Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annus) Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
Seeds Collection
2.2. Methodology
2.2.1. Oil Content Determination
Seed Preparation
Soxhlet Extraction
Fatty Acids Methyl Esters (FAMEs)
Chromatographic Identification
2.2.2. Microbial Characterization
Surface Sterilization
DNA Extraction, PCR Amplification and Sequencing
Data Processing and Statistical Evaluation
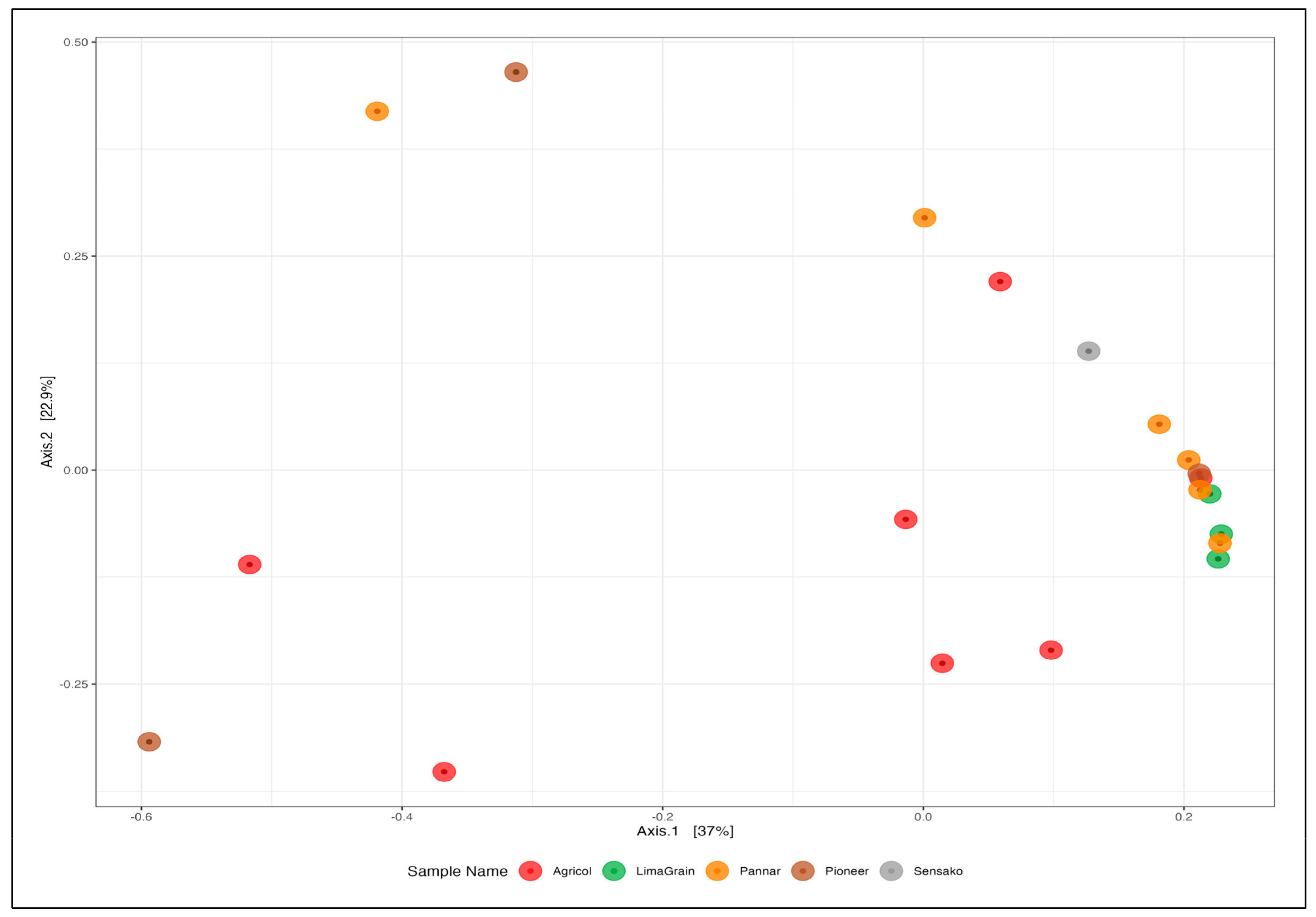
3. Results
3.1. Fats Composition
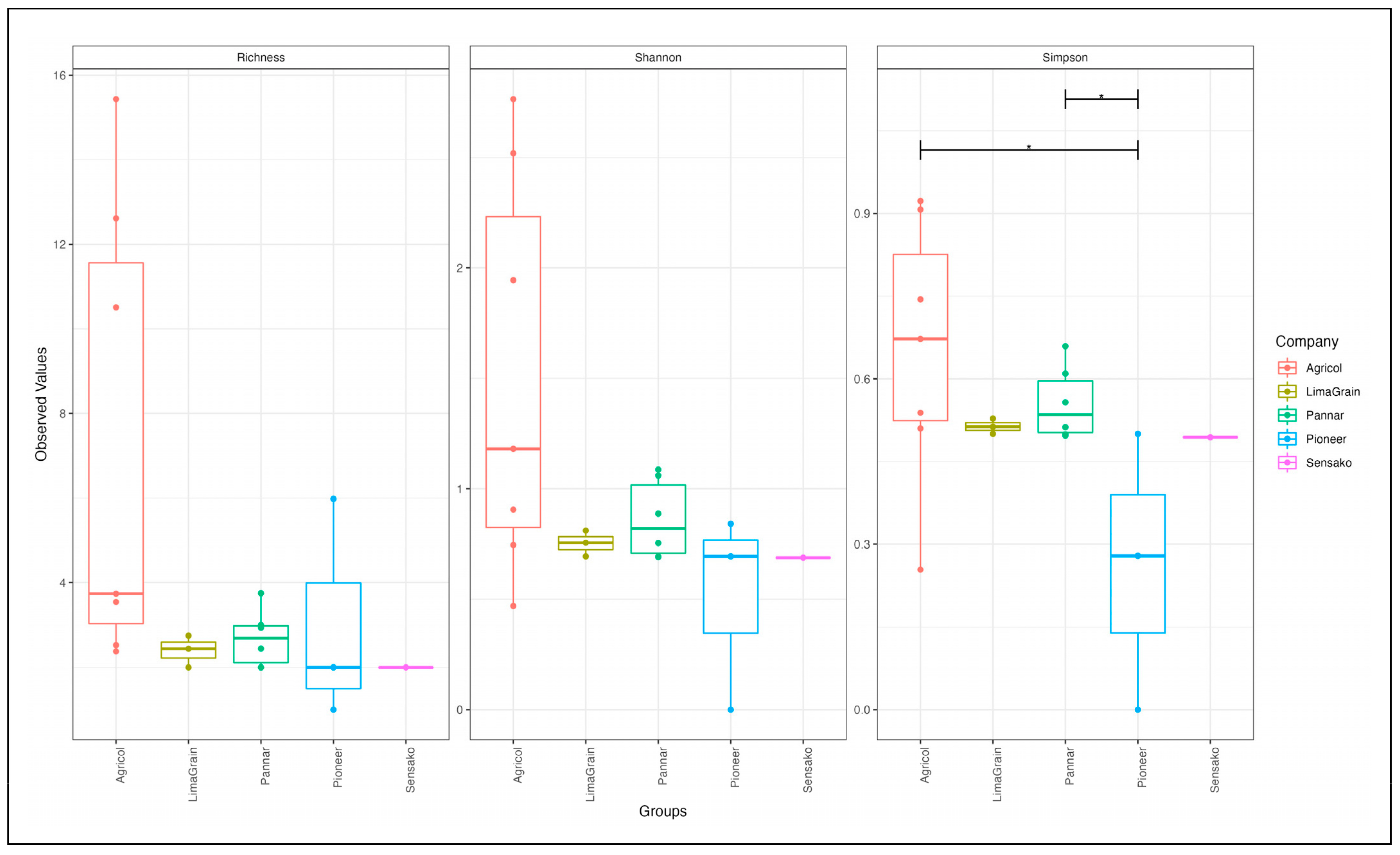
3.2. Fungal Diversity and Abundance
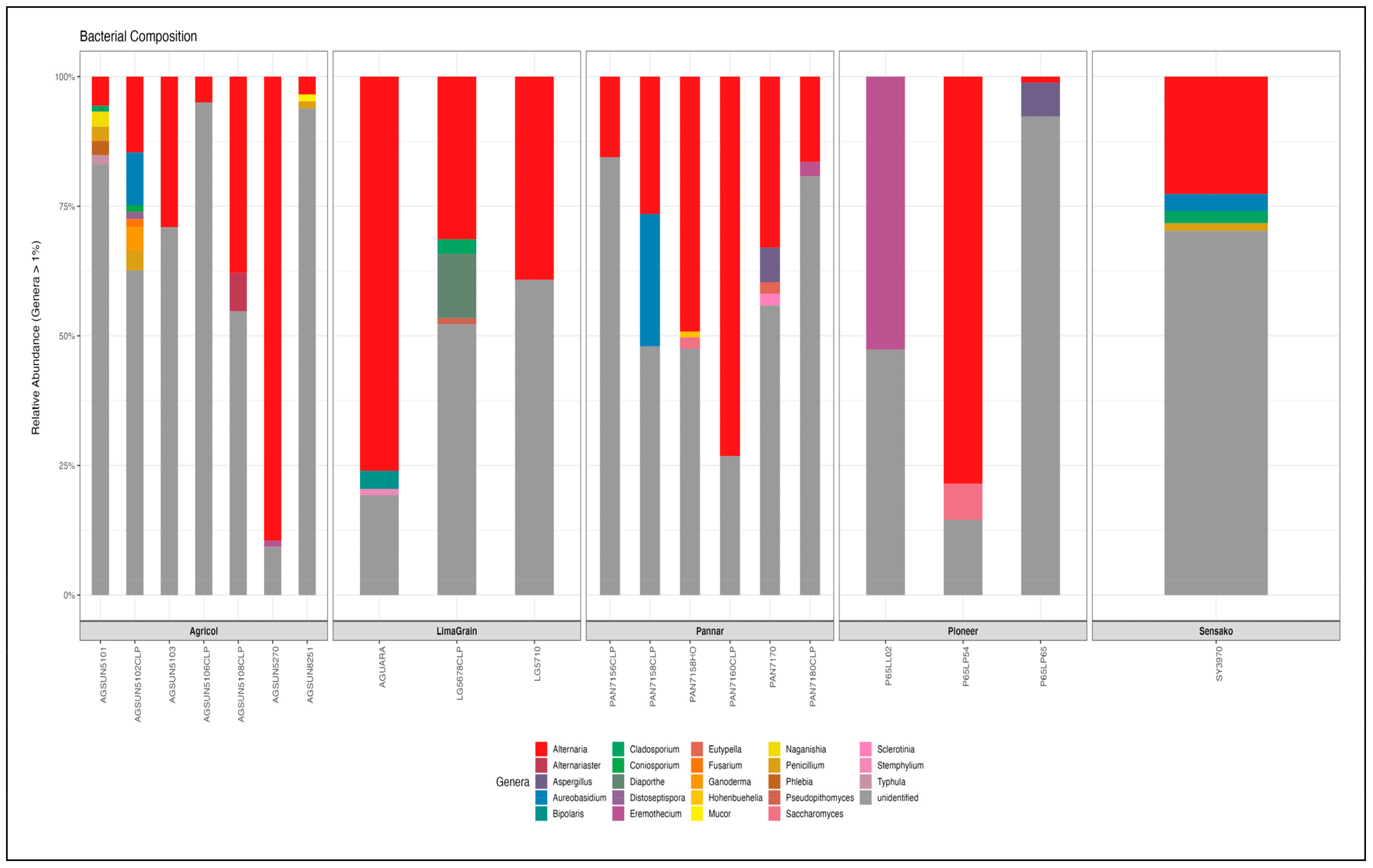
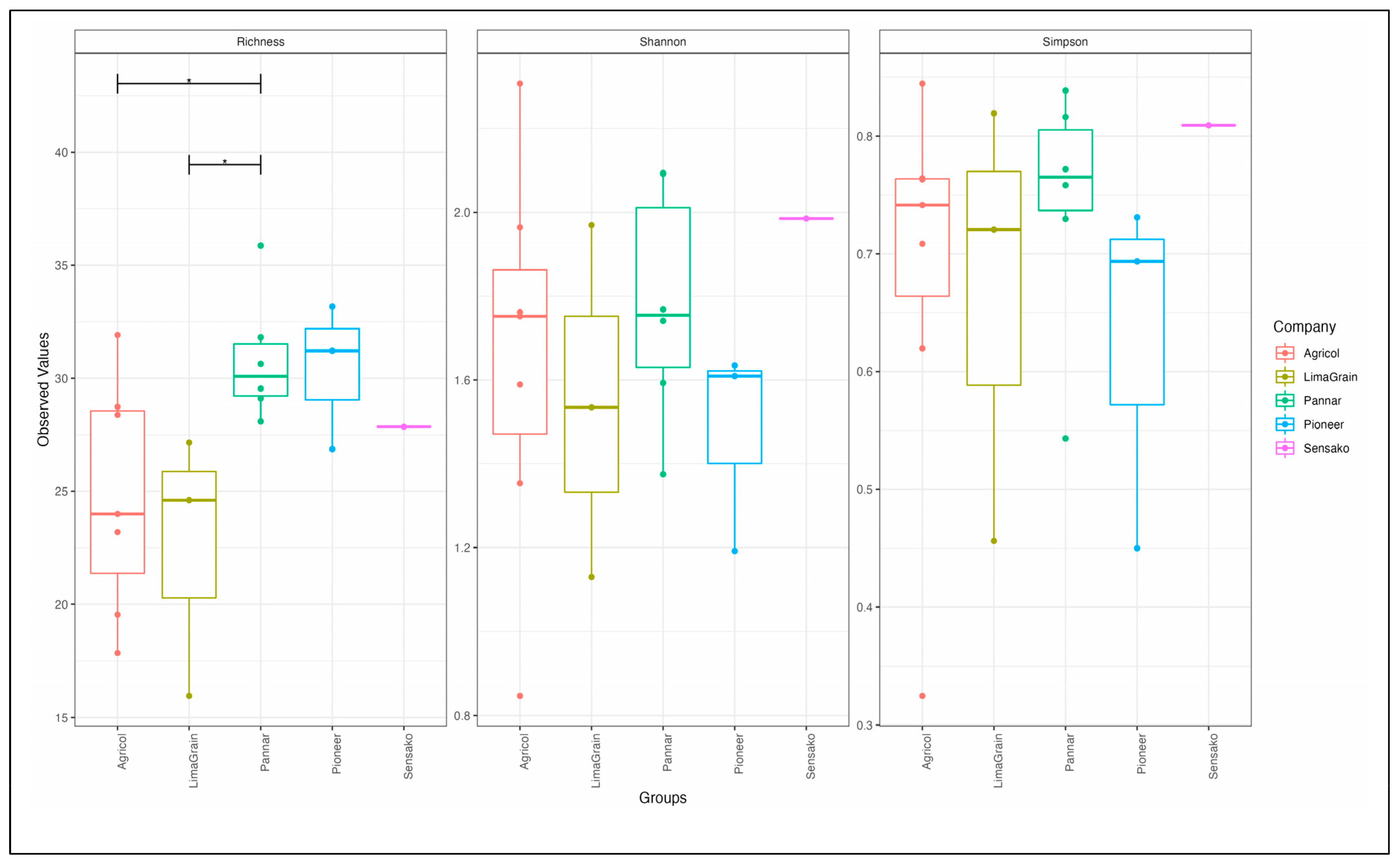
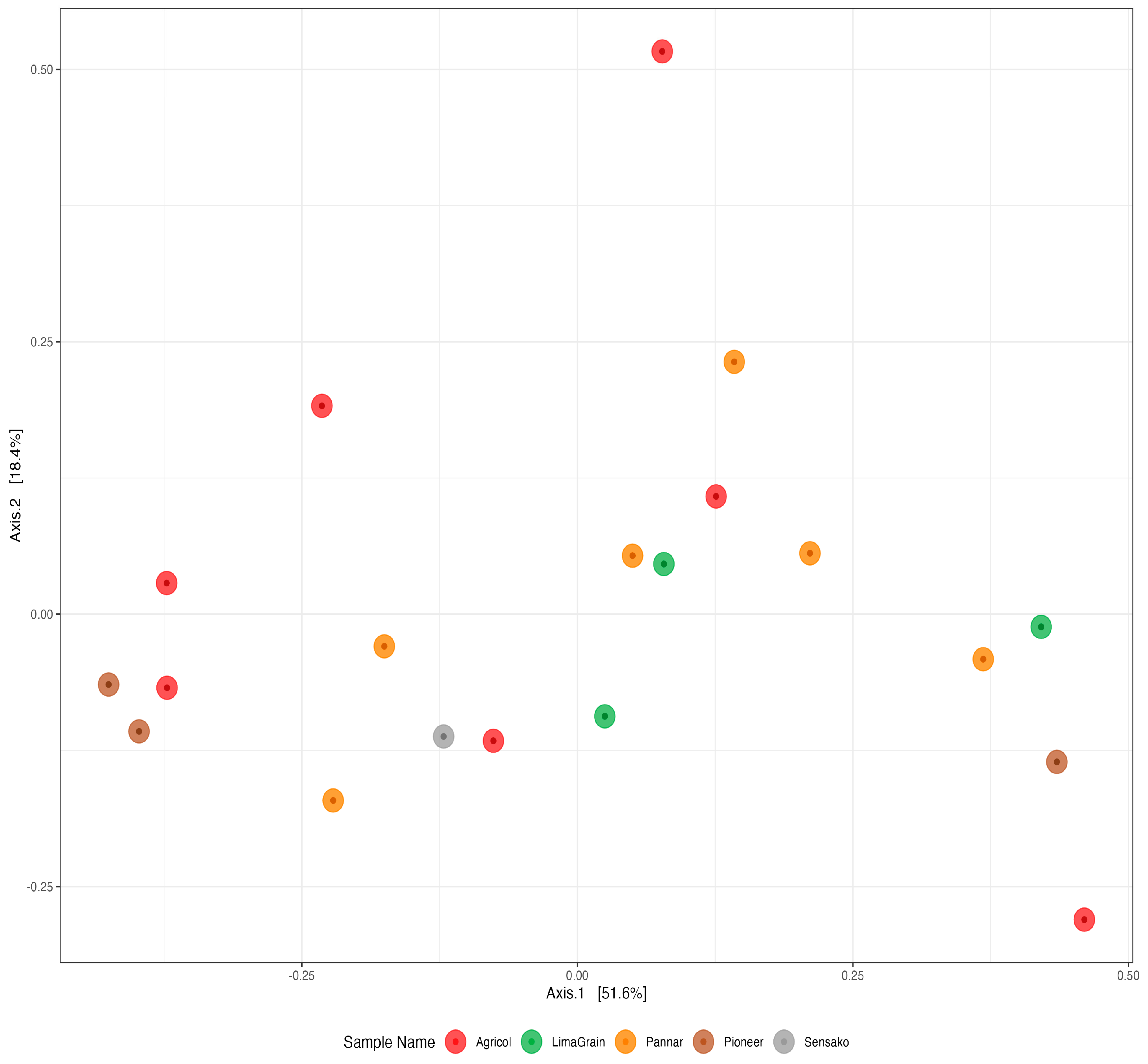
3.3. Bacteria Diversity and Abundance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panero, J.; Funk, V.A. Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae). Proc. Biol. Soc. Wash. 2002. [Google Scholar]
- Baron, N.C.; Rigobelo, E.C. Endophytic fungi: A tool for plant growth promotion and sustainable agriculture. Mycology 2022, 13, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.-S.; Patra, J.K. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Card, S.; Johnson, L.; Teasdale, S.; Caradus, J. Deciphering endophyte behaviour: The link between endophyte biology and efficacious biological control agents. FEMS Microbiol. Ecol. 2016, 92, fiw114. [Google Scholar] [CrossRef]
