Abstract
Edwardsiella piscicida is an important fish pathogen that causes substantial economic losses. In order to understand its pathogenic mechanism, additional new virulence factors need to be identified. The bacterial thioredoxin system is a major disulfide reductase system, but its function is largely unknown in E. piscicida. In this study, we investigated the roles of the thioredoxin system in E. piscicida (named TrxBEp, TrxAEp, and TrxCEp, respectively) by constructing a correspondingly markerless in-frame mutant strain: ΔtrxB, ΔtrxA, and ΔtrxC, respectively. We found that (i) TrxBEp is confirmed as an intracellular protein, which is different from the prediction made by the Protter illustration; (ii) compared to the wild-type strain, ΔtrxB exhibits resistance against H2O2 stress but high sensitivity to thiol-specific diamide stress, while ΔtrxA and ΔtrxC are moderately sensitive to both H2O2 and diamide conditions; (iii) the deletions of trxBEp, trxAEp, and trxCEp damage E. piscicida’s flagella formation and motility, and trxBEp plays a decisive role; (iv) deletions of trxBEp, trxAEp, and trxCEp substantially abate bacterial resistance against host serum, especially trxBEp deletion; (v) trxAEp and trxCEp, but not trxBEp, are involved in bacterial survival and replication in phagocytes; (vi) the thioredoxin system participates in bacterial dissemination in host immune tissues. These findings indicate that the thioredoxin system of E. piscicida plays an important role in stress resistance and virulence, which provides insight into the pathogenic mechanism of E. piscicida.
1. Introduction
The development of aquaculture plays a vital role in the rational utilization of land resources. Aquaculture accounts for approximately half of all fish production worldwide and remains one of the fastest-growing agricultural industries [1]. However, the emergence of edwardsiellosis has become one of the factors that jeopardize aquaculture, so there is an urgent need to combat this disease. Edwardsiella was initially described in 1962 and was established in 1965 by Ewing and his collaborators as a member of the Enterobacteriaceae family [2,3]. Currently, the Edwardsiella genus contains five species, including E. tarda, E. ictaluri, E. hoshinae, E. anguillarum, and E. piscicida [4]. E. piscicida, formerly known as E. tarda, is a Gram-negative, facultatively anaerobic, capsule-free, rod-shaped, motile, and intracellular pathogen that causes serious infections both in marine and freshwater fish, causing severe economic losses in the aquaculture industry [5,6]. In addition, E. piscicida has a wide host range, such as shellfish, amphibians, and reptiles. Diseases caused by E. piscicida infection have been reported in a lot of countries and regions [7,8,9].
Like other important pathogens, E. piscicida expresses many important virulence factors, which can help it escape the immune defenses of its host and achieve survival, reproduction, and infection in said host. The main virulence factors include a type III secretion system and a type VI secretion system, several two-component systems, an iron-absorption regulatory factor, hemolysin, a quorum-sensing system, and some environmental stress factors [4,7,10,11,12,13,14,15]. For example, E. piscicida adapts well to different pH, salinity, and temperatures [16,17,18]. All of these characteristics have made this bacterium a perfect model for elucidating the pathogenesis of intracellular pathogens [4]. Despite all of this, the pathogenesis of E. piscicida is still not very clear, and it is important and urgent to identify and analyze new unknown virulence factors.
The thioredoxin system is one of the two main antioxidant enzyme systems found in various organisms [19] and is ubiquitous in all living organisms [20]. The thioredoxin system is composed of thioredoxin (Trx), thioredoxin reductase (TrxR/TrxB), and reduced substrate nicotinamide adenine (NADPH). Trx, containing thioredoxin-A (TrxA) and thioredoxin-C (TrxC), is usually a 12 kDa thiol-disulfide oxidoreductase with a variety of functions, including reducing protein disulfide, maintaining proper functions of key regulators in motility, boosting the capability of pathogen to adhere to the epithelial cell, and affecting various virulence factors [21]. TrxR/TrxB is a dimeric enzyme belonging to the flavoprotein family of pyridine nucleotide–disulfide oxidoreductases. Members of this family are homodimeric proteins, each of which includes an FAD prosthetic group, an NADPH binding site, and an active site containing a redox-active disulfide [22]. Two forms of TrxR/TrxB have evolved, one in bacteria, archaea, and lower eukaryotes and another in higher eukaryotes [22]. In humans, TrxR is widely used as a target for cancer therapy, due to its role in regulating cellular redox balance as well as redox-mediated signal transduction [23], while in bacteria, TrxR/TrxB plays an important role by removing reactive oxygen species (ROS) in cells, thereby contributing to the resistance of bacteria to oxidative stress [24]; NADPH is responsible for transferring electrons to TrxR/TrxB to keep it in a reduced state [25]. The thioredoxin system is involved in enzyme function, DNA synthesis, gene transcription, cell growth and apoptosis, and the defense of oxidative stress [26]. However, the function of the thioredoxin system proteins is largely unknown in E. piscicida.
In this study, the thioredoxin system, TrxBEP, TrxAEp, and TrxCEp, of E. piscicida were identified, and their roles in adversity adaptation, motility, and pathogenicity were dissected. Our results present the first insights into the biological function of the thioredoxin system proteins in E. piscicida, which will contribute to comprehending E. piscicida pathogenesis.
2. Materials and Methods
2.1. Bioinformatics Analysis
Gene and protein information was obtained using NCBI (https://www.ncbi.nlm.nih.gov/ (accessed on 14 November 2022)). Jalview was used to analyze homologous sequence alignment. Protter (http://wlab.ethz.ch/protter/# (accessed on 14 November 2022)) was used to predict protein sequence features.
2.2. Bacteria, Plasmids and Cells
The bacteria and plasmids used in this study were described in Table S1. E. piscicida TX01 was isolated from diseased Japanese flounder at fish farms in northern China and was determined by 16S rRNA gene analysis [27]. E. coli D1000 S17-λpir was purchased from Biomedal (Sevilla, Spain). E. coli DH5α was purchased from TransGen Biotech (Beijing, China). Bacteria were grown in Luria–Bertani medium (LB) broth or on LB agar at 37 °C (for E. coli) or 28 °C (for E. piscicida). When antibiotics were needed, the final concentrations of tetracycline (TC), chloramphenicol (CAP), polymyxin B (PMB), ampicillin (AMP), and kanamycin (KAN) in the medium were 15, 30, 100, 100, and 50 μg/mL, respectively. The RAW264.7 cells were propagated at 37 °C in a 5% CO2 incubator in DMEM supplemented with 10% fetal bovine serum.
2.3. Animals and Ethics
Clinically healthy tilapias were purchased from local commercial fish farms and raised in aerated fresh water at about 26 °C as previously reported [28]. Clinically healthy female mice (4–6-week-old) were purchased from a local commercial mouse breeding base and housed at about 26 °C. No mouse died before meeting the criteria for humane endpoint euthanization during this study. The study was approved by the ethics committee of the Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences. Efforts were taken to ensure that all research animals received good care and humane treatment. Animals were euthanized followed by cervical dislocation when the experimental endpoints were reached.
