A Fatal Case of Pseudomonas aeruginosa Community-Acquired Pneumonia in an Immunocompetent Patient: Clinical and Molecular Characterization and Literature Review
Abstract
1. Introduction
2. Materials and Methods
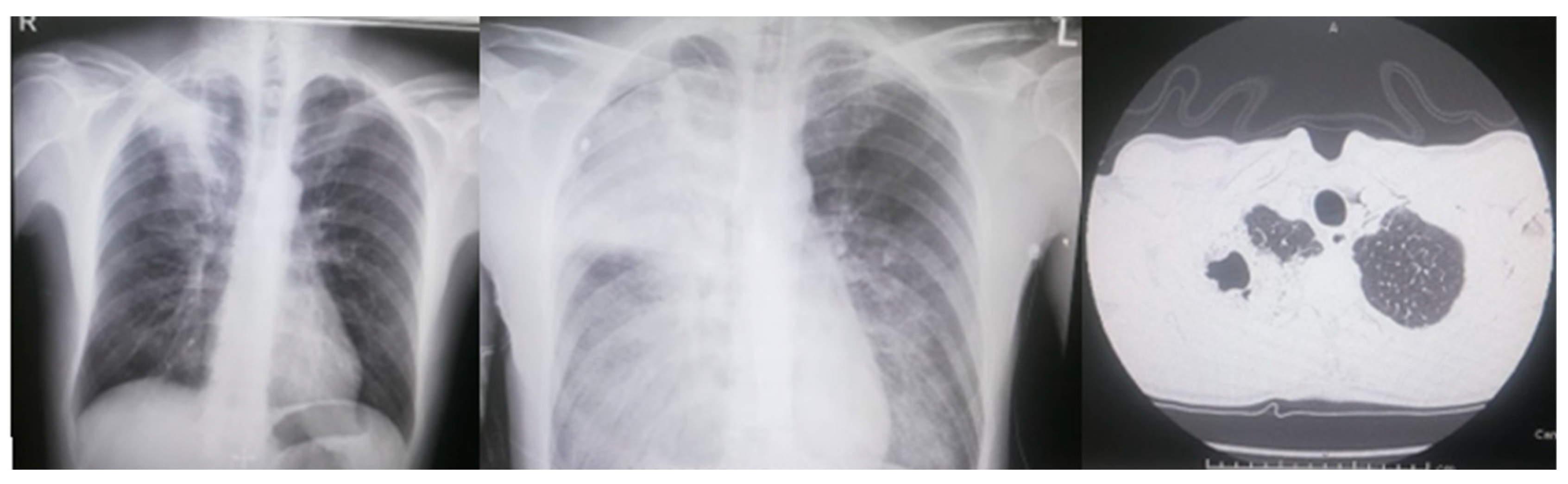
3. Case Report Description
4. Discussion and Literature Review
4.1. Previous PA-CAP Cases Description Review
4.2. Risk Factors and Score for PA-CAP Identification
4.3. Clinical Features of PA-CAP
4.4. Microbiology and Whole Genome Sequencing
4.5. Antibiotic Therapy
4.6. Should the Guidelines Be Changed?
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Woodhead, M.; Blasi, F.; Ewig, S.; Garau, J.; Huchon, G.; Ieven, M.; Ortqvist, A.; Schaberg, T.; Torres, A.; van der Heijden, G.; et al. Joint Taskforce of the European Respiratory Society and European Society for Clinical Microbiology and Infectious Diseases. Guidelines for the management of adult lower respiratory tract infections—Full version. Clin. Microbiol. Infect. 2011, 17, E1–E59. [Google Scholar] [CrossRef]
- Metlay, J.P.; Waterer, G.W.; Long, A.C.; Anzueto, A.; Brozek, J.; Crothers, K.; Cooley, L.A.; Dean, N.C.; Fine, M.J.; Flanders, S.A.; et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care Med. 2019, 200, e45–e67. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Niederman, M.S. Too Much or Too Little Empiric Treatment for Pseudomonas aeruginosa in Community-acquired Pneumonia? Ann. Am. Thorac. Soc. 2021, 18, 1456–1458. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, C.; Langille, M.G.; Fothergill, J.L.; Kukavica-Ibrulj, I.; Paradis-Bleau, C.; Sanschagrin, F.; Thomson, N.R.; Winsor, G.L.; Quail, M.A.; Lennard, N.; et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009, 19, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Riviere, P.; Patin, D.; Delaporte, E.; Mahfoudi, H.; Lecailtel, S.; Poher, F.; Villette, P.; Duclaux, J.; Jouault, P.; Brunin, G. Septic shock secondary to an acute necrotizing community-acquired pneumonia with bacteremia due to Pseudomonas aeruginosa. IDCases 2019, 17, e00563. [Google Scholar] [CrossRef]
- Ramirez, J.A.; Musher, D.M.; Evans, S.E.; Dela Cruz, C.; Crothers, K.A.; Hage, C.A.; Aliberti, S.; Anzueto, A.; Arancibia, F.; Arnold, F.; et al. Treatment of Community-Acquired Pneumonia in Immunocompromised Adults: A Consensus Statement Regarding Initial Strategies. Chest 2020, 158, 1896–1911. [Google Scholar] [CrossRef]
- Restrepo, M.I.; Babu, B.L.; Reyes, L.F.; Chalmers, J.D.; Soni, N.J.; Sibila, O.; Faverio, P.; Cilloniz, C.; Rodriguez-Cintron, W.; Aliberti, S. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: A multinational point prevalence study of hospitalised patients. Eur. Respir. J. 2018, 52, 1701190. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 December 2022).
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://pubmlst.org/bigsdb?db=pubmlst_paeruginosa_seqdef (accessed on 6 December 2022).
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Available online: https://github.com/tseemann/abricate (accessed on 6 December 2022).
