1. Introduction
Many studies have focused on investigating the relationship between pathogenicity in plants and their associated microorganisms. However, through various analyses and studies on the microbial diversity related to different plant species, it has been suggested that only a small proportion of the microorganisms that interact with plants are actually harmful [
1]. The bulk of the bacteria that reside inside plants contribute significantly to the health and development of the plant, even if they may sometimes be neutral [
2,
3].
Endophytes are microorganisms, such as fungi, actinomycetes, or mycoplasma, that reside within plant tissues. Over 200 genera and 16 phyla of bacterial species have been identified as associated with endophytes, with the majority belonging to the Actinobacteria, Proteobacteria, and Firmicutes phyla [
4].
Endophytic fungi have been found to produce compounds with antimicrobial, antitumor, and antioxidant activities [
5]. Moreover, endophytic fungi can also increase the host plant’s tolerance to abiotic and biotic stress, including drought, salinity, disease, and herbivores. Due to their potential applications in various fields, the study of endophytic fungi has received increasing attention in recent years [
6].
Endophytes have demonstrated the ability to produce a diverse array of bioactive metabolites with promising applications in the food, cosmetics, pharmaceutical, and agricultural industries. Furthermore, these metabolites hold potential as therapeutic agents for treating a wide range of diseases [
7,
8]. The characterized secondary metabolites included alkaloids, benzopyranones, chinones, flavonoids, phenolic acids, quinones, steroids, saponins, tannins, terpenoids, tetralones, and xanthones, as well as several other functional groups [
7]. Alzheimer’s disease (AD), a prevalent form of dementia, is a progressive neurological ailment that primarily affects the elderly [
9]. The medial temporal lobe and neocortical structures of the brain are heavily impacted by the accumulation of amyloid-beta peptide (Aβ) [
10,
11].
Mild cognitive impairment (MCI) is a stage between older adults and Alzheimer’s disease (AD) that is not significantly impaired in daily life. Although not all MCI patients develop AD and remain cognitively stable for years, the rate of advancement is estimated to be between 10% and 15% every year [
12]. The 2015 World Alzheimer Report revealed that approximately 46.8 million individuals worldwide suffered from dementia, with an estimated global social cost of
$818 billion USD. Alzheimer’s disease (AD) is the most prevalent type of dementia, accounting for 60–70% of all dementia cases. Degradation in areas rich in cholinergic neurons, such as the nucleus basalis of Meynert, frontal cortex, anterior cingulate cortex, and posterior cingulate cortex, has been linked to memory loss, irritability, and apathy in individuals with Alzheimer’s disease, according to pathological data. Acetylcholine (ACh), which is closely linked to memory function, including memory encoding, consolidation, and retrieval, has been strongly associated with this disease [
13].
The main objective of this study was to investigate the diversity of endophytic fungi in Juncus rigidus, a common shrub species found in the Wadi-El Natron Valley in Egypt, with the aim of identifying new species or strains that produce valuable bioactive compounds. This study focused on assessing the diversity of endophytic fungi found in various healthy tissues of Juncus rigidus and their potential anti-activities. This research is the first report to our knowledge to evaluate the biodiversity, phylogeny, and anti-acetylcholinesterase activity of endophytic fungi present in Juncus rigidus.
4. Discussion
Endophytic fungi are symbiotic microorganisms that live inside plants without causing any harm to the host plant. Endophytes may actively or passively stimulate plant development via a range of processes, resulting in higher host fitness and plant resilience to biotic and abiotic stressors. These fungi can produce a wide range of biologically active secondary metabolites that have various pharmacological properties. As a result, endophytic fungi have become an attractive source of novel bioactive compounds for drug discovery [
31]. By inducing the solubilization of phosphorus, potassium, and zinc and inducing the host plant’s defensive response against phytopathogens via a variety of methods, fungal endophytes either directly or indirectly support the development of plants. Auxin, abscisins, ethylene, and gibberellins are a few examples of plant hormones that may be changed by these mechanisms, along with competition, niche exclusion, and siderophore synthesis [
32]. Pathogens can also be directly antagonistic via antibiosis, parasitosis, or predation. The photochemistry of the host may be related to variations in endophytes throughout plants and tissues, according to prior research [
33]. Based on the current findings,
Juncus rigidus is a wild plant that harbors a significant number of endophytic fungi, which may be attributable to its greater active chemical concentration [
34].