2.4. Construction of Mutant Strain and Complementary Strain
The in-frame deletion mutant strains used in this study were constructed as described previously [29]. The primers used in this study were shown in Table S2. The in-frame deletion mutant was constructed by double-crossover allelic exchange using suicide vector pDM4. To construct the mutant strain ΔtrxA, primers TrxAKOF1/TrxAKOR1 and TrxAKOF2/TrxAKOR2 were used for the upstream and downstream PCR of trxA. The fusion PCR products amplified by primer pair TrxAKOF1/R2 were cloned into the pDM4 at the Bgl II site, in which a 1330 bp segment in-frame deletion was created, resulting in pDMTrxA. S17-1 λpir was transformed with pDMTrxA, and the transformants were conjugated with TX01. The transconjugants were selected on TSA plates supplemented with 10% sucrose. The mutations of ΔtrxB and ΔtrxC were made with the same operation application.
2.5. Expression of Recombinant TrxB and Determination of Enzyme Activity
In order to express the recombinant TrxB (rTrxB) protein, segments of trxB were amplified with the primer pair TrxBF/R, and then the PCR segments were cloned into pET28a with double-enzyme digestion (BamHI + HindIII), resulting in pET-TrxB. The recombinant plasmid pET-TrxB was extracted and transformed into E. coli BL21(DE3). rTrxBEp was expressed and purified as described previously [30]. In brief, E. coli BL21(DE3) carrying pET-TrxB was cultured to an OD600 of 0.5 in LB broth in the presence of 25 µg/mL kanamycin, and then 0.1 mM IPTG was added to the broth. After 6 h of induction, the preparation of the lysate and the purification and refolding of the recombinant protein were conducted as follows: the E. coli BL21(DE3) carrying pET-TrxB were incubated with Lysis Buffer B (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M urea [pH 8.0]) at 20 °C for 2 h and the protein extract was obtained after centrifugation. The recombinant protein was purified by Ni-NTA agarose (QIAGEN, Germany) under denaturing conditions. The purified recombinant protein was dialyzed in a buffer containing 50 mM Tris-Cl (pH = 8.5), 50 mM NaCl, 2 mM reduced glutathione, 5 mM EDTA, 5 mM β-mercaptoethanol, 20% glycerol, and gradually decreasing urea. At the same time, a control protein, rOmpREp (unpublished data), was obtained through the same experimental process.
The content of rTrxBEp and rOmpREp (control) was measured using the BCA Protein Assay Kit (Solarbio, Beijing, China). The thioredoxin reductase activity of the rOmpREp and rTrxBEp was determined by using Micro Oxidized Thioredoxin Reductase Assay Kit (Solarbio, Beijing, China) according to the manufacturer’s instructions and the description by Zheng et al. [31]. Thioredoxin reductase catalyzes the reduction of DTNB by NADPH to produce TNB and NADP+. TNB has a characteristic absorption peak at 412 nm, but reduced glutathione can also react with DTNB to produce TNB. Therefore, this kit uses 2-vinyl pyridine to inhibit reduced glutathione in the sample and calculates the activity of thioredoxin reductase by measuring the increase rate of TNB at 412 nm expressed as U/mg protein. The experiment was repeated three times.
2.6. Preparation of Antibody and ELISA
The antibody of TrxBEp was prepared as described previously [30]. Freund’s complete and incomplete adjuvant were purchased from Sigma-Aldrich. The detection of antibody titer was determined by indirect ELISA [32].
2.7. Preparation of Cellular Component Protein and Western Blotting
E. piscicida was inoculated into 50 mL DMEM containing polymyxin B at 1% inoculation and was cultured at 28 °C until OD600 = 0.5. Then, the bacteria were incubated at 0 °C for 10 min, and the final concentration of 1 mM phenylmethylsulfonyl fluoride (PMSF) was added. After mixing, the mixture was centrifuged by 5000× g for 10 min at 4 °C, and the supernatant and precipitation were collected, respectively. The supernatant was filtered by 0.22 μm microporous membrane and added into a 10 kDa ultrafiltration tube. The supernatant was concentrated to 200 μL by centrifugation, and finally, the extracellular protein was obtained. The extraction of whole-cell protein, periplasmic protein, and intracellular protein was conducted as described in a previous report [33]. The obtained proteins were subjected to SDS-PAGE. The antibody of TrxBEp and Goat anti-Mouse IgG HRP secondary antibody (TransGen Biotech, Beijing, China) were used for Western blotting as described previously [30].
2.8. Resistance to Environmental Stress
The bacterial growth curve was carried out in 96-well plates. E. piscicida strains were cultured in LB broth medium until the exponential phase, then bacteria were gradient diluted to 105 CFU/mL in fresh LB medium supplemented with 300 μM H2O2, or with 550 μM diamide, or with pH = 5, or with 100 μM 2,2’-Bipyridine and were cultured at 28 °C with gentle shaking. Using the Bioscreen C Automated Growth Curve System (OyGrowth Curves Ab Ltd., Finland), the absorbance at 600 nm was measured and the growth curve was monitored at 2 h intervals. Under the challenge of 300 μM H2O2 or 550 μM Diamide, E. piscicida strains were diluted in different concentrations, dropped on LB agar plates, and incubated at 28 °C for 48 h. For the survival experiment, bacteria in the logarithmic growth phase were diluted and treated with 300 μM H2O2 or 550 μM diamide for 1 h, and then the colony-forming units were counted. The survival rate was calculated as follows: (number of each strain with pressure treatment)/(number of each strain without pressure treatment) × 100%. The experiment was repeated three times.
2.9. Measurements of Intracellular Reductive Capacity in Cell Extracts
The thioredoxin reductase activity assay was referred to Ana Paunkov [34]. In short, E. piscicida strains were cultured in LB medium, supplemented with 30 μM H2O2 or 50 μM diamide overnight at 28 °C, and 3 mL of 5 h cultures were centrifuged in order to obtain the pellets. Pellets were resuspended in a 1 × PBS buffer containing 0.5% Triton X-100 and were lysed by an ultrasonic disrupter (SCIENTZ, China). Insoluble material was removed by centrifugation for 10 min at 12,000× g at 4 °C. The supernatants were collected and the protein concentrations were determined. The intracellular reductive capacity of cell extract was measured at OD412 by determining the reduction of DNTB in reaction buffer containing 100 mM KH2PO4, 0.2 mM NADPH, 1 mM DTNB, and 100 μL of cell extract. The experiment was repeated three times. The reductive capacity was calculated as follows: (data of each strain with pressure treatment)/(data of each strain without pressure treatment) × 100%.
2.10. Transmission Electron Microscopy
Bacteria grown to an OD600 of 0.5 were collected by centrifugation (3500× g for 10 min) and softly resuspended in PBS. Suspensions of different strains were dropped onto the copper grid and were allowed to stand for 20 min with the purpose of forming thin films on the copper grids. Subsequently, the excess solutions were removed, and the copper grids were dried naturally. Ultimately, all samples were tested by HT7700 biological TEM (Hitachi, Japan) [35].
2.11. Motility Assay
The motility assay was performed as described previously [36]. Briefly, 2 μL of bacteria suspension were spotted onto the center of fresh swimming plates (0.3% agar) or swarming plates (0.6% agar). Then, the plates were incubated at room temperature and bacterial motility was observed. The experiment was performed three times.
2.12. Bacterial Resistance to Non-Immune Fish Serum
The experiment on the bacterial resistance to non-immune fish serum was performed as described previously [37]. Briefly, bacteria in the exponential phase were washed three times with PBS. Next, 10 μL of bacteria (including approximately 1 × 105 CFU) were incubated with 50 μL non-immune fish serum or PBS (control) for 60 min. The mixtures were serially diluted and plated in triplicate on LB agar plates. The plates were incubated at 28 °C for 24 h, then the number of colonies was determined. The survival rate was calculated as follows: [(number of serum-treated cells)/(number of control cells)] × 100%. The experimental procedures were performed three times.