- Bamisile, B.; Dash, C.; Akutse, K.; Keppanan, R.; Wang, L. Fungal endophytes: Beyond herbivore management. Front. Microbiol. 2018, 9, 544. [Google Scholar] [CrossRef]
- Yan, K.; Pei, Z.; Meng, L.; Zheng, Y.; Wang, L.; Feng, R.; Li, Q.; Liu, Y.; Zhao, X.; Wei, Q. Determination of community structure and diversity of seed-vectored endophytic fungi in Alpinia zerumbet. Front. Microbiol. 2022, 13, 53. [Google Scholar] [CrossRef]
- Donoso, R.; Leiva-Novoa, P.; Zúñiga, A.; Timmermann, T.; Recabarren-Gajardo, G.; González, B. Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium Paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 2017, 83, e01991-16. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Babalola, O.O. Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi J. Biol. Sci. 2020, 27, 3622–3633. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.N.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial plant-microbes interactions: Biodiversity of microbes from diverse extreme environments and its impact for crop improvement. In Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts; Springer: Singapore, 2017; pp. 543–580. [Google Scholar]
- Berg, G.; Raaijmakers, J.M. Saving seed microbiomes. ISME J. 2018, 12, 1167–1170. [Google Scholar] [CrossRef]
- Kinge, T.; Cason, E.D.; Valverde Portal, Á.; Nyaga, M.; Gryzenhout, M. Endophytic seed mycobiome of six sorghum (Sorghum bicolor) cultivars from commercial seedlots using an Illumina sequencing approach. Mycosphere 2019, 10, 739–756. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef]
- Bhutani, N.; Maheshwari, R.; Kumar, P.; Suneja, P. Bioprospecting of endophytic bacteria from nodules and roots of Vigna radiata, Vigna unguiculata and Cajanus cajan for their potential use as bioinoculants. Plant Gene 2021, 28, 100326. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.; Reddy, M.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Eziashi, E.; Omamor, I.; Odigie, E. Antagonism of Trichoderma viride and effects of extracted water soluble compounds from Trichoderma species and benlate solution on Ceratocystis paradoxa. Afr. J. Biotechnol. 2007, 6, 388–392. [Google Scholar]
- Hosni, T.; Abbes, Z.; Abaza, L.; Medimagh, S.; Ben Salah, H.; Kharrat, M. Biochemical Characterization of Seed Oil of Tunisian Sunflower (Helianthus annuus L.) Accessions with Special Reference to Its Fatty Acid Composition and Oil Content. J. Food Qual. 2022, 2022, 2875072. [Google Scholar] [CrossRef]
- United States Department of Agriculture. Oilseeds: World Market and Trades; USDA: Washington, DC, USA, 2023. [Google Scholar]
- Louw, M. Sunflower Production Fields crops of South Africa. 2022. South Africa Online (Pty) Limited. Available online: https://southafrica.co.za/sunflower-production.html (accessed on 12 September 2022).