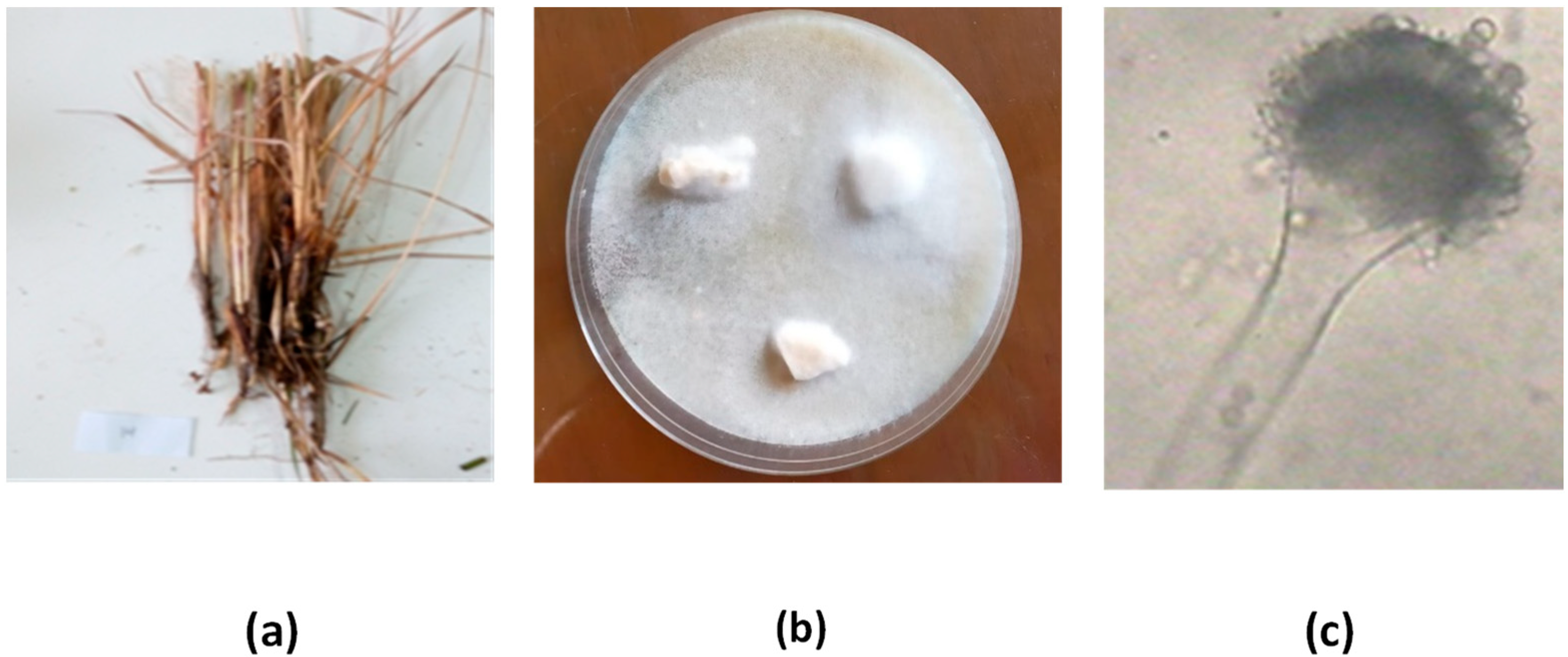
The goal of this research was to find a natural compound that could be used to treat Alzheimer’s disease. As a result of their unique living conditions, endophytic marine microorganisms frequently create bioactive compounds with novel activities and structures. Thus, a total of eight endophytic fungi were recovered from
Juncus rigidus. Due to their tremendous structural variety and complexity, endophyte-isolated bioactive natural chemicals have played a significant role in the search for new therapeutics [
35].
