MALDI-TOF MS Approaches for the Identification of the Susceptibility of Extended-Spectrum β-Lactamases in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Hydrolysis Assay
2.3. MALDI-TOF MS Analysis
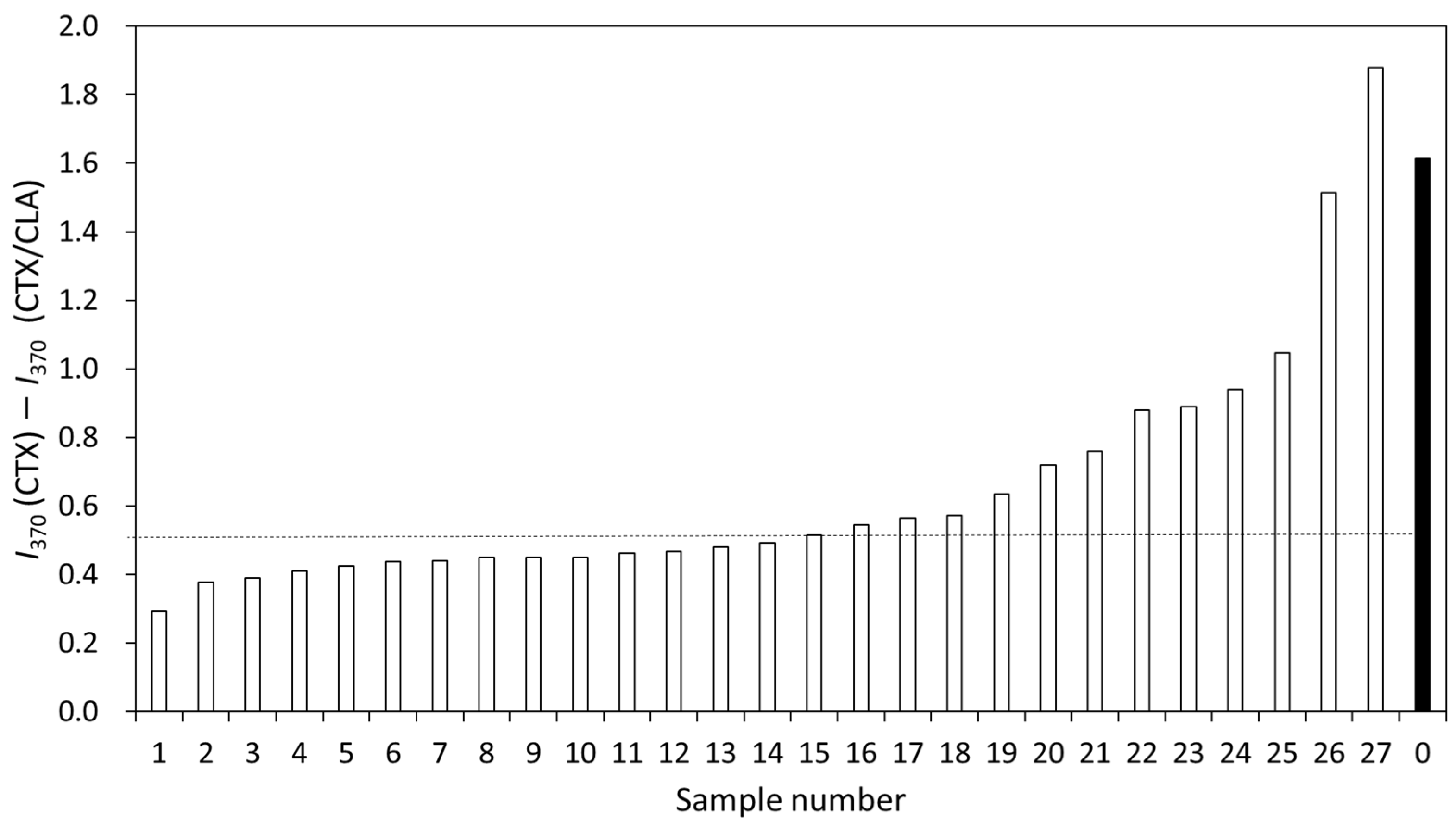
3. Results
3.1. Strain Characterization
3.2. Detection of CTX and Hydrolyzed CTX by VITEK® MS plus Using Standard Strains
3.3. Detection of Clinically Isolated Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shaikh, S.; Fatima, J.; Shakil, S.; Mohd, S.; Rizvi, D.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2014, 22, 90–101. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum b-lactamases (ESBLs) in the developed world. J. Travel. Med. 2017, 24, 44–51. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef]
- Yamasaki, K.; Komatsu, M.; Yamashita, T.; Shimakawa, K.; Ura, T.; Nishio, H.; Satho, K.; Washidu, R.; Kinoshita, S.; Aihara, M. Production of CTX-M-3 extended-spectrum β-lactamase and IMP-1 metallo β-lactamase by five gram-negative bacilli: Survey of clinical isolates from seven laboratories collected in 1998 and 2000, in Kinki region of Japan. J. Antimicrob. Chemother. 2003, 51, 631–638. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Moritake, J.; Suzuki, K.; Kira, S.; Koide, H.; Kiyota, H.; Hori, S. Prevalence and drug-susceptibilities of extended-spectrum β-lactamase producing Escherichia coli strains isolated from urine. Jpn. J. Chemother. 2014, 62, 198–203. [Google Scholar]
- Hrabák, J.; Chudackova, E.; Walkova, R. Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDITOF) Mass Spectrometry for Detection of Antibiotic Resistance Mechanisms: From Research to Routine Diagnosis. Clin. Microbiol. Rev. 2013, 26, 103–114. [Google Scholar] [CrossRef]