- Guo, S.; Ge, Y.; Na Jom, K. A review of phytochemistry, metabolite changes, and medicinal uses of the common sunflower seed and sprouts (Helianthus annuus L.). Chem. Cent. J. 2017, 11, 95. [Google Scholar] [CrossRef]
- Rauf, S.; Jamil, N.; Tariq, S.A.; Khan, M.; Kausar, M.; Kaya, Y. Progress in modification of sunflower oil to expand its industrial value. J. Sci. Food Agric. 2017, 97, 1997–2006. [Google Scholar] [CrossRef]
- Ferfuia, C.; Turi, M.; Vannozzi, G.P. Variability of seed fatty acid composition to growing degree-days in high oleic acid sunflower genotypes. Helia 2015, 38, 61–78. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. Oilseed crop sunflower (Helianthus annuus) as a source of food: Nutritional and health benefits. Food Sci. Nutr. 2020, 8, 4666–4684. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. The endosphere microbial communities, a great promise in agriculture. Int. Microbiol. 2021, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gontcharov, S.; Antonova, T.; Saukova, S. Sunflower breeding for resistance to fusarium/selección de girasol por resistencia a fusarium/sélection de tournesol pour la résistance au fongus fusarium. Helia 2006, 29, 49–54. [Google Scholar] [CrossRef]
- Encheva, J.; Christov, M.; Shindrova, P. Developing mutant sunflower line (Helianthus annuus L.) by combined used of classical method with induced mutagenesis and embryo culture method. Bulg. J. Agric. Sci. 2008, 14, 397–404. [Google Scholar]
- Gulya, T.; Mathew, F.; Harverson, R.; Markell, S.; Block, C. Handbook of Florists’ Crops Diseases; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Bacterial community structure of the sunflower (Helianthus annuus) endosphere. Plant Signal. Behav. 2021, 16, 1974217. [Google Scholar] [CrossRef]
- Larson, L. The effect soaking pea seeds with or without seedcoats has on seedling growth. Plant Physiol. 1968, 43, 255–259. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists. Extraction of Oil Content, Standard Method; Association of Official Analytical Chemists: Rockville, MD, USA, 1990. [Google Scholar]
- Fagbemi, K.O.; Aina, D.A.; Olajuyigbe, O.O. Soxhlet extraction versus hydrodistillation using the clevenger apparatus: A comparative study on the extraction of a volatile compound from Tamarindus indica seeds. Sci. World J. 2021, 2021, 5961586. [Google Scholar] [CrossRef]
- McCurry, J. GC Analysis of Total Fatty Acid Methyl Esters (FAME) and Methyl Linolenate in Biodiesel Using the Revised EN14103: 2011 Method; Agilent Technologies Inc.: Santa Clara, CA, USA, 2012. [Google Scholar]
- Usyk, M.; Zolnik, C.P.; Patel, H.; Levi, M.H.; Burk, R.D. Novel ITS1 fungal primers for characterization of the mycobiome. MSphere 2017, 2, e00488-17. [Google Scholar] [CrossRef]
- Eyre, A.W.; Wang, M.; Oh, Y.; Dean, R.A. Identification and characterization of the core rice seed microbiome. Phytobiomes J. 2019, 3, 148–157. [Google Scholar] [CrossRef]
- Yang, R.-H.; Su, J.-H.; Shang, J.-J.; Wu, Y.-Y.; Li, Y.; Bao, D.-P.; Yao, Y.-J. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS ONE 2018, 13, e0206428. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. Package Tidyverse. Easily Install and Load the ‘Tidyverse 2017. Available online: https://cran.r-project.org/web/packages/tidyverse/index.html (accessed on 8 March 2023).
- Oksanen, J.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Henry, M.; Stevens, H. Vegan: Community Ecology Package. R Package Version 2.2-0. 2014, p. 280. Available online: https://CRAN.Rproject.org/package=vegan (accessed on 8 March 2023).