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E.; et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.H.; Tetu, S.G.; Larouche, A.; Elbourne, L.; Tremblay, S.; Ren, Q.; Dodson, R.; Harkins, D.; Shay, R.; Watkins, K.; et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 2010, 5, e8842. [Google Scholar] [CrossRef] [PubMed]
- Pelegrin, A.C.; Palmieri, M.; Mirande, C.; Oliver, A.; Moons, P.; Goossens, H.; van Belkum, A. Pseudomonas aeruginosa: A clinical and genomics update. FEMS Microbiol. Rev. 2021, 45, fuab026. [Google Scholar] [CrossRef] [PubMed]
- Huber, P.; Basso, P.; Reboud, E.; Attree, I. Pseudomonas aeruginosa renews its virulence factors. Environ. Microbiol. Rep. 2016, 8, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Wiehlmann, L.; Cramer, N.; Tummler, B. Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ. Microbiol. Rep. 2015, 7, 955–960. [Google Scholar] [CrossRef]
- Hilker, R.; Munder, A.; Klockgether, J.; Losada, P.M.; Chouvarine, P.; Cramer, N.; Davenport, C.F.; Dethlefsen, S.; Fischer, S.; Peng, H.; et al. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ. Microbiol. 2015, 17, 29–46. [Google Scholar] [CrossRef]
- Grace, A.; Sahu, R.; Owen, D.R.; Dennis, V.A. Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review. Front. Microbiol. 2022, 13, 1023523. [Google Scholar] [CrossRef]
- Mikkelsen, H.; McMullan, R.; Filloux, A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS ONE 2011, 6, e29113. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Rother, C.; Salih, W.; Ewig, S. Healthcare-associated pneumonia does not accurately identify potentially resistant pathogens: A systematic review and meta-analysis. Clin. Infect. Dis. 2014, 58, 330–339. [Google Scholar] [CrossRef]
- Hatchette, T.F.; Gupta, R.; Marrie, T.J. Pseudomonas aeruginosa community-acquired pneumonia in previously healthy adults: Case report and review of the literature. Clin. Infect. Dis. 2000, 31, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Maharaj, S.; Isache, C.; Seegobin, K.; Chang, S.; Nelson, G. Necrotizing Pseudomonas aeruginosa Community-Acquired Pneumonia: A Case Report and Review of the Literature. Case Rep. Infect. Dis. 2017, 2017, 1717492. [Google Scholar] [CrossRef]
- Huhulescu, S.; Simon, M.; Lubnow, M.; Kaase, M.; Wewalka, G.; Pietzka, A.T.; Stöger, A.; Ruppitsch, W.; Allerberger, F. Fatal Pseudomonas aeruginosa pneumonia in a previously healthy woman was most likely associated with a contaminated hot tub. Infection 2011, 39, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Yamaoka, J.; Chikayama, S.; Ohishi, T.; Nakajima, T.; Nakashima, T. Successful treatment of a previously healthy woman with Pseudomonas aeruginosa community-acquired pneumonia with plasmapheresis. Intern. Med. 2012, 51, 2809–2812. [Google Scholar] [CrossRef] [PubMed]
- Kunimasa, K.; Ishida, T.; Kimura, S.; Tanaka, M.; Kouyama, Y.; Yamashita, S.; Morita, M.; Tachibana, H.; Tokioka, F.; Ito, A.; et al. Successful treatment of fulminant community-acquired Pseudomonas aeruginosa necrotizing pneumonia in a previously healthy young man. Intern. Med. 2012, 51, 2473–2478. [Google Scholar] [CrossRef]
- Gharabaghi, M.A.; Abdollahi, S.M.; Safavi, E.; Abtahi, S.H. Community acquired Pseudomonas pneumonia in an immune competent host. BMJ Case Rep. 2012, 2012, bcr0120125673. [Google Scholar] [CrossRef]