2.13. Bacterial Invasion of Host Cells
The bacterial replication in RAW264.7 cells was carried out as previously described [37]. RAW264.7 (1 × 105) cells were cultured in 96-well plates in a 5% CO2 incubator at 28 °C, and different E. piscicida strains, WT, ΔtrxA, ΔtrxB, and ΔtrxC, (1 × 106 CFU) were added to RAW264.7 cells, which were cultured in DMEM medium (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco, Grand Island, NY, USA). After incubation at 28 °C for 2 h, cells were gently washed with PBS and incubated with fresh DMEM containing 200 μg/mL gentamicin (the minimum inhibition concentration of gentamicin is 20 μg/mL) for 2 h in order to kill extracellular bacteria. After washing with PBS, the cells were cultured in fresh DMEM containing 10 μg/mL gentamicin for different time points. In order to detect the number of viable bacteria in RAW264.7 cells at different time points, as mentioned above, the cells were lysed and evenly coated on LB agar plates, and then cultured at 28 °C for 24 h. The experiment was repeated three times.
2.14. Bacterial Dissemination in Fish Tissues
Healthy tilapias (average weight 14.5 g) were purchased from a commercial fish farm in Haikou and fed in aerated water at 26 °C for 2 weeks. Before the experiment, fish were randomly sampled in order to examine whether there were bacteria in the blood, liver, spleen, and kidney. For tissue dissemination analysis, different E. piscicida strains were cultured in LB broth medium to an OD600 of 0.5, the cells were washed with PBS and resuspended in PBS to 107 CFU/mL. Tilapias were randomly divided into 5 groups with 10 fish per group, infected with 50 μL of each bacterial suspension or PBS (control) by intramuscular injection. At 24 h and 48 h post-infection, excessive MS-222 (Sigma-Aldrich, St. Louis, MO, USA) was used for euthanasia on fish. The spleen and kidney tissues were taken under sterile conditions and bacteria recoveries was analyzed by plate counting [38]. The experiment was repeated three times.
2.15. Statistical Analysis
Analysis of variance (ANOVA) was performed on all data using SPSS 23 software (SPSS Inc., Chicago, IL, USA). For multigroup comparisons with Gaussian distribution, one-way ANOVA with Tukey–Kramer’s multiple-comparison test was used after the confirmation of homogeneity of variance among the groups by Bartlett’s test. For multigroup comparisons with non-Gaussian distribution, a Kruskal–Wallis test with Dunn’s test was used. Data are presented as the means ± SEMs (N = 3). N, the number of times the experiment was performed. p values were obtained by analysis of variance using SPSS 23.
3. Results
3.1. Bioiformatics Analysis of Trx System in E. piscicida
E. piscicida TrxB, TrxBEp (ETAE_2202), is a 323-amino-acid protein encoded by a 972 bp open reading frame (ORF). Bioinformatics analysis reveals that the predicted molecular weight of TrxBEp is 34.59 kDa, and the isoelectric point of TrxBEp is 5.47. E. piscicida TrxA, TrxAEp (ETAE_0099) contains a 327 bp ORF, which codes 108 amino acid residues with a calculated molecular mass of 11.59 kDa and a theoretical pI of 5.04. E. piscicida TrxC, TrxCEp (ETAE_0559) has a 143 bp ORF, which codes 143 amino acid residues with a calculated molecular mass of 15.80 kDa and a theoretical pI of 5.31. As shown in Figure 1, TrxAEp, TrxBEp, and TrxCEp share high amino acid sequence identities with homologues of many Gram-negative bacteria, including H. alvei, E. coli, S. enterica, and L. richardisequence; however, they share low sequence identities (36.59%, 40.74%, and 19.58%, respectively) with the Gram-positive stain L. monocytogenes.
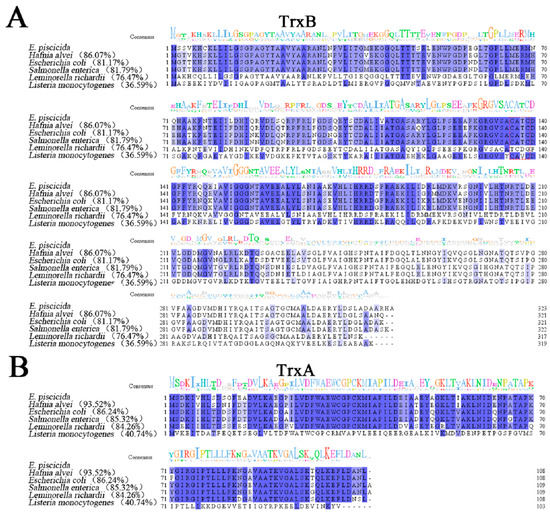

Figure 1.
Multiple sequence alignments of TrxB (A), TrxA (B), and TrxC (C) with their homologues. The consensus amino acid residues are shown in blue, and the amino acid residues with conservative degree higher than 75% are shown in light blue. A consensus sequence logo has been created using Jalview. The GenBank accession numbers of the TrxB homologues are as follows: Edwardsiella piscicida, WP_012849033.1; Hafnia alvei, WP_149226215.1; Escherichia coli, NP_415408.1; Salmonella enterica, AGK67561.1; Leminorella richardii, SQI39560.1; Listeria monocytogenes, WP_003722610.1. The GenBank accession numbers of the TrxA homologues are as follows: E. piscicida, WP_012846980.1; H. alvei, WP_115349310.1; Escherichia coli, HAW3242924.1; S. enterica, AGK68906.1; L. richardii, WP_111741836.1; L. monocytogenes, NP_464758.1. The GenBank accession numbers of the TrxC homologues are as follows: E. piscicida, WP_012847430.1; H. alvei, WP_130998671.1; E. coli, NP_417077.1; S. enterica, NP_461584.1; L. richardii, WP_111739434.1; L. monocytogenes, WP_003723835.1.
3.2. The Location of Trx System and the Enzyme Activity of TrxBEP
The thioredoxin system plays a central role in intracellular redox maintenance in most cells [39]. However, the Protter illustration shows that TrxBEp has a signal peptide and is predicted to be extracellularly located, although TrxAEp and TrxCEp are predicted to be intracellular (Figure 2A). In order to observe the distribution of TrxBEp, whole-cell protein, secreted protein, periplasmic protein, and the intracellular protein of E. piscicida were prepared (Figure 2B); meanwhile, rTrxBEp was expressed and purified (Figure S1) and antiserum against rTrxBEp was prepared (Figure S2). Western blot analysis displayed that the target band was observed in the whole-cell protein and intracellular protein but not in the secreted protein and periplasmic protein (Figure 2C). These results suggest that TrxBEp is an intracellular protein.
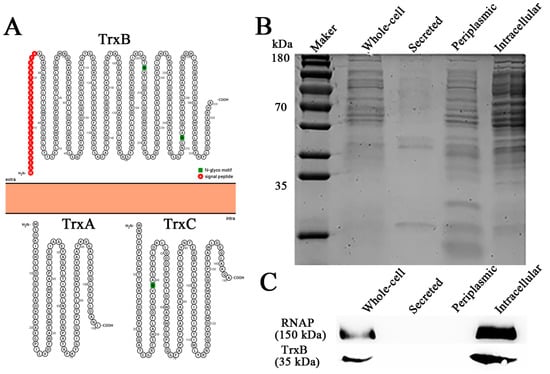
Figure 2.