- McMurdie, P.; Paulson, J. Biomformat: An Interface Package for the BIOM File Format. R/Bioconductor Package, Version 1.0 2016. Available online: https://github.com/joey711/biomformat/ (accessed on 20 January 2023).
- Rauf, S.; Sadaqat, H.; Khan, I.; Ahmed, R. Genetic analysis of leaf hydraulics in sunflower (Helianthus annuus L.) under drought stress. Plant Soil Environ. 2009, 55, 62–69. [Google Scholar] [CrossRef]
- Ceccarini, L.; Macchia, M.; Flamini, G.; Cioni, P.; Caponi, C.; Morelli, I. Essential oil composition of Helianthus annuus L. leaves and heads of two cultivated hybrids “Carlos” and “Florom 350”. Ind. Crops Prod. 2004, 19, 13–17. [Google Scholar] [CrossRef]
- Rubiolo, P.; Sgorbini, B.; Liberto, E.; Cordero, C.; Bicchi, C. Essential oils and volatiles: Sample preparation and analysis. A review. Flavour Fragr. J. 2010, 25, 282–290. [Google Scholar] [CrossRef]
- Akkaya, M. Fatty acid compositions of sunflowers (Helianthus annuus L.) grown in east Mediterranean region. Riv. Ital. Sostanze Grasse 2018, 95, 239–247. [Google Scholar]
- Gavrilova, V.; Shelenga, T.; Porokhovinova, E.; Dubovskaya, A.; Kon’kova, N.; Grigoryev, S.; Podolnaya, L.; Konarev, A.; Yakusheva, T.; Kishlyan, N. The diversity of fatty acid composition in traditional and rare oil crops cultivated in Russia. Biol. Commun. 2020, 65, 68–81. [Google Scholar] [CrossRef]
- Roche, J.; Bouniols, A.; Mouloungui, Z.; Barranco, T.; Cerny, M. Management of environmental crop conditions to produce useful sunflower oil components. Eur. J. Lipid Sci. Technol. 2006, 108, 287–297. [Google Scholar] [CrossRef]
- Ismail, A.I.; Arafat, S.M. Quality characteristics of high-oleic sunflower oil extracted from some hybrids cultivated under Egyptian conditions. J. Food Technol. Res. 2014, 1, 73–83. [Google Scholar] [CrossRef]
- Hernández-Martínez, M.; Gallardo-Velázquez, T.; Osorio-Revilla, G.; Castañeda-Pérez, E.; Uribe-Hernández, K. Characterization of Mexican fishes according to fatty acid profile and fat nutritional indices. Int. J. Food Prop. 2016, 19, 1401–1412. [Google Scholar] [CrossRef]
- Puri, R.R.; Adachi, F.; Omichi, M.; Saeki, Y.; Yamamoto, A.; Hayashi, S.; Itoh, K. Culture-dependent analysis of endophytic bacterial community of sweet potato (Ipomoea batatas) in different soils and climates. J. Adv. Microbiol. 2018, 13, 1–12. [Google Scholar] [CrossRef]
- David, V.; Terrat, S.; Herzine, K.; Claisse, O.; Rousseaux, S.; Tourdot-Maréchal, R.; Masneuf-Pomarede, I.; Ranjard, L.; Alexandre, H. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 2014, 41, 811–821. [Google Scholar] [CrossRef]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [CrossRef]
- Price, L.B.; Liu, C.M.; Melendez, J.H.; Frankel, Y.M.; Engelthaler, D.; Aziz, M.; Bowers, J.; Rattray, R.; Ravel, J.; Kingsley, C. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: Impact of diabetes and antibiotics on chronic wound microbiota. PLoS ONE 2009, 4, e6462. [Google Scholar] [CrossRef]
- Heeger, F.; Bourne, E.C.; Baschien, C.; Yurkov, A.; Bunk, B.; Spröer, C.; Overmann, J.; Mazzoni, C.J.; Monaghan, M.T. Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments. Mol. Ecol. Resour. 2018, 18, 1500–1514. [Google Scholar] [CrossRef]