Isolation of endophytic fungi from plants is a crucial step in exploring the diversity of endophytic fungi and their potential as sources of novel bioactive compounds. The process of isolation involves surface sterilization of the plant samples to eliminate any epiphytic fungi or bacteria that may contaminate the samples. After surface sterilization, different plant parts, such as leaves, stems, and roots, are aseptically removed and cultured on suitable media. Several studies have reported the isolation of endophytic fungi from various plant species. For example, one study isolated endophytic fungi from the leaves and stems of the medicinal plant
Carpesium abrotanoides. The study identified 41 endophytic fungal strains belonging to 18 different genera, and many of these strains exhibited antimicrobial and antioxidant activities [
36]. Another study isolated endophytic fungi from the bark of the medicinal plant
Maytenus ilicifolia. The study identified 102 fungal strains belonging to 28 different genera, and many of these strains exhibited cytotoxic and antifungal activities [
37].
Furthermore, a study isolated endophytic fungus from the roots of the medicinal plant
Panax notoginseng. The study identified 116 fungal strains belonging to 22 different genera, and many of these strains exhibited antioxidant and anti-inflammatory activities [
37] Overall, the isolation of endophytic fungi from plants has the potential to lead to the discovery of novel bioactive compounds with various pharmacological properties. The diversity of endophytic fungi found in different plant species highlights the importance of exploring different plant sources for the isolation of these microorganisms.