Cellular localization of TrxBEp. (A) Protter illustration of TrxBEp, TrxAEp and TrxCEp. (B) protein components of Edwardsiella piscicida cells. (C) The distribution of TrxBEp tested by Western blot.
Next, the thioredoxin reductase activity of rTrxBEp was determined using the Micro Oxidized Thioredoxin Reductase Assay Kit. The analysis of thioredoxin reductase activity based on OD412 showed that the absorbance induced by 20 μL of rOmpREp and rTrxBEp was 0.0000 and 0.0066, which corresponded to the thioredoxin reductase activity of 0.00 and 2.54 U/mg protein, indicating that TrxBEp is a thioredoxin reductase (Figure S3).
3.3. Construction of Mutant Strains of Trx System
In order to further study the function of the thioredoxin system, we constructed three mutants—ΔtrxA, ΔtrxB, and ΔtrxC—by deleting the gene segment from 64 to 234 of trxAEp, 115 to 858 of trxBEp, and 103 to 319 bp of trxCEp (Figure S4).
3.4. The Trx System Is Involved in the Resistance against Oxidative Stress, Acid Stress, and Iron Deficiency Stress
Under normal conditions, ΔtrxB grows a little slower than WT, and the growths of ΔtrxA and ΔtrxC have no significant difference with WT (Figure 3[A1]). The intracellular reductive capacities of WT and three mutants have no significant differences when cultured in normal LB broth medium (Figure 3[A2]). These results suggest that the deletion of trxAEp, trxBEp, or trxCEp basically does not affect the growth and reductive capacity of E. piscicida under normal conditions. As the Trx system plays a central role in maintaining intracellular redox, we compared the roles of the Trx system in resistance H2O2 and diamide. When grown in solid LB medium containing H2O2, ΔtrxB appeared to have similar growth, but ΔtrxA and ΔtrxC presented sluggish growth compared to WT (Figure 3[B1]). However, when grown in LB liquid medium containing H2O2, the growth of the four strains is similar (Figure 3[B3]). When temporarily exposed to a high concentration of H2O2, the intracellular reductive capacities of the three mutants were basically equal but were significantly lower than the WT (Figure 3[B2]); the survival rate of ΔtrxA (55%) was comparative to ΔtrxC (62%), and both were significantly lower than the WT (73%), which is also significantly lower than ΔtrxB (109%) (Figure 3[B4]). When the oxidant was diamide, ΔtrxB hardly grew in LB broth medium or LB agar medium, while ΔtrxC was basically similar to WT, and ΔtrxA was slightly slower than WT (Figure 3[C1,C3]). When temporarily exposed to a high concentration of diamide, the intracellular reductive capacities of all three mutants were lower than the WT (Figure 3[C2]), the survival rates of ΔtrxB (58%) were comparative to ΔtrxC (55%), and both were significantly lower than the WT (72%), though that of ΔtrxA (14%) was dramatically lower than those of ΔtrxB and ΔtrxC (Figure 3[C4]). These results indicate that TrxBEp, TrxAEp, and TrxCEp play an important role in bacterial resistance against different types of oxidation stress.
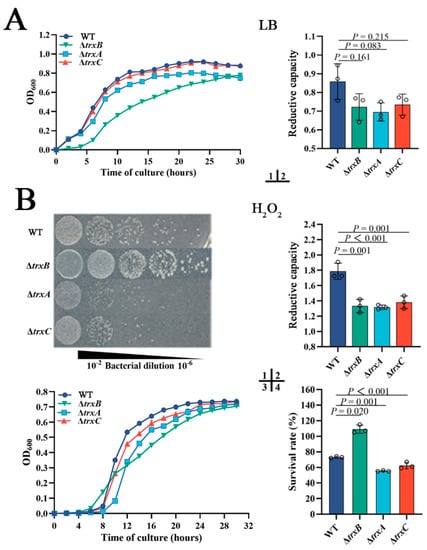
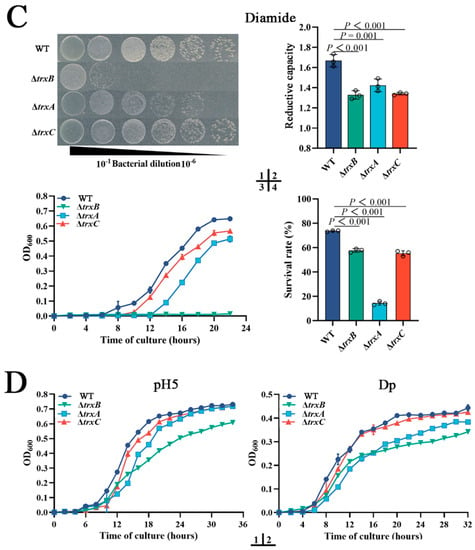
Figure 3.
The roles of the Trx system in bacterial adversity resistance. (A) WT, ΔtrxB, ΔtrxA, and ΔtrxC were cultured in LB broth, and then the cell density was measured at OD600, and the intracellular reductive capacity was detected. (B) Bacterial growths on LB agar plates (1) and in liquid LB (3) with 300 μM H2O2; bacteria in the logarithmic growth phase were challenged with 30 mM H2O2 for 5 h, and the intracellular reductive capacity was determined (2). Bacteria in the logarithmic growth phase were diluted to 1:1000 and treated with PBS containing 300 mM H2O2 for 1 h, then the amount of viable bacteria was determined (4). (C) bacterial growths on LB agar plates (1) and in liquid LB (3) with 550 mM diamide; bacteria in the logarithmic growth phase were challenged with 50 mM diamide for 5 h, and intracellular reductive capacity was detected (2). Bacteria in the logarithmic growth phase were diluted to 1:1000 and treated with PBS containing 550 mM diamide for 1 h, then the content of viable bacteria was determined (4). (D) Strains were cultured in LB broth containing acid pressure (pH = 5) (1) or iron deficiency stress (100 μM Dp) (2) and incubated at 28 °C for 48 h. Data are expressed as means ± SEM (N = 3). N, the number of experiments performed. p values were obtained by analysis of variance using SPSS 23.
In addition to oxidation pressure, acid stress and iron deficiency stress were also used in the resistance experiment, and the results indicated that ΔtrxB and ΔtrxA exhibit weaker growth than WT under acid conditions (pH = 5) and iron deficiency conditions (Figure 3D), while ΔtrxC is basically similar to WT. The results collectively suggest that the Trx system of E. piscicida participates in various adverse circumstances, especially oxidative stress.
3.5. The Trx System Is Essential for Bacterial Motility and Flagellum Formation
In this study, we explored the involvement of the thioredoxin system in bacterial mobility by detected bacterial swimming, and the result showed that the motility zone diameters of ΔtrxB (average diameter 19 ± 3 mm), ΔtrxA (average diameter 16 ± 1 mm), and ΔtrxC (average diameter 22 ± 1 mm) were significantly smaller than that of the WT (average diameter 27 ± 1 mm) (Figure 4A). At the same time, bacterial swarming was examined, and the result showed that the branching motions of ΔtrxA and ΔtrxB were obviously weaker than that of the WT, but ΔtrxB completely lost such motion (Figure 4B). In order to explore whether decreased motility was correlated with flagellum formation, morphological observations were performed, and the results showed that there were a great quantity of flagella around WT and ΔtrxC, a few flagella around ΔtrxA, and no flagella around ΔtrxB (Figure 4C). These results indicate that the Trx system participates in E. piscicida’s flagella formation and motility, and trxBEp plays a decisive role.