- Cazabonne, J.; Bartrop, L.; Dierickx, G.; Gafforov, Y.; Hofmann, T.A.; Martin, T.E.; Piepenbring, M.; Rivas-Ferreiro, M.; Haelewaters, D. Molecular-based diversity studies and field surveys are not mutually exclusive: On the importance of integrated methodologies in mycological research. Front. Fungal Biol. 2022, 3, 860777. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, E.; Hong, M.; Yin, X.; Cai, H.; Yuan, L.; Yuan, F.; Li, L.; Zhao, K.; Lan, X. Analysis of endophytic and rhizosphere bacterial diversity and function in the endangered plant Paeonia ludlowii. Arch. Microbiol. 2020, 202, 1717–1728. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Pem, D.; Rathnayaka, A.R.; Wijesinghe, S.N.; Tibpromma, S.; Wanasinghe, D.N.; Phookamsak, R.; Kularathnage, N.D.; Gomdola, D.; Harishchandra, D.; et al. Predicting global numbers of teleomorphic ascomycetes. Fungal Divers. 2022, 114, 237–278. [Google Scholar] [CrossRef]
- Rim, S.O.; Roy, M.; Jeon, J.; Montecillo, J.A.V.; Park, S.-C.; Bae, H. Diversity and communities of fungal endophytes from four Pinus species in Korea. Forests 2021, 12, 302. [Google Scholar] [CrossRef]
- Kinge, T.R.; Ghosh, S.; Cason, E.D.; Gryzenhout, M. Characterization of the endophytic mycobiome in cowpea (Vigna unguiculata) from a single location using illumina sequencing. Agriculture 2022, 12, 333. [Google Scholar] [CrossRef]
- Zhao, Y.; Fu, W.; Hu, C.; Chen, G.; Xiao, Z.; Chen, Y.; Wang, Z.; Cheng, H. Variation of rhizosphere microbial community in continuous mono-maize seed production. Sci. Rep. 2021, 11, 1544. [Google Scholar] [CrossRef]
- Pambuka, G.T.; Kinge, T.R.; Ghosh, S.; Cason, E.D.; Nyaga, M.M.; Gryzenhout, M. Baseline Data of the Fungal Phytobiome of Three Sorghum (Sorghum bicolor) Cultivars in South Africa using Targeted Environmental Sequencing. J. Fungi 2021, 7, 978. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef]
- Cason, E.D.; Mahlomaholo, B.J.; Taole, M.M.; Abong, G.O.; Vermeulen, J.-G.; de Smidt, O.; Vermeulen, M.; Steyn, L.; Valverde, A.; Viljoen, B. Bacterial and fungal dynamics during the fermentation process of Sesotho, a traditional beer of Southern Africa. Front. Microbiol. 2020, 11, 1451. [Google Scholar] [CrossRef]
- Stewart, G.G. Saccharomyces species in the Production of Beer. Beverages 2016, 2, 34. [Google Scholar] [CrossRef]
- Leff, J.W.; Lynch, R.C.; Kane, N.C.; Fierer, N. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 2017, 214, 412–423. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of endophytic Bacillus cereus T4S and its plant growth-promoting traits. Plants 2021, 10, 1776. [Google Scholar] [CrossRef]
- Babin, D.; Deubel, A.; Jacquiod, S.; Sørensen, S.J.; Geistlinger, J.; Grosch, R.; Smalla, K. Impact of long-term agricultural management practices on soil prokaryotic communities. Soil Biol. Biochem. 2019, 129, 17–28. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Adam, E.; Wicaksono, W.A.; Bernhart, M.; Olimi, E.; Müller, H.; Berg, G. Microbiome-assisted breeding to understand cultivar-dependent assembly in Cucurbita pepo. Front. Plant Sci. 2021, 12, 642027. [Google Scholar] [CrossRef] [PubMed]
- Kutzman, C.P.; Fell, J.W.; Boekhout, T. The Yeast, a Taxonomic Study; Elsevier Publishers: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Tozlu, E.; Tekiner, N.; Örtücü, S. Investigation on the biological control of Alternaria alternata. Indian J. Agric. Sci. 2018, 88, 1241–1248. [Google Scholar] [CrossRef]
- Islam, T.; Gupta, D.R.; Surovy, M.Z.; Mahmud, N.U.; Mazlan, N.; Islam, T. Identification and application of a fungal biocontrol agent Cladosporium cladosporioides against Bemisia tabaci. Biotechnol. Biotechnol. Equip. 2019, 33, 1698–1705. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Boughalleb-M’Hamdi, N.; Salem, I.B.; M’Hamdi, M. Evaluation of the efficiency of Trichoderma, Penicillium, and Aspergillus species as biological control agents against four soil-borne fungi of melon and watermelon. Egypt. J. Biol. Pest Control 2018, 28, 1–12. [Google Scholar] [CrossRef]
