | − | + | − |
| MYC221 | + | − | − | − | − | − | − | + | − |
| MYC223 | + | − | − | − | − | − | − | + | − |
| General Features | MYC038 | MYC040 | MYC211 | MYC221 | MYC223 |
|---|---|---|---|---|---|
| Genome size (MB) | 6.26 | 6.03 | 6.18 | 6.28 | 6.17 |
| Genome size (bp) | 626,079 | 603,814 | 618,955 | 628,763 | 617,427 |
| Paired-end reads (Illumina/MiSeq) | 1,854,071 | 2,272,631 | 1,733,214 | 2,438,739 | 13,486,564 |
| Number of contigs | 99 | 113 | 95 | 121 | 79 |
| N50 (bp) | 149,987 | 118,372 | 147,758 | 139,998 | 133,202 |
| Completeness (%) | 100 | 100 | 100 | 100 | 100 |
| G + C (%) | 67.9 | 67.7 | 67.9 | 67.8 | 67.9 |
| Number of protein-coding genes | 5.883 | 5.628 | 5.751 | 5.853 | 5.755 |
| Number of rRNA operons | 2 | 2 | 2 | 2 | 4 |
| Number of tRNAs | 47 | 47 | 49 | 49 | 50 |
| Sequence coverage (x) | 70 | 91 | 68 | 96 | 510 |
| ANI % | GGD % | |
|---|---|---|
| MYC038/M. vanbaalenii PYR-1 | 98.89 | 90.30 |
| MYC040/M. vanbaalenii PYR-1 | 95.77 | 64.60 |
| MYC211/M. vanbaalenii PYR-1 | 98.61 | 87.60 |
| MYC221/M. vanbaalenii PYR-1 | 98.62 | 87.70 |
| MYC223/M. vanbaalenii PYR-1 | 98.72 | 88.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, N.M.; Romagnoli, C.L.; Santiago, C.R.d.N.; de Lacerda, J.P.A.; Leão, S.C.; Digiampietri, L.A.; Viana-Niero, C. Multi-Approach Characterization of Novel Pyrene-Degrading Mycolicibacterium austroafricanum Isolates Lacking nid Genes. Microorganisms 2023, 11, 1413. https://doi.org/10.3390/microorganisms11061413
Silva NM, Romagnoli CL, Santiago CRdN, de Lacerda JPA, Leão SC, Digiampietri LA, Viana-Niero C. Multi-Approach Characterization of Novel Pyrene-Degrading Mycolicibacterium austroafricanum Isolates Lacking nid Genes. Microorganisms. 2023; 11(6):1413. https://doi.org/10.3390/microorganisms11061413
Chicago/Turabian StyleSilva, Natalia Maria, Camila Lopes Romagnoli, Caio Rafael do Nascimento Santiago, João Paulo Amorim de Lacerda, Sylvia Cardoso Leão, Luciano Antonio Digiampietri, and Cristina Viana-Niero. 2023. "Multi-Approach Characterization of Novel Pyrene-Degrading Mycolicibacterium austroafricanum Isolates Lacking nid Genes" Microorganisms 11, no. 6: 1413. https://doi.org/10.3390/microorganisms11061413
APA StyleSilva, N. M., Romagnoli, C. L., Santiago, C. R. d. N., de Lacerda, J. P. A., Leão, S. C., Digiampietri, L. A., & Viana-Niero, C. (2023). Multi-Approach Characterization of Novel Pyrene-Degrading Mycolicibacterium austroafricanum Isolates Lacking nid Genes. Microorganisms, 11(6), 1413. https://doi.org/10.3390/microorganisms11061413