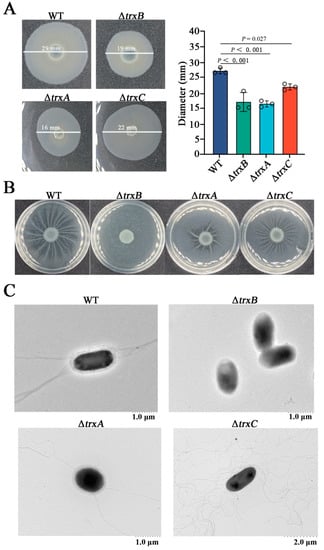
Figure 4.
The effects of the thioredoxin system mutations on bacterial motility and flagellum formation. (A) The swimming of Edwardsiella piscicida. WT, ΔtrxB, ΔtrxA, and ΔtrxC were cultured in LB medium to an OD600 of 0.6, and 1 μL cell suspensions were spotted onto the center of swimming plates containing LB medium plus 0.3% (w/v) agar. The plates were incubated at 28 °C for 24 h, and the motility zone diameter was measured. (B) The swarming of E. piscicida. Bacteria as described above were spotted onto the center of swimming plates containing LB medium plus 0.6% (w/v) agar and were incubated at 28 °C for 24 h. (C), the flagellum observation of E. piscicida. WT, ΔtrxB, ΔtrxA and ΔtrxC were grown in LB medium, and the flagella were observed using the TEM. Data are presented as the means ± SEM (N = 3). N, the number of times the experiments were performed. p values were obtained by analysis of variance using SPSS 23.
3.6. The Trx System Is Involved in Bacterial Resistance against Non-Immune Fish Serum and Bacterial Survival in Host Phagocytes
Resistance against host serum killing is an important virulence characteristic of E. piscicida [40], thus we examined the effect of the thioredoxin system. The results show that three Trx mutants exhibited remarkably lower survival rates than the WT, with the lowest-ranking strain being ΔtrxB (27%), which was much lower than ΔtrxA and ΔtrxC (about 50%) (Figure 5A). This result suggests that the thioredoxin system is involved in bacterial resistance against serum killing.
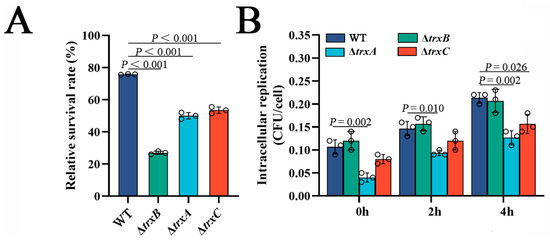
Figure 5.
The effects of thioredoxin system mutations on bacterial survival in host serum and host macrophages. (A) The survival rate of WT, ΔtrxB, ΔtrxA and ΔtrxC against non-immune fish serum. Strains in the early logarithmic phase were incubated with non-immune tilapia serum or PBS (control) for 1 h. The number of viable bacteria was determined. (B) Edwardsiella piscicida replication in macrophages. The murine macrophage cell line RAW264.7 was incubated with WT, ΔtrxB, ΔtrxA and ΔtrxC for 2 h. After killing and washing extracellular bacteria, the macrophages were cultured for different lengths of time. At each time point, the viable intracellular bacteria were determined. Data are presented as the means ± SEM (N = 3). N, the number of times the experiments were performed. p values were obtained by analysis of variance using SPSS 23.
Next, we investigated the role of Trx system in the survival and replication of E. piscicida in host phagocytes. The result showed that the amounts of WT and ΔtrxB from RAW264.7 cells was basically the same at the three examined time points, but the amounts of ΔtrxA were significantly less than WT at these three examined time points, while ΔtrxC was less than WT only at 4 h post-infection (Figure 5B). This result indicates that the thioredoxin system of E. piscicida, except trxBEp, is involved in bacterial survival and replication in host phagocytes.
3.7. The Trx System Participates in Bacterial Dissemination in Host Tissues
In order to explore the involvement of the thioredoxin system in bacterial virulence in vivo, tilapia were infected by different strains, then the invasion of immune tissues by bacteria was determined. The results showed that the counts of the bacteria from the spleen of three mutant-strain-infected tilapia were significantly lower than those of WT-infected tilapia at two examined time points, and a similar result was observed from the kidney of treated tilapia (Figure 6). Furthermore, we did not observe fish mortality during the experiment because of the short duration. According to our laboratory data, infections with the same amount of WT generally show mortality 4–5 days after injection [28,37]. These results suggest that the Trx system of E. piscicida is correlated with bacterial dissemination in host tissues.
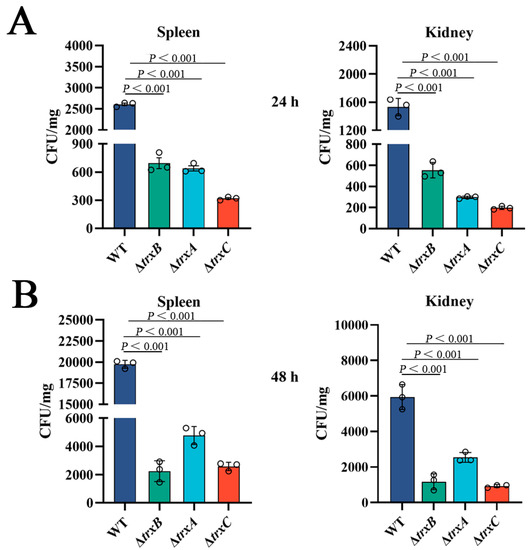
Figure 6.
Effects of the thioredoxin system mutations on bacterial virulence. (A) Edwardsiella piscicida strains (WT, ΔtrxB, ΔtrxA, and ΔtrxC) were used to infect tilapias, then, after 24 h, bacteria recovery from immune tissues (spleen and kidney) was analyzed by plate counting. (B) 48 h post-infection, bacteria were counted by plate counting. Data are presented as the means ± SEM (N = 3). N, the number of times the experiments were performed. p values were obtained by analysis of variance using SPSS 23.
4. Discussion
Oxidative stress is one of the most susceptible pressures during bacterial growth and reproduction [41]. The latest research reveals that ROS in E. tarda are related to bacterial resistance to antibiotics and antibiotic-mediated killing efficacy [42]. The thioredoxin system is one of the major disulfide reductase systems used by bacteria against oxidative stress [43,44]. The function of the thioredoxin system in E. piscicida remains mostly unknown so far. In this study, several proteins (TrxAEp, TrxBEp, and TrxCEp) included in the thioredoxin system of E. piscicida were identified, and their functions were investigated. Sequence analysis indicated that TrxAEp, TrxBEp, and TrxCEp all share a substantial portion of their identities with many homologous Gram-negative bacteria. It should be emphasized that those three proteins possess the CXXC motif, which is a characteristic active site employed by many redox proteins for the formation, isomerization, and reduction of disulfide bonds and for other redox functions [45]. TrxBEp is predicted to be extracellular, but our results confirm that it is an intracellular protein, which is consistent with the results of other microorganisms [46].