- Fujii, A.; Seki, M.; Higashiguchi, M.; Tachibana, I.; Kumanogoh, A.; Tomono, K. Community-acquired, hospital-acquired, and healthcare-associated pneumonia caused by Pseudomonas aeruginosa. Respir. Med. Case Rep. 2014, 12, 30–33. [Google Scholar] [CrossRef]
- Takajo, D.; Iwaya, K.; Katsurada, Y.; Miyai, K.; Takasu, A.; Matsubara, O.; Sakamoto, T.; Tamai, S.; Tsuda, H. Community-acquired lobar pneumonia caused by Pseudomonas aeruginosa infection in Japan: A case report with histological and immunohistochemical examination. Pathol. Int. 2014, 64, 224–230. [Google Scholar] [CrossRef]
- de Campos, F.P.; Felipe-Silva, A.; de Melo, A.C.; Passadore, L.F.; Guida, S.M.; Balabakis, A.J.; dos Santos Martines, J.A. Community-acquired Pseudomonas aeruginosa-pneumonia in a previously healthy man occupationally exposed to metalworking fluids. Autops. Case Rep. 2014, 4, 31–37. [Google Scholar] [CrossRef]
- Woods, E.; Cohen, G.; Bressman, E.; Lin, D.; Zeitouni, N.E.; Beckford, C.; Hamula, C.; van Bakel, H.; Sullivan, M.; Altman, D.R.; et al. Community-acquired cavitary Pseudomonas pneumonia linked to use of a home humidifier. Case Rep. Infect. Dis. 2017, 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Su, I.C.; Lee, K.L.; Liu, H.Y.; Chuang, H.C.; Chen, L.Y.; Lee, Y.J. Severe community-acquired pneumonia due to Pseudomonas aeruginosa coinfection in an influenza A(H1N1)pdm09 patient. J. Microbiol. Immunol. Infect. 2019, 52, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hou, Y.; Wang, R. A case report of community-acquired Pseudomonas aeruginosa pneumonia complicated with MODS in a previously healthy patient and related literature review. BMC Infect. Dis. 2019, 19, 130. [Google Scholar]
- Dong, C.; Shen, F.; Dong, H.; Dong, L.; Fu, Y.; Xu, Y.; Ning, J. Community-acquired Pseudomonas aeruginosa pneumonia manifested by bloody pleural effusion in a previously healthy infant: A case report. J. Clin. Lab. Anal. 2022, 36, e24466. [Google Scholar] [CrossRef]
- Wang, J.; Yun, L.; Zhao, H.; Li, X. Combination Therapy of Polymyxin B and Amikacin for Community-Acquired Pseudomonas aeruginosa Pneumonia with MODS in a Previously Healthy Patient: A Case Report. Infect. Drug Resist. 2021, 14, 2895–2900. [Google Scholar] [CrossRef]
- Arancibia, F.; Bauer, T.T.; Ewig, S.; Mensa, J.; Gonzalez, J.; Niederman, M.S.; Torres, A. Community-acquired pneumonia due to gram-negative bacteria and Pseudomonas aeruginosa: Incidence, risk, and prognosis. Arch. Intern. Med. 2002, 162, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.H.; Shorr, A.; Tabak, Y.P.; Gupta, V.; Liu, L.Z.; Johannes, R.S. Epidemiology and outcomes of health-care-associated pneumonia: Results from a large US database of culture-positive pneumonia. Chest 2005, 128, 3854–3862. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Gabarrús, A.; Ferrer, M.; de la Bellacasa, J.P.; Rinaudo, M.; Mensa, J.; Niederman, M.S.; Torres, A. Community-acquired pneumonia due to multidrug- and non-multidrug-resistant Pseudomonas aeruginosa. Chest 2016, 150, 415–425. [Google Scholar] [CrossRef]