- Kostrzewa, M.; Sparbier, K.; Maier, T.; Schubert, S. MALDI-TOF MS: An upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteom. Clin. Appl. 2013, 7, 767–778. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Susceptibility testing of bacteria using Maldi-Tof Mass spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef]
- Belén, R.S.; Emilia, C.; Coste, A.T.; Gilbert, G. Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Euro Surveill. 2019, 24, 1800193. [Google Scholar] [CrossRef]
- Oviaño, M.; Bou, G. Matrixassisted laser desorption ionization–time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin. Microbiol. Rev. 2019, 32, e00037-18. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of antibiotic-resistance by MALDI-TOF Mass spectrometry: An expanding area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef] [PubMed]
- Oviaño, M.; Belén, R.S. MALDI-TOF mass spectrometry in the 21st century clinical microbiology laboratory. Enferm. Infect. Microbiol. Clin. 2021, 39, 192–200. [Google Scholar] [CrossRef]
- Ikegaya, K.; Matsumura, Y.; Iwasawa, A.; Kimura, S.; Tsuchiya, K. Examination of ESBLs detection method by mass spectrometry. Nikka Med. Assoc. Mag. 2018, 67, 29–37. [Google Scholar]
- Kaji, D.; Matsumura, Y.; Iwasawa, A.; Kimura, S.; Iwama, A. Rapid detection of bacteria that produce extended-spectrum β-lactamase by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Glob. Antimicrob. Resist. 2021, 27, 309–314. [Google Scholar] [CrossRef]
- Naas, T.; Oxacelay, C.; Nordmann, P. Identification of CTX-M-Type Extended-Spectrum-β-Lactamase Genes Using Real-Time PCR and Pyrosequencing. Antimicrob. Agents Chemother. 2007, 51, 223–230. [Google Scholar] [CrossRef]
- KEGG. DRUG: Cefotaxime. Available online: https://www.genome.jp/entry/D07647 (accessed on 24 April 2023).
- KEGG. DRUG: Cefotaxime Sodium. Available online: https://www.kegg.jp/entry/D00919 (accessed on 24 April 2023).