The thioredoxin system plays a crucial role in maintaining internal redox homeostasis [26]. For example, in E. coli, the trxB, trxA, and trxC mutants exhibit increased sensitivity compared with the wild-type strain when exposed to direct oxidant hydrogen peroxide in the stationary phase [47,48]. In L. monocytogenes, although hydrogen peroxide does not affect bacterial growth when trxA is deleted, trxB is significantly induced when bacteria are treated with H2O2 [49]. Consistently, our results showed that the deletion of trxA or trxC negatively affects the growth and survival rate of E. piscicida when in the presence of hydrogen peroxide. However, trxB mutation exhibits a contrary result, namely better growth and higher survival rate than the wild strain under H2O2 stress. Similar results have been reported in Neisseria gonorrhoeae and E. coli, in which the trxB mutant strains exhibit significantly higher resistance to H2O2 stress [47,48]. While the intracellular reductive capacity of the trxB mutant is similar to ΔtrxA and ΔtrxC, which is to say that it is less than the wild strain, we speculate that the deficiency of thioredoxin reductase TrxB probably activates an unknown catalase. Such an interesting result inspires us to explore another oxidant, diamide, which is a thiol-specific oxidant. For example, in Bacillus subtilis and Staphylococcus aureus, diamide has been shown to induce the expression of trxB [40,50]. In E. coli, S. aureus, and L. monocytogenes, the expressions of trxA and trxC also rise when challenged with diamide [39,49,51]. Diamide is shown to oxidize glutathione (GSH) to the disulfide (GSSG) [52]. As expected, we found that under diamide stress, the deletion of trxBEp tremendously reduced bacterial growth, whether in solid or liquid medium. In the presence of diamide, the intracellular reductive capacities of three mutants were consistent and were all lower than the wild strain, while the survival rate of three mutants were inconsistent—ΔtrxA was remarkably lower than ΔtrxB and ΔtrxC, despite the latter two being lower than WT. The difference between the growth curve and survival rate may be caused by the discrepancy between continuous and transient oxidation stress. These findings illustrate that the thioredoxin system of E. piscicida is an important factor in bacterial stress tolerance, especially to oxidative stress. However, the mechanism through which the thioredoxin system responds to different types of oxidants should be explored in the future.
Studies of the effect of the thioredoxin system on motility have mainly focused on trxB [25,53]. In this study, three components of the Trx system were investigated, and the results showed that the deletion of trxBEp, trxAEp, and trxCEp all weaken the motility of E. piscicida. In particular, the swarming of ΔtrxB is almost completely irradicated, and, consistently, ΔtrxB has lost its flagella, which indicates that trxBEp is indispensable for flagellar formation and motility in E. piscicida. Similarly, in E. coli, a motility assay has shown that the mutation of trxB caused a strong inhibition of swarming but not of swimming [53]. It is unlikely that, in L. monocytogenes, trxA is essential for bacterial motility by maintaining the reduced intracellular monomer status of MogR, the key regulator for flagellar formation [49]. These reports and our results indicate that the Trx system is requisite for bacterial motility.
Reports indicate that the thioredoxin system plays an important role in bacterial virulence [54,55,56]. For example, in the plant pathogen Fusarium graminearum, thioredoxin reductase is required for virulence [57]. In the human pathogen Aspergillus fumigatus, thioredoxin reductase is also important for its successful infection [58]. Likely, the lack of TrxA in Acinetobacter baumannii is associated with decreased expression of type IV pili-related genes and attenuated virulence [21]. Similarly, in our case, the mutation of trxAEp or trxCEp lead to reduced survival and replication in host phagocyte, despite the mutation of trxBEp not having any effect. At the individual level, the deletions of three genes led to a significant decrease in the ability of E. piscicida to infect tilapia tissues. One of the important reasons for this is that the deficiency of the thioredoxin system led to a considerable reduction in resistance to bacterial serum and host oxidative killing. These results illustrate that the thioredoxin system plays an important role in the virulence of E. piscicida.
In conclusion, we investigated for the first time the function of the thioredoxin system in E. piscicida. The system is essential to resisting the oxidative stress induced by a direct oxidant such as hydrogen peroxide and the thiol-specific oxidizing agent diamide. The thioredoxin system also plays a role in bacterial motility and flagellum formation. More importantly, the thioredoxin system is required for the pathogenicity of E. piscicida. These findings support the conclusion that the thioredoxin system is a stress resistance factor and virulence factor of E. piscicida, which provides insights into the pathogenic mechanism of E. piscicida.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11040827/s1, Figure S1: Expression and purification of rTrxBEp. The rTrxBEp carrying His-Tag is highly purified by affinity chromatography with Ni-NTA. The band indicated by the red arrow is rTrxBEp protein, which mainly appears in the SDS-PAGE gel stained with Coomassie blue. Lane M: PageRuler Prestained Protein Ladder (Thermo Scientific, US); lane 1: non-induced sample; lane 2: IPTG-induced sample; lane 3: purified sample; lane 4: renatured sample; Figure S2: Determination of antiserum titer by ELISA. (A) the antibody reacted with purified rTrxBEp protein by continuous dilution (from 1000-fold, 2000-fold, 5000-fold, 10,000-fold to 50,000-fold). The unimmunized mice serum was used as a negative control. (B) the ratio of absorbance between anti-rTrxBEp serum and negative control serum. When the dilution ratio was 1:50,000, the absorbance ratio of anti-rTrxBEp serum to negative control serum was 4.5; Figure S3: The thioredoxin reductase activity of the rTrxBEp. (A) the content of rOmpREp (control) and rTrxBEp was measured using the BCA Protein Assay. (B) based on OD412 analysis the activity of thioredoxin reductase compared with the control rOmpREp; Figure S4: The sequences knockout location map of trxA, trxB, trxC. Retained sequences are indicated by red shapes and knocked-out regions are indicated by blue shapes; Table S1: Strains, plasmids used in this study; Table S2: Oligonucleotide primers used in this study.
Author Contributions
Conceptualization, Y.H. and H.G.; investigation, J.H. and S.L.; data curation, Q.F.; writing—original draft preparation, H.G.; writing—review and editing, Y.H.; funding acquisition, Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was jointly supported by the grants from the National Natural Science Foundation of China (32273184), Hainan Provincial Natural Science Foundation of China (ZDYF2022XDNY246; 420QN339), the Central Public-Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (19CXTD-32), and the Shandong Provincial Natural Science Foundation of China (ZR2019BC102).