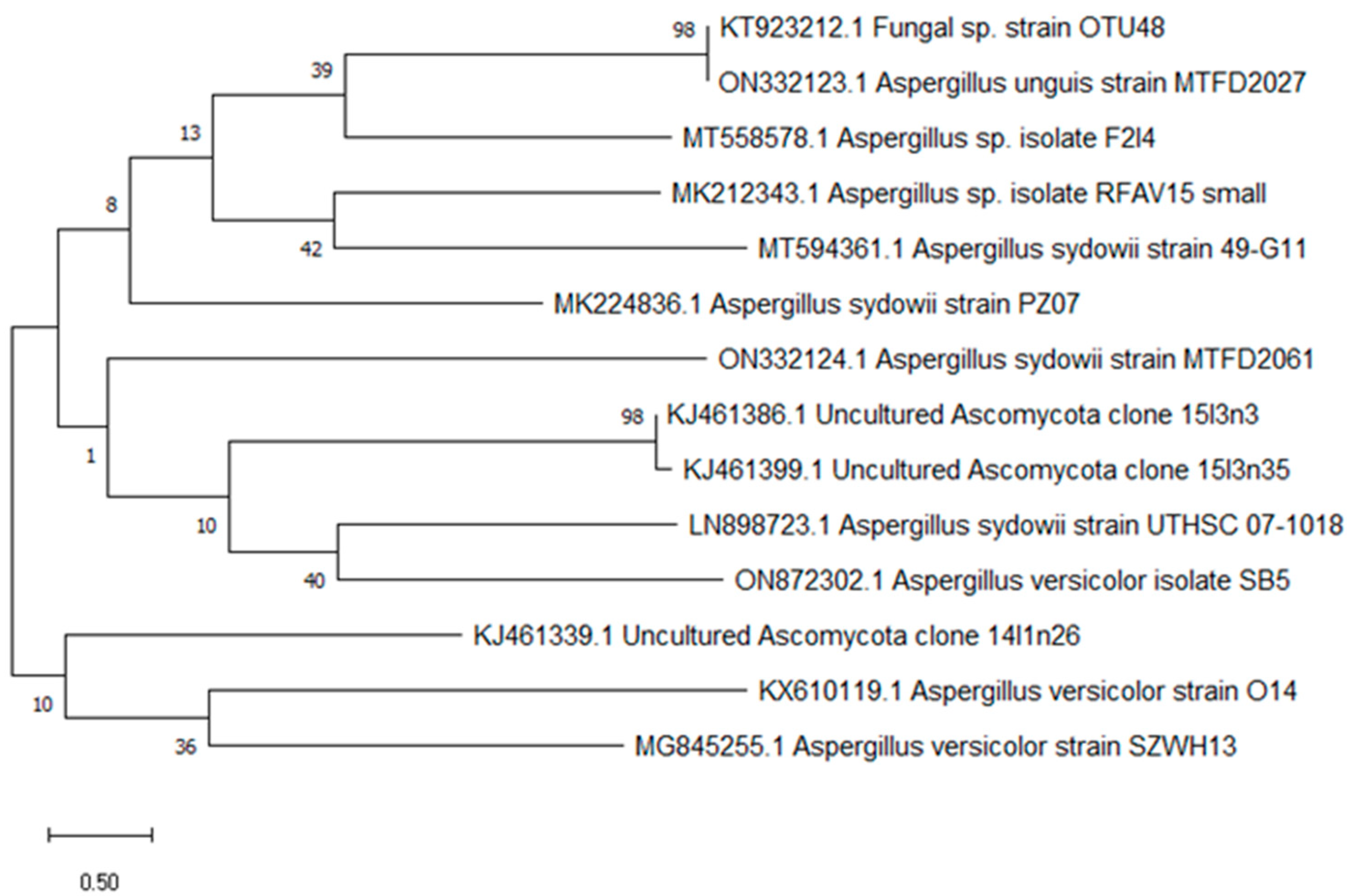
The ability of ethyl acetate extracts from an isolated fungus to produce AChEIs was evaluated. Multiple isolates showed anticholinesterase activity, but only one endophytic fungal strain (SB5) was found to be the most effective, suppressing AChE at a rate of 79.5 ± 1.39. Fungal endophytes produce more secondary metabolites than other types of endophytic microbes. To identify the most potent endophyte, isolate SB5, the strain SB5 was identified visually and genetically using the 18s rRNA gene approach and was identified as Aspergillus versicolor SB5. It has been deposited in GenBank with accession number ON872302.1.
Fractionation and separation of the crude extract of
Aspergillus versicolor SB5 was done by flash column chromatography and sephadex LH20 according to the AChE activity of the obtained fractions (
Table 2 and
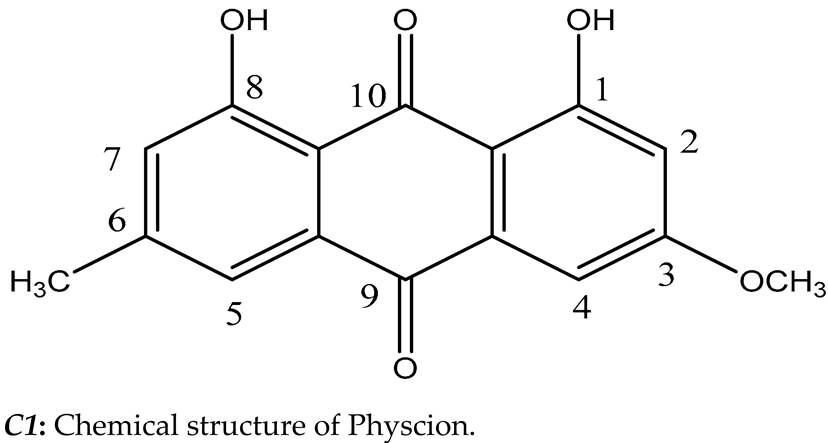
Table 3). Then, the most potent compound was identified as Physcion (1,8-dihydroxy-3-methoxy-6-methylanthracene-9,10-dione) based on its chromatographic properties, mass analysis, and available reported data (
Figure 3). Endophytic fungi have been shown to be a perennial and abundant source of antioxidant, antimicrobial, anticancer, and anticholinesterase compounds [
5]. C1 constitutes an important class of natural compounds with a broad scope of pharmacological properties, including anti-bacterial, antioxidant, laxative, anti-tumor, and other activities. They may have also held a variety of pharmacological properties, including laxative, anti-tumor, anti-inflammatory, antibacterial, antioxidant, anti-injury, and acetylcholinesterase inhibitory properties, according to recent medical studies [
38].