- Sibila, O.; Laserna, E.; Maselli, D.J.; Fernandez, J.F.; Mortensen, E.M.; Anzueto, A.; Waterer, G.; Restrepo, M.I. Risk factors and antibiotic therapy in P. aeruginosa community-acquired pneumonia. Respirology 2015, 20, 660–666. [Google Scholar] [CrossRef]
- Crnich, C.J.; Gordon, B.; Andes, D. Hot tub-associated necrotizing pneumonia due to Pseudomonas aeruginosa. Clin. Infect. Dis. 2003, 36, e55–e57. [Google Scholar] [CrossRef]
- Fujitani, S.; Sun, H.Y.; Yu, V.L.; Weingarten, J.A. Pneumonia due to Pseudomonas aeruginosa: Part I: Epidemiology, clinical diagnosis, and source. Chest 2011, 139, 909–919. [Google Scholar] [CrossRef]
- Brito, V.; Niederman, M.S. Healthcare-associated pneumonia is a heterogeneous disease, and all patients do not need the same broad-spectrum antibiotic therapy as complex nosocomial pneumonia. Curr. Opin. Infect. Dis. 2009, 22, 316–325. [Google Scholar] [CrossRef]
- Govan, J.; Reiss-Levy, E.; Bader, L.; Schonell, M. Pseudomonas pneumonia with bacteraemia. Med. J. Aust. 1977, 1, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Hoogwerf, B.J.; Khan, M.Y. Community-acquired bacteremic Pseudomonas pneumonia in a health adult. Am. Rev. Respir. Dis. 1981, 123, 132–134. [Google Scholar] [PubMed]
- Jayatilleke, K.; Bandara, P.; Satharasinghe, R. A case of community acquired pneumonia caused by Pseudomonas aeruginosa. Sri. Lanka. J. Infect. Dis. 2012, 2, 53–54. [Google Scholar] [CrossRef]
- Zell, L.; Mack, U.; Sommerfeld, A.; Buchter, A.; Sybrecht, G.W. Abszedierende Pneumonie durch Pseudomonas aeruginosa als Berufskrankheit bei einem Bohrwerksdreher. Pneumologie 1999, 53, 620–625. [Google Scholar] [CrossRef]
- Lee, B.; Robinson, K.M.; McHugh, K.J.; Scheller, E.V.; Mandalapu, S.; Chen, C.; Di, Y.P.; Clay, M.E.; Enelow, R.I.; Dubin, P.J.; et al. Influenza-induced type I interferon enhances susceptibility to gram-negative and gram-positive bacterial pneumonia in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 309, L158–L167. [Google Scholar] [CrossRef]
- Prina, E.; Ranzani, O.T.; Polverino, E.; Cillóniz, C.; Ferrer, M.; Fernandez, L.; Puig de la Bellacasa, J.; Menéndez, R.; Mensa, J.; Torres, A. Risk factors associated with potentially antibiotic-resistant pathogens in community-acquired pneumonia. Ann. Am. Thorac. Soc 2015, 12, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Von Baum, H.; Welte, T.; Marre, R.; Suttorp, N.; Ewig, S.; CAPNETZ Study Group. Community-acquired pneumonia through Enterobacteriaceae and Pseudomonas aeruginosa: Diagnosis, incidence and predictors. Eur. Respir. J. 2010, 35, 598–605. [Google Scholar] [CrossRef]
- Taniguchi, T.; Tsuha, S.; Shiiki, S.; Narita, M. Gram-stain-based antimicrobial selection reduces cost and overuse compared with Japanese guidelines. BMC Infect. Dis. 2015, 15, 458. [Google Scholar] [CrossRef]
- Cillóniz, C.; Dominedò, C.; Nicolini, A.; Torres, A. PES Pathogens in Severe Community-Acquired Pneumonia. Microorganisms 2019, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, K.; Lee, J.; Kim, J.; Jo, Y.H.; Lee, J.H.; Hwang, J.E. Impact of bacteremia prediction rule in CAP: Before and after study. Am. J. Emerg. Med. 2018, 36, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.M.; Wechsler, R.; Salazar, A.M.; Spirn, P.W. S pectrum of CT findings in nosocomial Pseudomonas aeruginosa pneumonia. J. Thorac. Imaging 2002, 17, 53–57. [Google Scholar] [CrossRef]