Data Availability Statement
The data in this study are readily available upon reasonable request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Napier, J.A.; Haslam, R.P.; Olsen, R.E.; Tocher, D.R.; Betancor, M.B. Agriculture can help aquaculture become greener. Nature Food 2020, 1, 680–683. [Google Scholar] [CrossRef]
- Hoshina, T. On a new bacterium, Paracolobactrum anguillimortiferum n. sp. Bull. Jpn. Soc. Sci. Fish 1962, 28, 162–164. [Google Scholar] [CrossRef]
- Ewing, W.H.; Mcwhorter, A.C.; Escobar, M.R.; Lubin, A.H. Edwardsiella, a new genus of Enterobacteriaceae based on a new species, E. tarda. Int. Bull. Bacteriol. Nomencl. Taxon. 1965, 15, 33–38. [Google Scholar] [CrossRef]
- Leung, K.Y.; Wang, Q.; Yang, Z.; Siame, B.A. Edwardsiella piscicida: A versatile emerging pathogen of fish. Virulence 2019, 10, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Peng, Y.; Ahmed, M.A.H.; Ma, J.; Xu, R.; Zhang, Y.; Ma, Y.; Wang, Q. PepA binds to and negatively regulates esrB to control virulence in the fish pathogen Edwardsiella piscicida. Microbiol. Res. 2020, 232, 126349. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Lai, Q.; Liu, Q.; Wu, H.; Xiao, J.; Shao, Z.; Wang, Q.; Zhang, Y. Phylogenomics characterization of a highly virulent Edwardsiella strain ET080813(T) encoding two distinct T3SS and three T6SS gene clusters: Propose a novel species as Edwardsiella anguillarum sp. nov. Syst. Appl. Microbiol. 2015, 38, 36–47. [Google Scholar]
- Park, S.B.; Aoki, T.; Jung, T.S. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 2012, 43, 67. [Google Scholar] [CrossRef]
- Jung, W.J.; Kwon, J.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kang, J.W.; Lee, S.B.; Lee, Y.M.; Oh, W.T.; Jun, J.W.; et al. Isolation and characterization of a highly virulent Edwardsiella piscicida strain responsible for mass mortality in marbled eel (Anguilla marmorata) cultured in Korea. Aquaculture 2022, 555, 738199. [Google Scholar] [CrossRef]
- Yan, M.; Liu, J.; Li, Y.; Wang, X.; Jiang, H.; Fang, H.; Guo, Z.; Sun, Y. Different concentrations of Edwardsiella tarda ghost vaccine induces immune responses in vivo and protects Sparus macrocephalus against a homologous challenge. Fish Shellfish. Immunol. 2018, 80, 467–472. [Google Scholar] [CrossRef]
- Chen, H.; Yang, D.; Han, F.; Tan, J.; Zhang, L.; Xiao, J.; Zhang, Y.; Liu, Q. The Bacterial T6SS Effector EvpP Prevents NLRP3 Inflammasome Activation by Inhibiting the Ca2+-Dependent MAPK-Jnk Pathway. Cell Host Microbe 2017, 21, 47–58. [Google Scholar] [CrossRef]
- Du, C.M.; Huo, X.P.; Gu, H.J.; Wu, D.; Hu, Y.H. Acid resistance system CadBA is implicated in acid tolerance and biofilm formation and is identified as a new virulence factor of Edwardsiella tarda. Vet. Res. 2021, 52, 117. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.D.; Zhang, M.; Cheng, S.; Sun, L. Analysis of Edwardsiella tarda DegP, a serine protease and a protective immunogen. Fish Shellfish Immunol. 2010, 28, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.Y.; Siame, B.A.; Tenkink, B.J.; Noort, R.J.; Mok, Y.K. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012, 14, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yu, T.; Dong, X.; Zhang, Z.; Song, L.; Xu, Y.; Zhang, X.H. Edwardsiella tarda invasion of fish cell lines and the activation of divergent cell death pathways. Vet. Microbiol. 2013, 163, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; He, T.T.; Li, D.Y.; Liu, L.Y.; Nie, P.; Xie, H.X. The Edwardsiella piscicida Type III Effector EseJ Suppresses Expression of Type 1 Fimbriae, Leading to Decreased Bacterial Adherence to Host Cells. Infect. Immun. 2019, 87, e00187-e19. [Google Scholar] [CrossRef]
- Leung, K.Y.; Wang, Q.; Zheng, X.; Zhuang, M.; Yang, Z.; Shao, S.; Achmon, Y.; Siame, B.A. Versatile lifestyles of Edwardsiella: Free-living, pathogen, and core bacterium of the aquatic resistome. Virulence 2022, 13, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.J.; Soto, E.; Wise, D.J. Edwardsiellosis. In Climate Change and Infectious Fish Diseases; Woo, P.T.K., Leong, J.A., Buchmann, K., Eds.; CABI: Wallingford, UK, 2020; pp. 235–264. [Google Scholar]
- Ahmed, M.A.H.; Ma, J.; Shao, S.; Wang, Q.; Xu, R.; Yin, K. Regulation mechanism of virulence by environmental acidic stress mediated by Prc in Edwardsiella piscicida. Aquaculture 2023, 565, 739092. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.M.; Lee, R.T. Thioredoxin and thioredoxin target proteins: From molecular mechanisms to functional significance. Antioxid. Redox Signal. 2013, 18, 1165–1207. [Google Scholar] [CrossRef]
- Ren, X.; Zou, L.; Lu, J.; Holmgren, A. Selenocysteine in mammalian thioredoxin reductase and application of ebselen as a therapeutic. Free Radic. Biol. Med. 2018, 127, 238–247. [Google Scholar] [CrossRef]
- May, H.C.; Yu, J.J.; Zhang, H.; Wang, Y.; Cap, A.P.; Chambers, J.P.; Guentzel, M.N.; Arulanandam, B.P. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS ONE 2019, 14, e0218505. [Google Scholar] [CrossRef]
- Tang, J.; Wang, X.; Yin, J.; Han, Y.; Yang, J.; Lu, X.; Xie, T.; Akbar, S.; Lyu, K.; Yang, Z. Molecular characterization of thioredoxin reductase in waterflea Daphnia magna and its expression regulation by polystyrene microplastics. Aquat. Toxicol. 2019, 208, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, I.; Ponniresan, V.K.; Govindasamy, K.; Prasad, N.R. Understanding the survival mechanisms of Deinococcus radiodurans against oxidative stress by targeting thioredoxin reductase redox system. Arch. Microbiol. 2020, 202, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Yang, J.; Ji, S.; Li, T.; Gao, S.Q.; Xu, H.P. Cancer Therapy by Targeting Thioredoxin Reductase Based on Selenium-Containing Dynamic Covalent Bond. CCS Chem. 2020, 2, 225–235. [Google Scholar] [CrossRef]
- Ju, H.Q.; Lin, J.F.; Tian, T.; Xie, D.; Xu, R.H. NADPH homeostasis in cancer: Functions, mechanisms and therapeutic implications. Signal Transduct. Target. Ther. 2020, 5, 231. [Google Scholar] [CrossRef]
- Balsera, M.; Buchanan, B.B. Evolution of the thioredoxin system as a step enabling adaptation to oxidative stress. Free Radic. Biol. Med. 2019, 140, 28–35. [Google Scholar] [CrossRef]
- Zhang, W.W.; Sun, K.; Cheng, S.; Sun, L. Characterization of DegQVh, a serine protease and a protective immunogen from a pathogenic Vibrio harveyi strain. Appl. Environ. Microbiol. 2008, 74, 6254–6262. [Google Scholar] [CrossRef]
- Xie, J.H.; Zhao, Q.; Huang, H.; Fang, Z.; Hu, Y.H. Edwardsiella piscicida HigB: A type II toxin that is essential to oxidative resistance, biofilm formation, serum survival, intracellular propagation, and host infection. Aquaculture 2021, 535, 736382. [Google Scholar] [CrossRef]
- Fang, Q.J.; Han, Y.X.; Shi, Y.J.; Huang, H.Q.; Fang, Z.G.; Hu, Y.H. Universal stress proteins contribute Edwardsiella piscicida adversity resistance and pathogenicity and promote blocking host immune response. Fish Shellfish Immunol. 2019, 95, 248–258. [Google Scholar] [CrossRef]