C1 obtained from the fungus
Aspergillus versicolor SB5 was assayed for its inhibitory action against COX-2 and LOX-1. C1 inhibited COX-2 and LOX-1 with 74.80 ± 1.40% and 91.06 ± 1.74% at 200 µg/mL. Physcion being the main component of
Reynoutria elliptica was likely to reduce the synthesis of NO and PGE2 via attenuating iNOS and COX-2 expression and inhibit TNF-α secretion by suppressing mitogen-activated protein kinases (MAPKs) and NF-κB activation [
39]. By blocking the MEK/ERK pathway, Physcion was able to decrease Jurkat E6.1 cells’ CXCR4-mediated chemotaxis. It also reduced HSC-T6 cells’ chemotactic migration [
40].
Additionally, endophytic fungi have recently been recognized for their ability to produce antioxidant compounds that have an anti-aging effect, protect hepatic cells from damage, improve the body’s defense mechanism, offer protection against digestive disorders, and reduce obesity. Antioxidants are a family of compounds considered to be the most effective against a variety of age-related conditions, such as Alzheimer’s disease. C1 capacity to scavenge free radicals was tested using different assays at different concentration (10–50 µg/mL) in comparison to Ascorbic acid as a reference drug at the same concentrations. The main pigments of
Stemphylium lycopersici, Physcion, may be able to scavenge hydroxyl and ABTS radicals [
41]. In contrast, it has been found by several studies that C1 exhibits clear activity in the ABTS and DPPH radical-scavenging assays [
42]. It is interesting to note that C1 demonstrates anti-tumor action via causing ROS production in tumor cells, which is the reverse of its impact on free radicals. Furthermore, according to certain research, C1 produced pro-oxidant effects on human neutrophil cells and possessed DPPH and superoxide radical scavenging properties [
43].
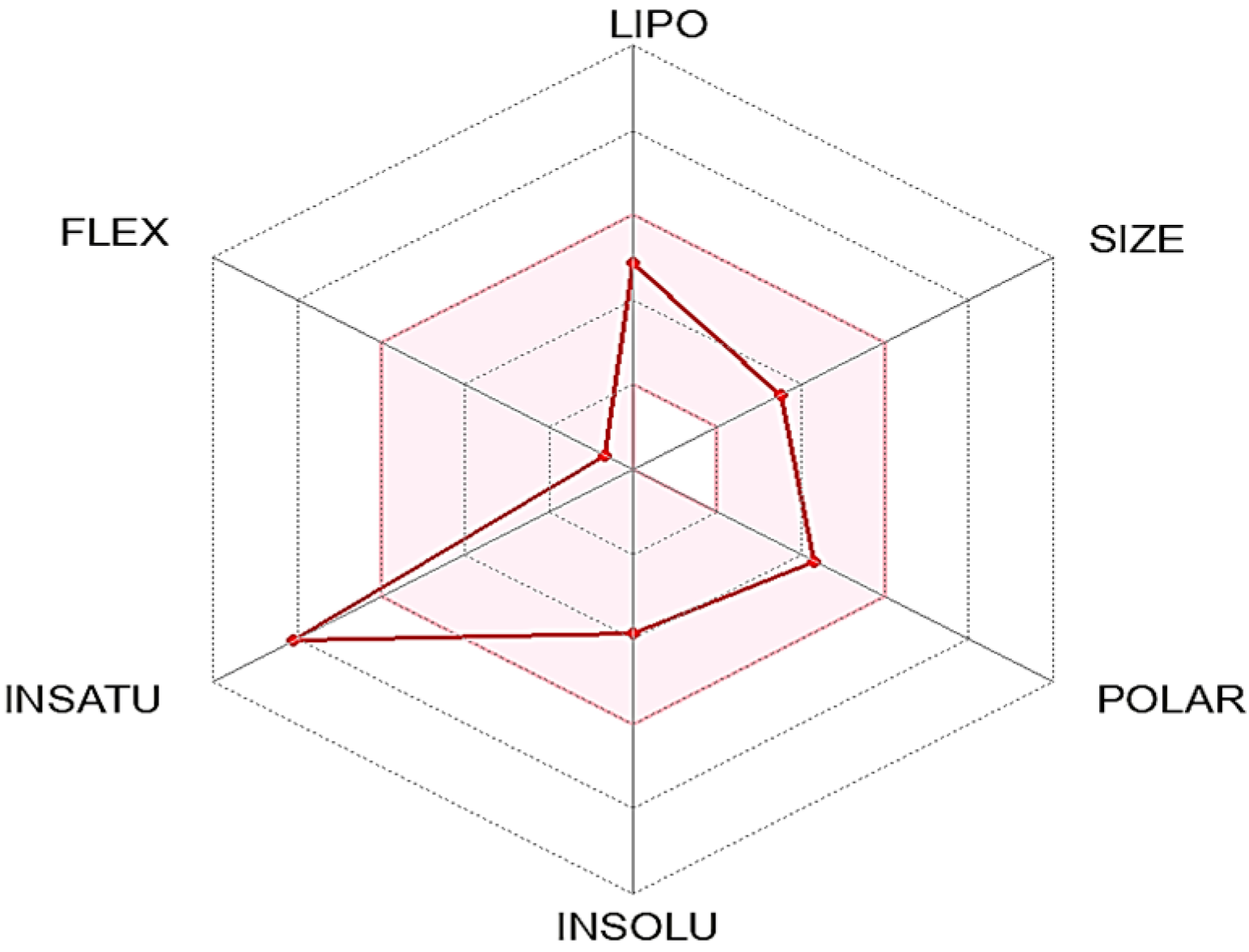