- Atallah, O.O.; Mazrou, Y.S.; Atia, M.M.; Nehela, Y.; Abdelrhim, A.S.; Nader, M.M. Polyphasic characterization of four Aspergillus species as potential biocontrol agents for white mold disease of bean. J. Fungi 2022, 8, 626. [Google Scholar] [CrossRef]
- Abdel-Baky, N.; Arafat, N.S.; Abdel-Salam, A. Three Cladosporium spp. as promising biological control candidates for controlling whiteflies (Bemisia spp.) in Egypt. Pak. J. Biol. Sci. 1998, 1, 188–195. [Google Scholar] [CrossRef]
- Damasceno, C.L.; De Sa, J.O.; Da Silva, R.M.; do Carmo, C.O.; Haddad, L.; Soares, A.C.F.; das Almas, C. Penicillium citrinum as a potential biocontrol agent for sisal bole rot disease. J. Agric. Sci. 2019, 11, 206–216. [Google Scholar] [CrossRef]
- Ahmed, H.; Strub, C.; Hilaire, F.; Schorr-Galindo, S. First report: Penicillium adametzioides, a potential biocontrol agent for ochratoxin-producing fungus in grapes, resulting from natural product pre-harvest treatment. Food Control 2015, 51, 23–30. [Google Scholar] [CrossRef]
- Abdullah, S.; Al-Mosawi, K. Fungi associated with seeds of sunflower (Helianthus annuus) cultivars grown in Iraq. Phytopathologia 2010, 57, 11–20. [Google Scholar]
- De Souza, R.S.C.; Okura, V.K.; Armanhi, J.S.L.; Jorrín, B.; Lozano, N.; Da Silva, M.J.; González-Guerrero, M.; de Araújo, L.M.; Verza, N.C.; Bagheri, H.C. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; He, H.; Long, C.-A.; Jiang, F. Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control 2020, 151, 104419. [Google Scholar] [CrossRef]
- Woudenberg, J.; Groenewald, J.; Binder, M.; Crous, P. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef]
- Gryzenhout, M.; Cason, E.D.; Vermeulen, M.; Kloppers, G.A.; Bailey, B.; Ghosh, S. Fungal community structure variability between the root rhizosphere and endosphere in a granite catena system in Kruger National Park, South Africa. Koedoe Afr. Prot. Area Conserv. Sci. 2020, 62, 1–11. [Google Scholar] [CrossRef]
- Woudenberg, J.; Seidl, M.; Groenewald, J.; De Vries, M.; Stielow, J.; Thomma, B.; Crous, P. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef]
- Ivanova, L.; Petersen, D.; Uhlig, S. Phomenins and fatty acids from Alternaria infectoria. Toxicon 2010, 55, 1107–1114. [Google Scholar] [CrossRef]
- Meena, M.; Gupta, S.K.; Swapnil, P.; Zehra, A.; Dubey, M.K.; Upadhyay, R.S. Alternaria toxins: Potential virulence factors and genes related to pathogenesis. Front. Microbiol. 2017, 8, 1451. [Google Scholar] [CrossRef]
- Vujanovic, V.; Hamel, C.; Yergeau, E.; St-Arnaud, M. Biodiversity and biogeography of Fusarium species from Northeastern North American asparagus fields based on microbiological and molecular approaches. Microb. Ecol. 2006, 51, 242–255. [Google Scholar] [CrossRef]
- Singh, S.; Singh, R.P.; Singh, H.K.; Singh, R.B. Alternaria Blight Severity and Fatty Acid Composition in Some Promising Genotypes of Indian Mustard. 2020. Available online: https://www.researchgate.net/publication/343239441_Alternaria_blight_severity_and_fatty_acid_composition_in_some_promising_genotypes_of_Indian_Mustard (accessed on 9 April 2023).
- Chen, J.; Ferris, H.; Scow, K.; Graham, K. Fatty acid composition and dynamics of selected fungal-feeding nematodes and fungi. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 130, 135–144. [Google Scholar] [CrossRef]