- Fetzer, A.E.; Werner, A.S.; Hagstrom, J.W. Pathologic features of pseudomonal pneumonia. Am. Rev. Respir. Dis. 1967, 96, 1121–1130. [Google Scholar]
- Opperman, C.J.; Moodley, C.; Lennard, K.; Smith, M.; Ncayiyana, J.; Vulindlu, M.; Gafoor, M.; Govender, N.; Ismail, H.; Bamford, C.; et al. A citywide, clonal outbreak of Pseudomonas aeruginosa. Int. J. Infect. Dis. 2022, 117, 74–86. [Google Scholar] [CrossRef]
- Reboud, E.; Basso, P.; Maillard, A.P.; Huber, P.; Attrée, I. Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers. Toxins 2017, 9, 364. [Google Scholar] [CrossRef]
- Prina, E.; Ranzani, O.T.; Torres, A. Community-acquired pneumonia. Lancet 2015, 386, 1097–1108. [Google Scholar] [CrossRef]
- Ferreira-Coimbra, J.; Tejada, S.; Campogiani, L.; Rello, J. Levels of evidence supporting European and American community-acquired pneumonia guidelines. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Pneumonia (Community-Acquired): Antimicrobial PrescribingNICE Guideline. 16 September 2019. Available online: https://www.nice.org.uk/guidance/ng138 (accessed on 6 December 2022).
| LEU /mmc | N % | PLT /mmc | CRP mg/dL | PCT µg/L | ALT U/L | CREA mg/dL | eGFR mL/min | LDH U/L | XDP ng/mL | CK U/L | TRO ng/L | LIP U/L | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0 | 4250 | 91 | 175,000 | 2.0 | 51 | 19 | 0.95 | 91 | 328 | 3608 | - | - | - |
| T1 | 1020 | 35 | 56,000 | 7.9 | 147 | 1609 | 4.54 | 13.7 | 12 052 | 2646 | 80,000 | 1070 | 425 |
| Reference Site (Year) | Exposure | Sex Age | Smoke Alcohol | Symptoms on admission | Radiology | WBC (/µL) CRP (mg/dL) PCT (ng/mL) | Positive colture for PA Other Infection | Antibiotic Therapy | Course and Outcome |
|---|---|---|---|---|---|---|---|---|---|
| Huhulescu et al [28] Austria (2010) | Hot tub water | F 49 | Yes - | Cough Pleuritic chest pain | Left lung infiltration | WBC 3 940 CRP 38.1 | Respiratory sample Blood |
|
|
| Okamoto et al [29] Japan (2011) | Water in rearing beetles cage | F 39 | Yes Yes | Cough Pleuritic chest pain Hemoptysis Dyspnea Diarrhea | Right upper consolidation Pleural effusion | WBC 21 500 CRP 6.61 | Respiratory sample Blood Pleural fluid |
|
|
| Kunimasa et al. [30] Japan (2012) | Washrooms, bathroom in care facility | M 25 | Yes - | Cough Fever Right back pain Hemoptysis | Right upper consolidation, cavitation | WBC 7 800 CRP 14 | Respiratory sample |
|
|
| Gharabaghi et al. [31] Iran (2012) | Not known exposure | M 26 | No - | Cough Fever Bone pain | Left upper consolidation, cavitation, left lower lobe mass with necrosis | WBC normal | Respiratory sample Lung biopsy Klebsiella pneumoniae |
| - |
| Fujii et al. [32] Japan (2012) | Not known exposure | M 29 | No | Fever Right back pain | Right upper consolidation, cavitation | WBC 26 400 CRP 20 | Respiratory sample Blood |
|
|