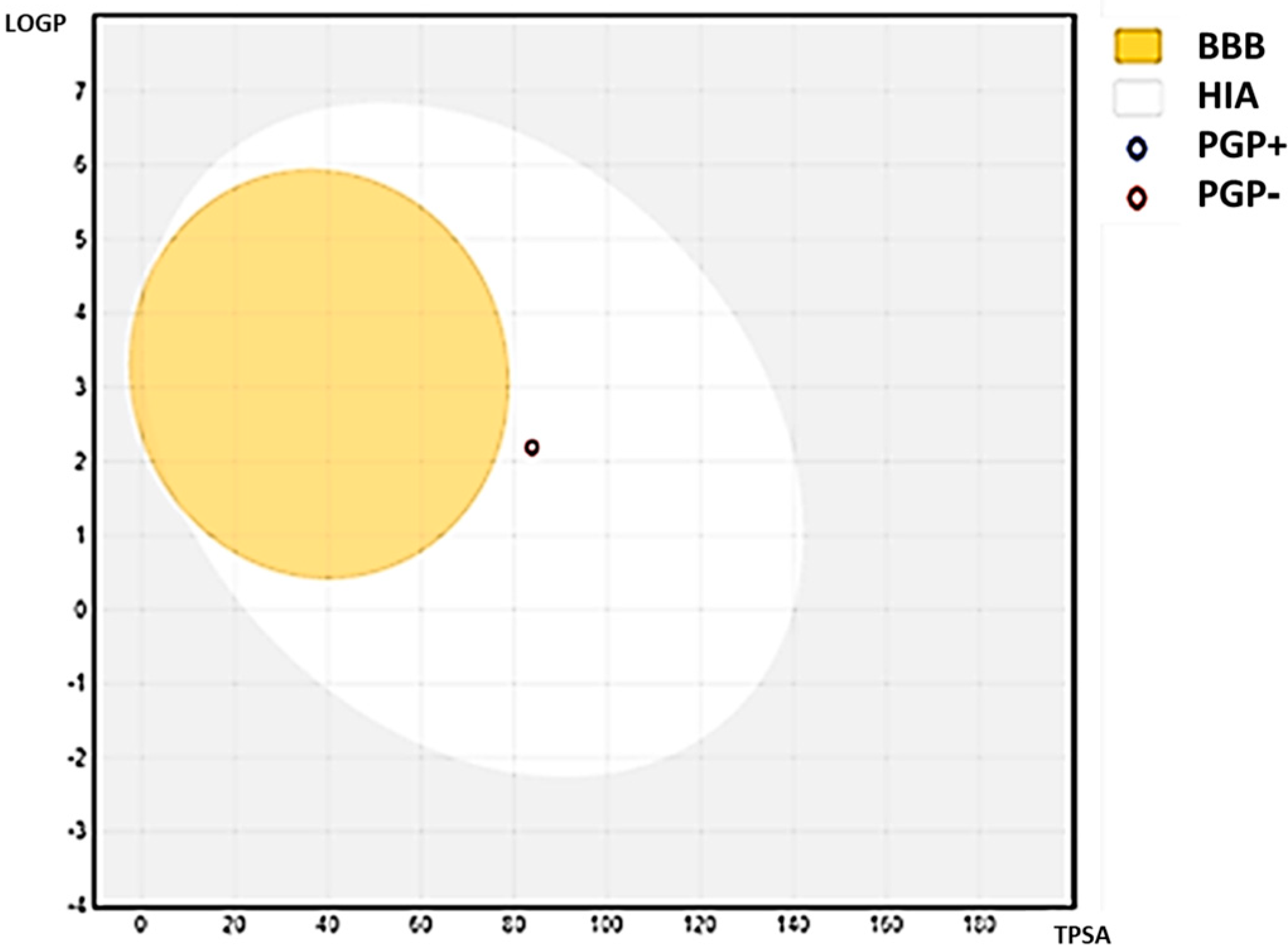
ADME properties are crucial in drug development, as they determine the drug’s effectiveness and safety in the body. Absorption refers to how a drug enters the bloodstream, and drugs can be formulated as prodrugs to optimize absorption. Distribution determines how the drug is transported to its target site and can be enhanced through targeted drug delivery systems. Metabolism involves how a drug is broken down by enzymes in the body and can be affected by genetic variations and drug interactions. Excretion refers to how the drug is eliminated from the body, primarily through the kidneys or liver, and can be influenced by the drug’s molecular weight, polarity, and ionization. By optimizing these ADME properties, drug developers can increase their chances of success in drug development and provide safe and effective treatments to patients [
44]. In this study, the ADME properties of C1 were examined and analyzed.