- Fan, D.; Schwinghamer, T.; Smith, D.L. Isolation and diversity of culturable rhizobacteria associated with economically important crops and uncultivated plants in Québec, Canada. Syst. Appl. Microbiol. 2018, 41, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Zhang, T.; Li, G.; Mu, Y.; Lv, X.; Wang, Z.; Zhuang, L. Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol. 2020, 20, 291. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Kumar, V.; Dhaliwal, H.S.; Prasad, R.; Saxena, A.K. Microbiome in crops: Diversity, distribution, and potential role in crop improvement. In Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–332. [Google Scholar]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J. Balešević-Tubić, S. The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef]
- Aloo, B.N.; Makumba, B.; Mbega, E.R. The potential of Bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiol. Res. 2019, 219, 26–39. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Albayrak, Ç.B. Bacillus species as biocontrol agents for fungal plant pathogens. In Bacilli and Agrobiotechnology: Phytostimulation and Biocontrol; Springer: Cham, Switzerland, 2019; Volume 2, pp. 239–265. [Google Scholar]
- Steglińska, A.; Kołtuniak, A.; Motyl, I.; Berłowska, J.; Czyżowska, A.; Cieciura-Włoch, W.; Okrasa, M.; Kręgiel, D.; Gutarowska, B. Lactic Acid Bacteria as Biocontrol Agents against Potato (Solanum tuberosum L.) Pathogens. Appl. Sci. 2022, 12, 7763. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, S.; Fernández-Pacheco, P.; Seseña, S.; Pintado, C.; Palop, M.L. Selection of probiotic Lactobacillus strains with antimicrobial activity to be used as biocontrol agents in food industry. LWT 2021, 143, 111142. [Google Scholar] [CrossRef]
- Cebrián, E.; Núñez, F.; Gálvez, F.J.; Delgado, J.; Bermúdez, E.; Rodríguez, M. Selection and evaluation of Staphylococcus xylosus as a biocontrol agent against toxigenic moulds in a dry-cured ham model system. Microorganisms 2020, 8, 793. [Google Scholar] [CrossRef]
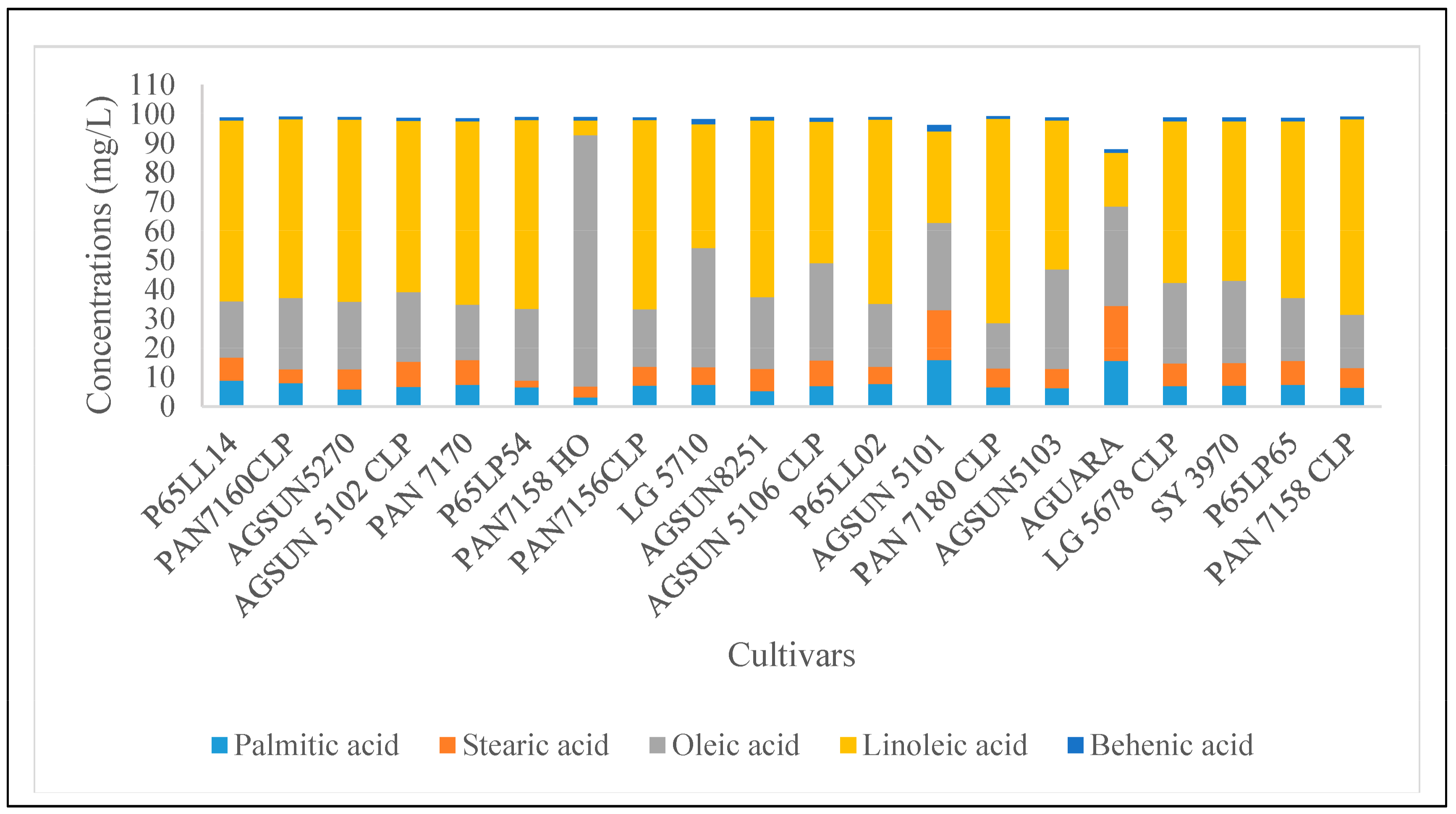
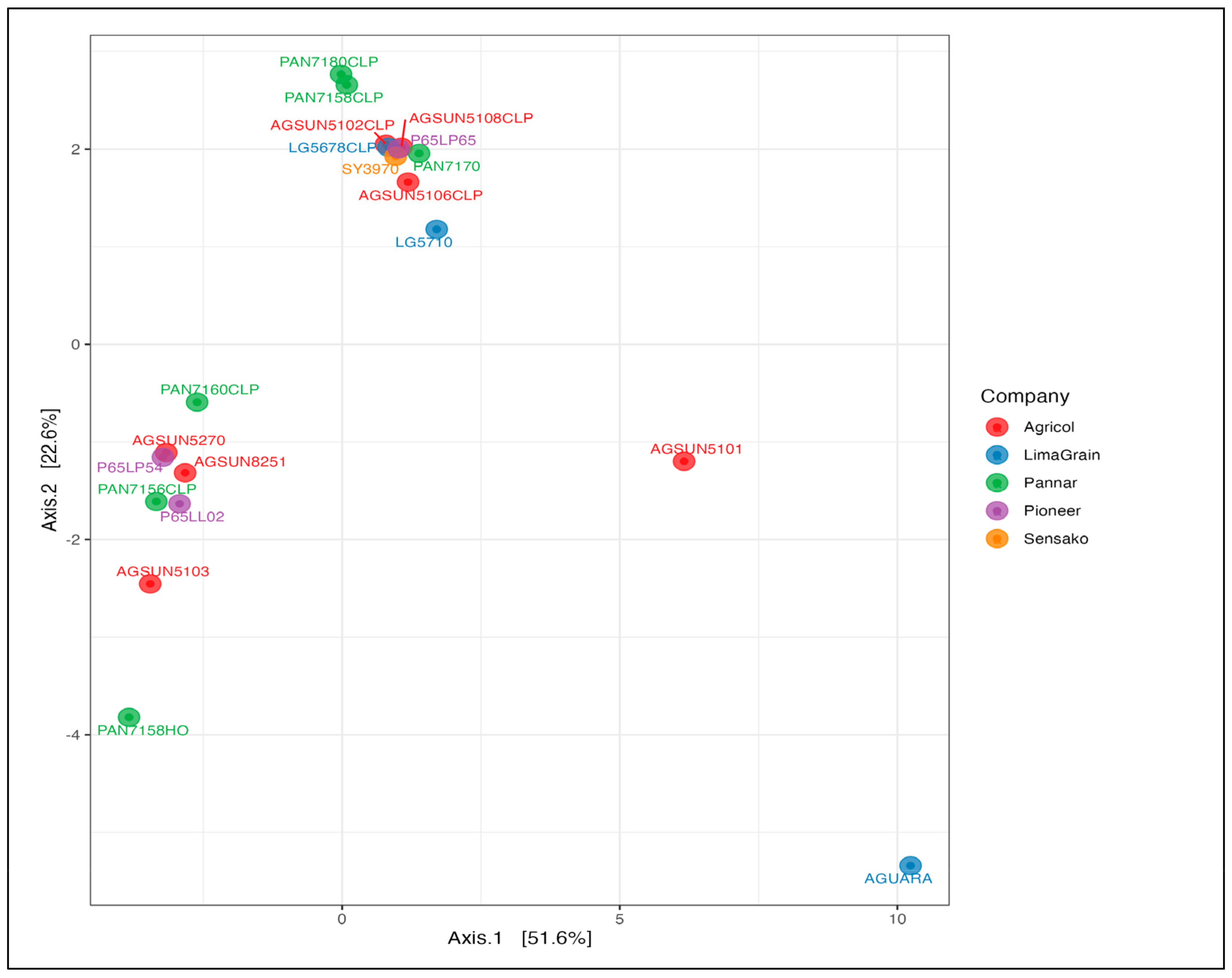
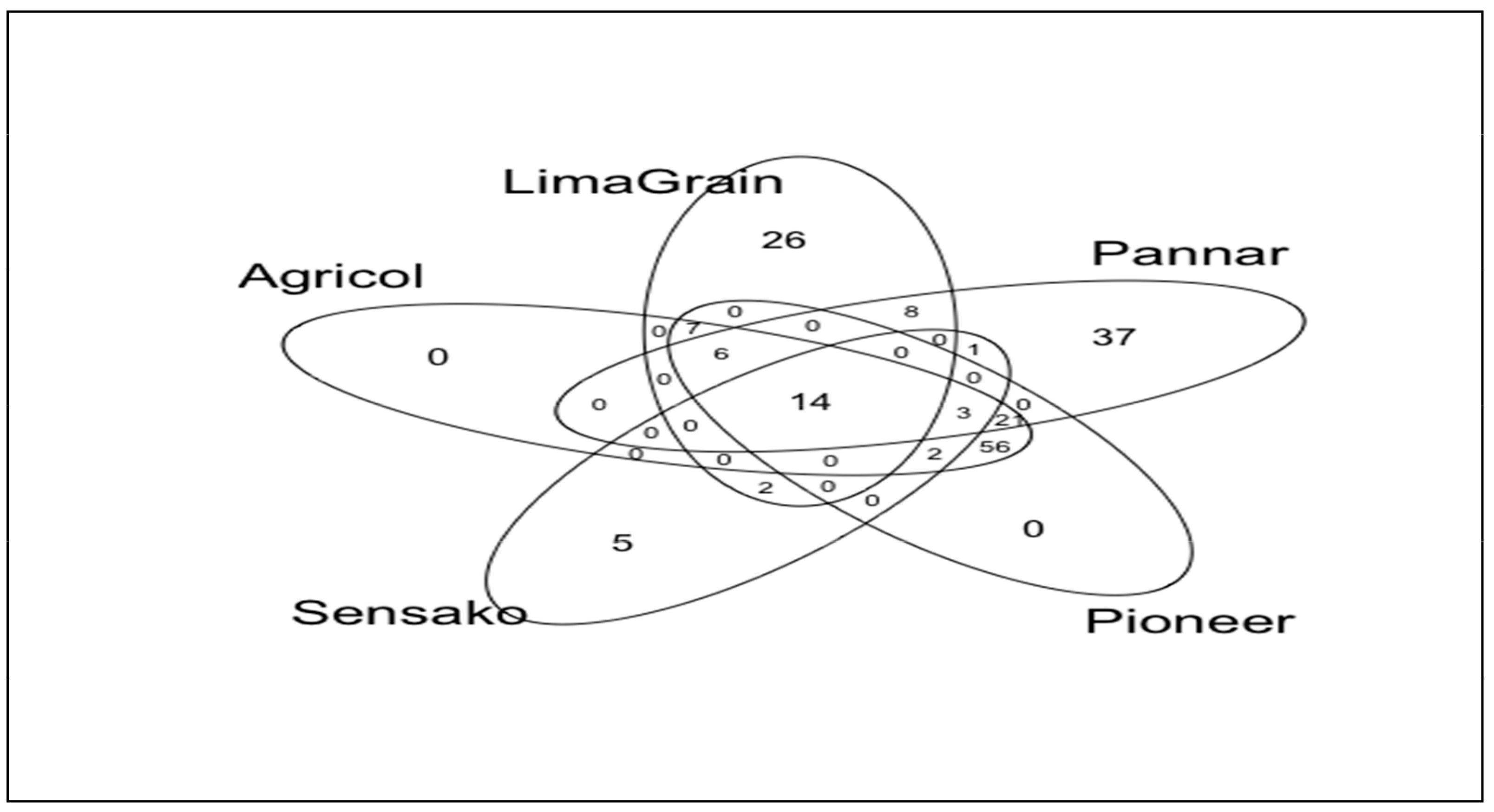
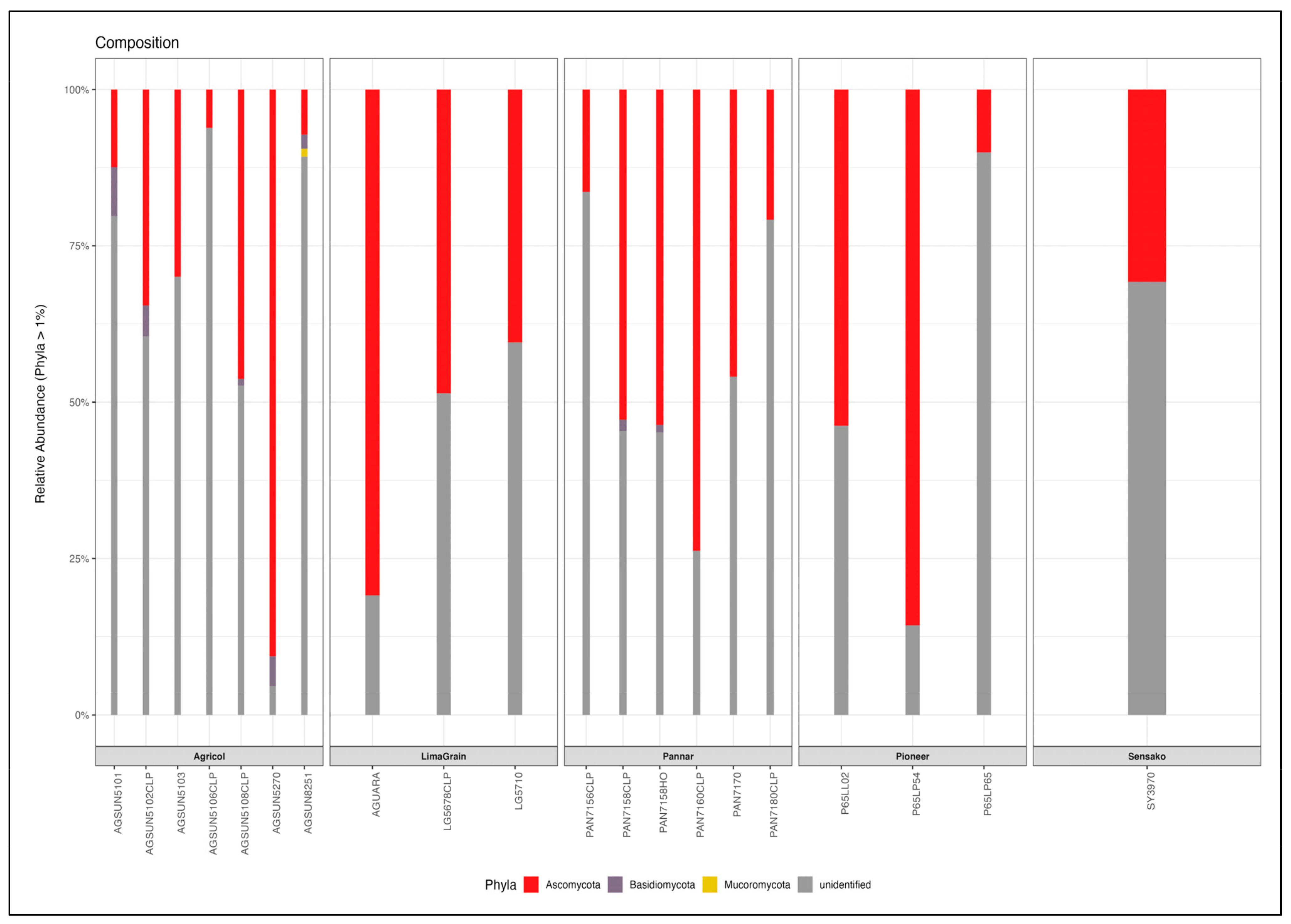



| Brands | Assign. No | Cultivar Names | Oil Contents (%) |
|---|---|---|---|
| AGRICOL | 4 | AGSUN5270 | 52.8 |
| 5 | AGSUN 5102 CLP | 45.0 | |
| 11 | AGSUN8251 | 44.9 | |
| 12 | AGSUN 5106 CLP | 50.0 | |
| 14 | AGSUN 5101 | 40.8 | |
| 16 | AGSUN5103 | 48.0 | |
| PANNAR | 3 | PAN7160CLP | 51.7 |
| 6 | PAN 7170 | 52.8 | |
| 8 | PAN7158 HO | 42.4 | |
| 9 | PAN7156CLP | 52.0 | |
| 15 | PAN 7180 CLP | 46.9 | |
| 21 | PAN 7158 CLP | 42.4 | |
| PIONEER | 2 | P65LL14 | 47.0 |
| 7 | P65LP54 | 43.0 | |
| 13 | P65LL02 | 42.9 | |
| 20 | P65LP65 | 41.0 | |
| LIMAGRAIN | 10 | LG 5710 | 50.5 |
| 17 | AGUARA | 44.0 | |
| 18 | LG 5678 CLP | 47.9 | |
| SENSAKO | 19 | SY 3970 | 41.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balogun, F.O.; Abdulsalam, R.A.; Ojo, A.O.; Cason, E.; Sabiu, S. Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annus) Seeds. Microorganisms 2023, 11, 988. https://doi.org/10.3390/microorganisms11040988
Balogun FO, Abdulsalam RA, Ojo AO, Cason E, Sabiu S. Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annus) Seeds. Microorganisms. 2023; 11(4):988. https://doi.org/10.3390/microorganisms11040988
Chicago/Turabian StyleBalogun, Fatai Oladunni, Rukayat Abiola Abdulsalam, Abidemi Oluranti Ojo, Errol Cason, and Saheed Sabiu. 2023. "Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annus) Seeds" Microorganisms 11, no. 4: 988. https://doi.org/10.3390/microorganisms11040988
APA StyleBalogun, F. O., Abdulsalam, R. A., Ojo, A. O., Cason, E., & Sabiu, S. (2023). Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annus) Seeds. Microorganisms, 11(4), 988. https://doi.org/10.3390/microorganisms11040988