| Takajo et al. [33] Japan (2013) | Not known exposure | F 50 | - - | Cough Fever Right back pain Dyspnea Hemoptysis | Right upper lobe consolidation Pneumothorax | WBC 2 100 | Respiratory sample Blood |
|
|
| Campos et al. [34] Brazil (2014) | Metalworking fluid | M 44 | Yes - | Cough Fever Right back pain Hemoptysis | Right upper and lower consolidation Left consolidation | WBC 2 880 | Respiratory sample Blood |
|
|
| Woods et al. [35] USA (2017) | Home humidifier water | M 30 | No No | Cough Fever Right back pain Weight loss | Right upper consolidation, cavitation | WBC 11 300 | Respiratory sample |
|
|
| Su et al. [36] Taiwan (2018) | Not known exposure | F 39 | - | Cough Fever Dyspnea | Bilateral patchy infiltrates | WBC 810 | Respiratory sample Blood Influenza A(H1N1)pdm09 |
|
|
| Wang et al. [37] China (2019) | Not known exposure | M 25 | No No | Cough Dyspnea Hemoptysis Brain abscess | Multiple right upper and lower consolidations Pleural effusion | WBC 390 PCT 100 | Respiratory sample Blood |
|
|
| Dong et al. [38] China (2019) | Not known exposure | M 1 | No No | Cough Fever Dyspnea Hemoptysis Bloody pleural effusion | Right middle and lower lobe consolidation, cavitation Pleural effusion | WBC 41 780 CRP 102 PCT 4.49 | Respiratory sample Pleural fluid Influenza A |
|
|
| Wang et al. [39] China (2019) | Not known exposure | M 67 | Yes (past) - | Cough Fever Dyspnea | Right upper consolidation, cavitation | WBC 20 360 | Respiratory sample Acinetobacter baumanii |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barp, N.; Marcacci, M.; Biagioni, E.; Serio, L.; Busani, S.; Ventura, P.; Franceschini, E.; Orlando, G.; Venturelli, C.; Menozzi, I.; et al. A Fatal Case of Pseudomonas aeruginosa Community-Acquired Pneumonia in an Immunocompetent Patient: Clinical and Molecular Characterization and Literature Review. Microorganisms 2023, 11, 1112. https://doi.org/10.3390/microorganisms11051112
Barp N, Marcacci M, Biagioni E, Serio L, Busani S, Ventura P, Franceschini E, Orlando G, Venturelli C, Menozzi I, et al. A Fatal Case of Pseudomonas aeruginosa Community-Acquired Pneumonia in an Immunocompetent Patient: Clinical and Molecular Characterization and Literature Review. Microorganisms. 2023; 11(5):1112. https://doi.org/10.3390/microorganisms11051112
Chicago/Turabian StyleBarp, Nicole, Matteo Marcacci, Emanuela Biagioni, Lucia Serio, Stefano Busani, Paolo Ventura, Erica Franceschini, Gabriella Orlando, Claudia Venturelli, Ilaria Menozzi, and et al. 2023. "A Fatal Case of Pseudomonas aeruginosa Community-Acquired Pneumonia in an Immunocompetent Patient: Clinical and Molecular Characterization and Literature Review" Microorganisms 11, no. 5: 1112. https://doi.org/10.3390/microorganisms11051112
APA StyleBarp, N., Marcacci, M., Biagioni, E., Serio, L., Busani, S., Ventura, P., Franceschini, E., Orlando, G., Venturelli, C., Menozzi, I., Tambassi, M., Scaltriti, E., Pongolini, S., Sarti, M., Pietrangelo, A., Girardis, M., Mussini, C., & Meschiari, M. (2023). A Fatal Case of Pseudomonas aeruginosa Community-Acquired Pneumonia in an Immunocompetent Patient: Clinical and Molecular Characterization and Literature Review. Microorganisms, 11(5), 1112. https://doi.org/10.3390/microorganisms11051112