- Hu, Y.H.; Liu, C.S.; Hou, J.H.; Sun, L. Identification, characterization, and molecular application of a virulence-associated autotransporter from a pathogenic Pseudomonas fluorescens strain. Appl. Environ. Microbiol. 2009, 75, 4333–4340. [Google Scholar] [CrossRef]
- Zheng, L.; Zhu, H.Z.; Wang, B.T.; Zhao, Q.H.; Du, X.B.; Zheng, Y.; Jiang, L.; Ni, J.Z.; Zhang, Y.; Liu, Q. Sodium selenate regulates the brain ionome in a transgenic mouse model of Alzheimer’s disease. Sci. Rep. 2016, 6, 39290. [Google Scholar] [CrossRef]
- Cai, N.L.; Lau, A.T.Y.; Yu, F.Y.; Wu, D.D.; Dai, L.J.; Mo, H.Y.; Lin, C.M.; Xu, Y.M. Purification and characterization of a highly specific polyclonal antibody against human extracellular signal-regulated kinase 8 and its detection in lung cancer. PLoS ONE 2017, 12, e0184755. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fu, X.; Shen, J.; Zhang, H.; Hong, W.; Chang, Z. Periplasmic proteins of Escherichia coli are highly resistant to aggregation: Reappraisal for roles of molecular chaperones in periplasm. Biophys. Chem. 2004, 316, 795–801. [Google Scholar] [CrossRef]
- Paunkov, A.; Kupc, M.; Soki, J.; Leitsch, D. Characterization of the components of the thioredoxin system in Bacteroides fragilis and evaluation of its activity during oxidative stress. Anaerobe 2022, 73, 102507. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, B.; Liu, Y.; Shi, C.; Fratamico, P.M.; Zhang, L.; Wang, D.; Zhang, J.; Cui, Y.; Xu, P. Two homologous Salmonella serogroup C1-specific genes are required for flagellar motility and cell invasion. BMC Genom. 2021, 22, 507. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, R.; Pan, W.; Xu, Z.; Yang, H.; Luo, Q.; Ye, X.; Cheng, X. Effects of L-carnitine combined with pancreatic kininogenase on thioredoxin 2, thioredoxin reductase 1, and sperm quality in patients with oligoasthenospermia. Transl. Androl. Urol. 2021, 10, 3515–3523. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Gu, H.; Shi, Y.; Huang, H.; Sun, D.; Hu, Y. Edwardsiella piscicida YefM-YoeB: A Type II Toxin-Antitoxin System That Is Related to Antibiotic Resistance, Biofilm Formation, Serum Survival, and Host Infection. Front. Microbiol. 2021, 12, 646299. [Google Scholar] [CrossRef]
- Wang, D.; Gong, C.; Gu, H.; Huang, H.; Xian, J.; Hu, Y. Bicistronic operon YhaO-YhaM contributes to antibiotic resistance and virulence of pathogen Edwardsiella piscicida. Aquaculture 2021, 541, 736849. [Google Scholar] [CrossRef]
- Uziel, O.; Borovok, I.; Schreiber, R.; Cohen, G.; Aharonowitz, Y. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 2004, 186, 326–334. [Google Scholar] [CrossRef]
- Sroka, J.; Antosik, A.; Czyz, J.; Nalvarte, I.; Olsson, J.M.; Spyrou, G.; Madeja, Z. Overexpression of thioredoxin reductase 1 in-hibits migration of HEK-293 cells. Biol. Cell 2007, 99, 677–687. [Google Scholar] [CrossRef]
- Mohamed, F.A.; Shaker, G.H.; Askoura, M.M. Oxidative Stress Influences Pseudomonas aeruginosa Susceptibility to Antibiotics and Reduces Its Pathogenesis in Host. Curr. Microbiol. 2020, 77, 479–490. [Google Scholar] [CrossRef]
- Ye, J.; Su, Y.; Peng, X.; Li, H. Reactive Oxygen Species-Related Ceftazidime Resistance Is Caused by the Pyruvate Cycle Perturbation and Reverted by Fe3 + in Edwardsiella tarda. Front. Microbiol. 2021, 12, 654783. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Yang, F.; Li, H.; So, P.K.; Yao, Z.; Xia, W.; Sun, H. Targeting the Thioredoxin Reductase-Thioredoxin System from Staphylococcus aureus by Silver Ions. Inorg. Chem. 2017, 56, 14823–14830. [Google Scholar] [CrossRef] [PubMed]
- Lata, S.; Akif, M. Structure-based identification of natural compound inhibitor against M. tuberculosis thioredoxin reductase: Insight from molecular docking and dynamics simulation. J. Biomol. Struct. Dyn. 2021, 39, 4480–4489. [Google Scholar] [CrossRef] [PubMed]
- Fomenko, D.E.; Gladyshev, V.N. Identity and Functions of CxxC-Derived Motifs. Biochemistry 2003, 42, 11214–11225. [Google Scholar] [CrossRef]
- Missirlis, F.; Ulschmid, J.K.; Hirosawa-Takamori, M.; Grönke, S.; Schäfer, U.; Becker, K.; Phillips, J.P.; Jäckle, H. Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J. Biol. Chem. 2002, 277, 11521–11526. [Google Scholar] [CrossRef]
- Takemoto, T.; Zhang, Q.M.; Yonei, S. Different Mechanisms of Thioredoxin in its Reduced and Oxidized Forms in Defense Against Hydrogen Peroxide in Escherichia coli. Free Radic. Biol. Med. 1998, 24, 556–562. [Google Scholar] [CrossRef]
- Ritz, D.; Patel, H.; Doan, B.; Zheng, M.; Åslund, F.; Storz, G.; Beckwith, J. Thioredoxin 2 Is Involved in the Oxidative Stress Response in Escherichia coli. J. Biol. Chem. 2000, 275, 2505–2512. [Google Scholar] [CrossRef]
- Cheng, C.; Dong, Z.; Han, X.; Wang, H.; Jiang, L.; Sun, J.; Yang, Y.; Ma, T.; Shao, C.; Wang, X. Thioredoxin A Is Essential for Motility and Contributes to Host Infection of Listeria monocytogenes via Redox Interactions. Front. Cell Infect. Microbiol. 2017, 7, 287. [Google Scholar] [CrossRef]
- Leichert, L.I.; Scharf, C.; Hecker, M. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 2003, 185, 1967–1975. [Google Scholar] [CrossRef]
- Zeller, T.; Klug, G. Thioredoxins in bacteria: Functions in oxidative stress response and regulation of thioredoxin genes. Sci. Nat. 2006, 93, 259–266. [Google Scholar] [CrossRef]
- Kosower, N.S.; Kosower, E.M.; Wertheim, B.; Correa, W.S. Diamide, a new reagent for the intracellular oxidation of glutathione to the disulfide. Biochem. Biophys. Res. Commun. 1969, 37, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Shingaki, R.; Hirose, S.; Waki, K.; Mori, H.; Fukui, K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 2007, 189, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.R.; Tzianabos, A.O.; Smith, C.J. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J. Bacteriol. 2007, 189, 8015–8023. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Potter, A.J.; Kidd, S.P.; Edwards, J.L.; Falsetta, M.L.; Apicella, M.A.; Jennings, M.P.; McEwan, A.G. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J. Infect. Dis. 2009, 199, 227–235. [Google Scholar] [CrossRef]
- Sem, X.; Rhen, M. Pathogenicity of Salmonella enterica in Caenorhabditis elegans relies on disseminated oxidative stress in the infected host. PLoS ONE 2012, 7, e45417. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Du, J.; Huang, Z.; Fang, A.; Yang, Y.; Bi, C.; Qing, L.; Yu, Y. Sclerotinia sclerotiorum Thioredoxin Reductase Is Required for Oxidative Stress Tolerance, Virulence, and Sclerotial Development. Front. Microbiol. 2019, 10, 233. [Google Scholar] [CrossRef]
- Binder, J.; Shadkchan, Y.; Osherov, N.; Krappmann, S. The Essential Thioredoxin Reductase of the Human Pathogenic Mold Aspergillus fumigatus Is a Promising Antifungal Target. Front. Microbiol. 2020, 11, 1383. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).