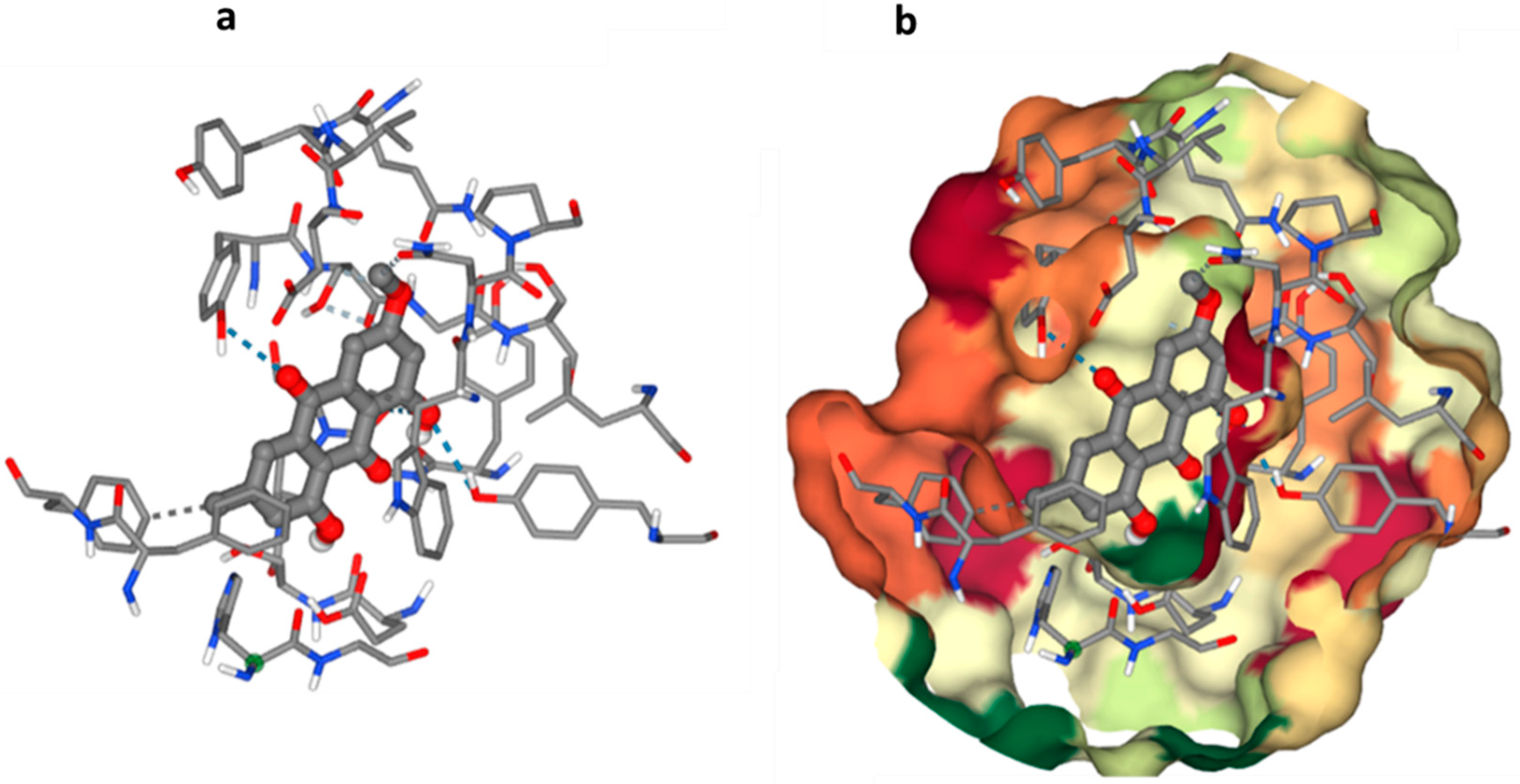
Molecular docking studies have become increasingly important in drug discovery and development, allowing for the evaluation of ligand–receptor interactions at a molecular level. In the case of cholinesterase enzymes, the goal is to identify compounds that can act as inhibitors of these enzymes. This information can then be used to identify potential inhibitors for further study and optimization. However, it is important to note that molecular docking results should be validated through experimental methods, such as in vitro and in vivo assays, to confirm the compound’s effectiveness and safety. Overall, molecular docking studies provide a valuable tool in the early stages of drug discovery, aiding in the identification of potential drug candidates for further development [
45].