- Sparbier, K.; Schubert, S.; Weller, U.; Boogen, C.; Kostrzewa, M. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 2012, 50, 927–937. [Google Scholar] [CrossRef]
- Jung, J.S.; Popp, C.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J. Clin. Microbiol. 2014, 52, 924–930. [Google Scholar] [CrossRef]
- Oviaño, M.; Fernandez, B.; Fernandez, A.; Barba, M.J.; Mourino, C.; Bou, G. Rapid detection of enterobacteriaceae producing extended spectrum beta-lactamases directly from positive blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Clin. Microbiol. Infect. 2014, 20, 1146–1157. [Google Scholar] [CrossRef]
- Foschi, C.; Compri, M.; Smirnova, V.; Denicolò, A.; Nardini, P.; Tamburini, M.V.; Lombardo, D.; Landini, M.P.; Ambretti, S. Ease-of-use protocol for the rapid detection of third-generation cephalosporin resistance in Enterobacteriaceae isolated from blood cultures using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Hosp. Infect. 2016, 93, 206–210. [Google Scholar] [CrossRef]
- De Carolis, E.; Paoletti, S.; Nagel, D.; Vella, A.; Mello, E.; Palucci, I.; De Angelis, G.; D’Inzeo, T.; Sanguinetti, M.; Posteraro, B.; et al. A rapid diagnostic workflow for cefotaxime resistant Escherichia coli and Klebsiella pneumoniae detection from blood cultures by MALDI-TOF mass spectrometry. PLoS ONE 2017, 12, e0185935. [Google Scholar] [CrossRef] [PubMed]
- Dixon, B.; Ahmed, W.M.; Felton, T.; Fowler, S.J. Molecular phenotyping approaches for the detection and monitoring of carbapenem-resistant Enterobacteriaceae by mass spectrometry. J. Mass Spectrom. Adv. Clin. Lab. 2022, 26, 9–19. [Google Scholar] [CrossRef] [PubMed]
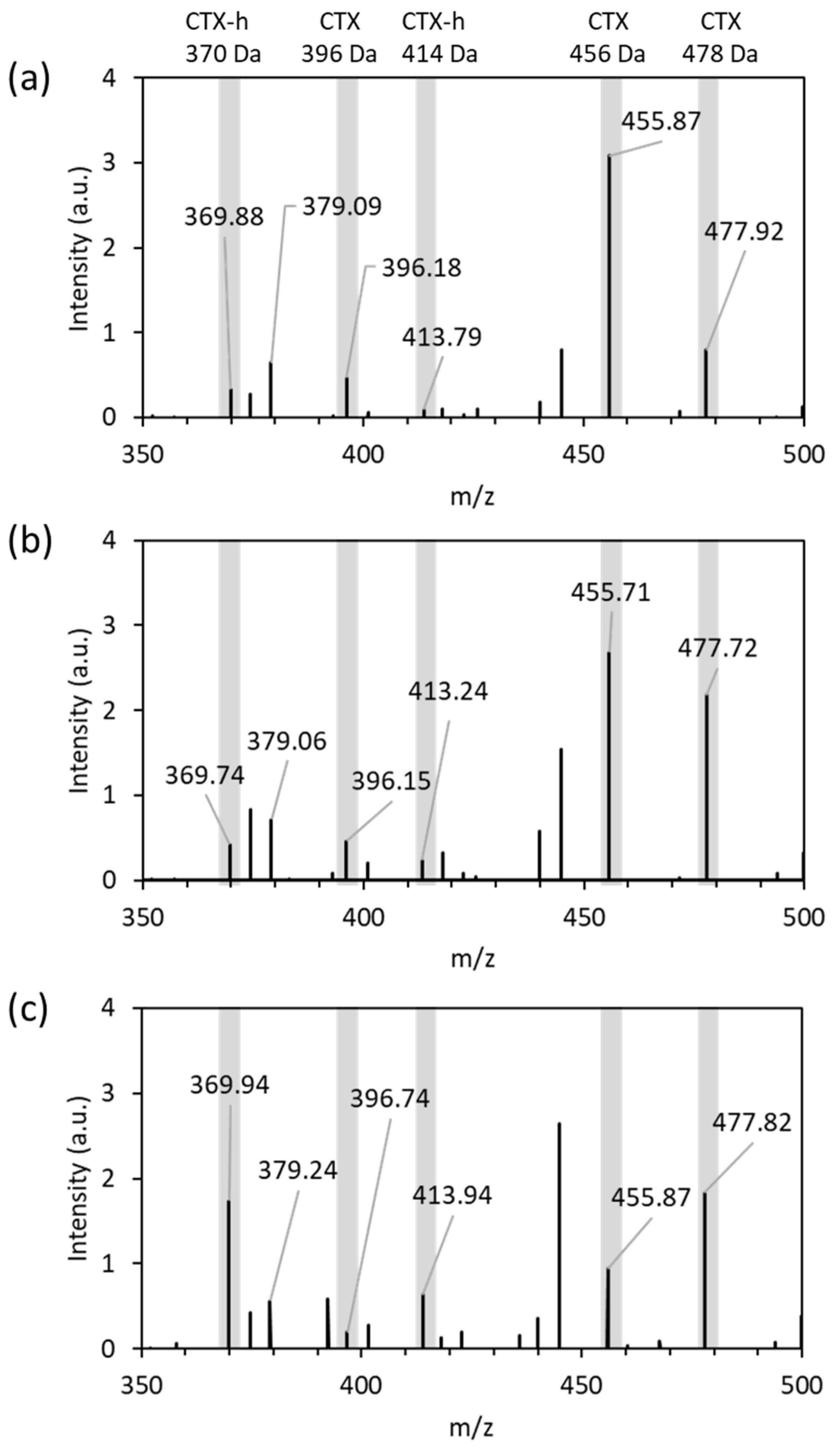
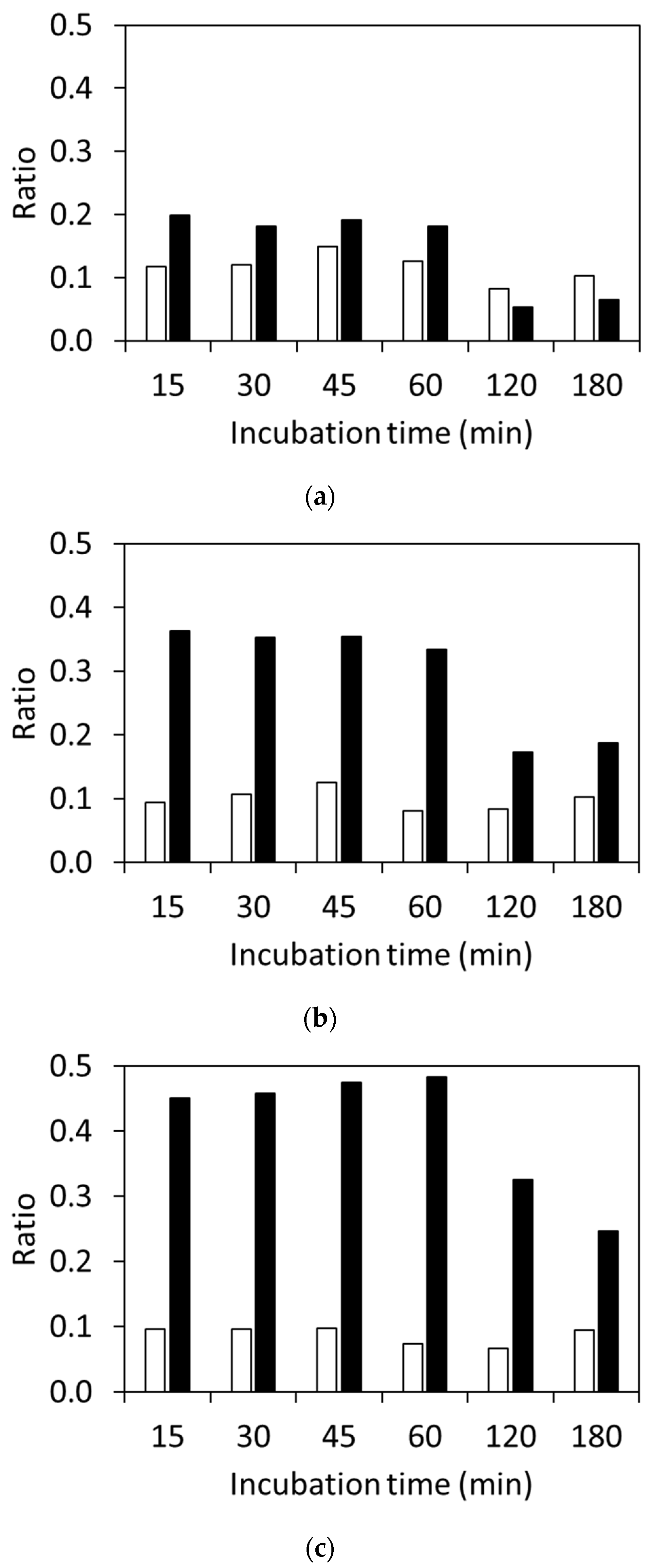
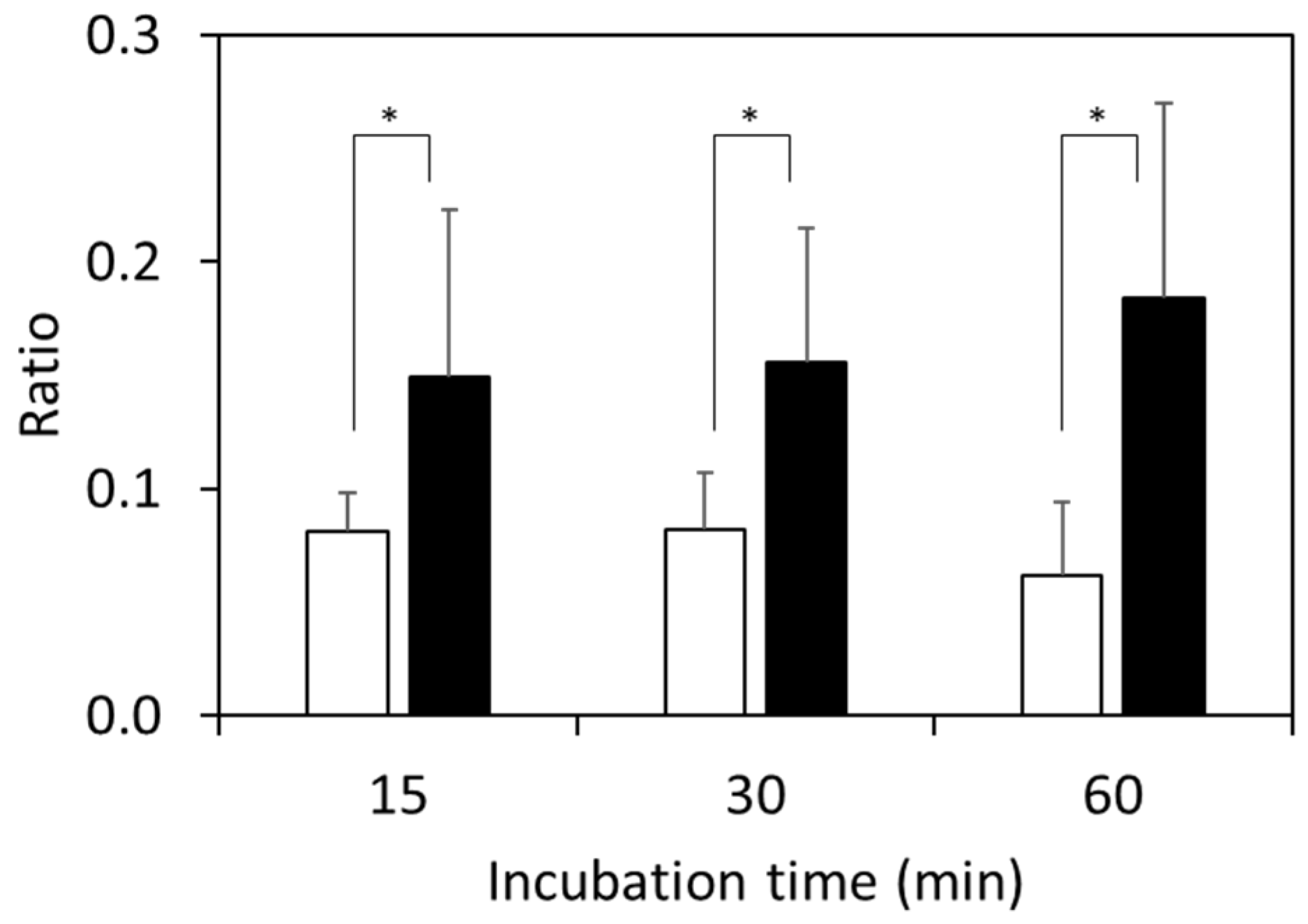
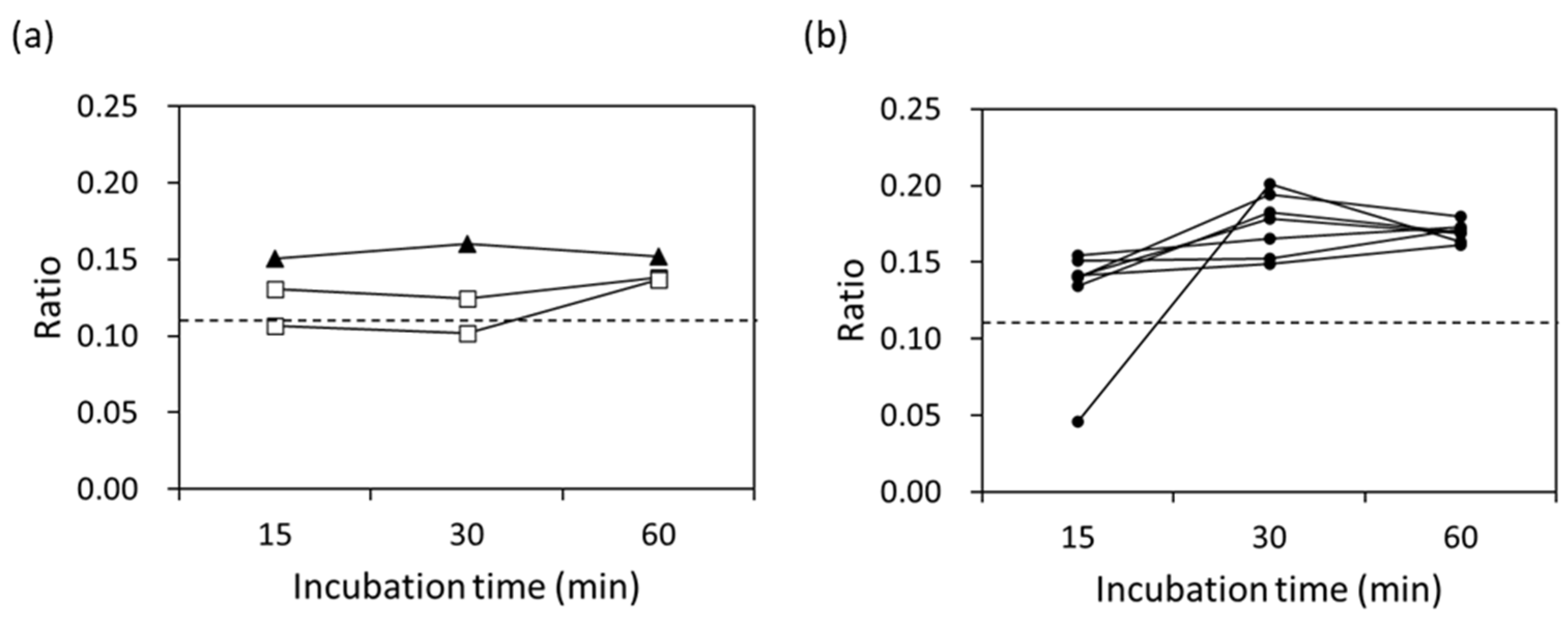
| Primer Name | Sequence (5′ to 3′) |
|---|---|
| CTX-MA1 bio (forward, biotinylated) | biotin-[C/G]C[G/A/C]ATGTGCAG[C/T]ACCAGTAA |
| CTX-MA5 (reverse) | TG[A/G]G[A/C]AATCA[A/G][C/T]TT[A/G]TTCAT |
| Bacterial Isolates | Number of Strains |
|---|---|
| Isolated E. coli | 414 |
| The strain used for the test | 96 |
| blaCTX-M detection using real-time PCR assay | 40 |
| MIC value for CTX | |
| <1 μg/mL | 1 |
| 2 μg/mL | 1 |
| 4 μg/mL | 2 |
| 8 μg/mL | 7 |
| 32 μg/mL | 2 |
| ≥64 μg/mL | 27 |
| Susceptibility to specified antibiotics 1 | 57 |
| MIC (μg/mL) | Number of Strains | I370(CTX) − I370(CTX/CLA) |
|---|---|---|
| 2 | 1 | 0.53 |
| 4 | 2 | 0.43 |
| 8 | 7 | 0.42 ± 0.12 |
| 32 | 2 | 0.50 |
| 64 | 25 1 | 0.56 ± 0.19 |
| 27 | 0.70 ± 0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsumura, Y.; Ikegaya, K. MALDI-TOF MS Approaches for the Identification of the Susceptibility of Extended-Spectrum β-Lactamases in Escherichia coli. Microorganisms 2023, 11, 1250. https://doi.org/10.3390/microorganisms11051250
Matsumura Y, Ikegaya K. MALDI-TOF MS Approaches for the Identification of the Susceptibility of Extended-Spectrum β-Lactamases in Escherichia coli. Microorganisms. 2023; 11(5):1250. https://doi.org/10.3390/microorganisms11051250
Chicago/Turabian StyleMatsumura, Yuriko, and Kazuko Ikegaya. 2023. "MALDI-TOF MS Approaches for the Identification of the Susceptibility of Extended-Spectrum β-Lactamases in Escherichia coli" Microorganisms 11, no. 5: 1250. https://doi.org/10.3390/microorganisms11051250
APA StyleMatsumura, Y., & Ikegaya, K. (2023). MALDI-TOF MS Approaches for the Identification of the Susceptibility of Extended-Spectrum β-Lactamases in Escherichia coli. Microorganisms, 11(5), 1250. https://doi.org/10.3390/microorganisms11051250






