Blood Stream Microbiota Dysbiosis Establishing New Research Standards in Cardio-Metabolic Diseases, A Meta-Analysis Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility and Exclusion Criteria
2.3. Data Extraction and Quality Analysis
2.4. Statistical Analysis
3. Results
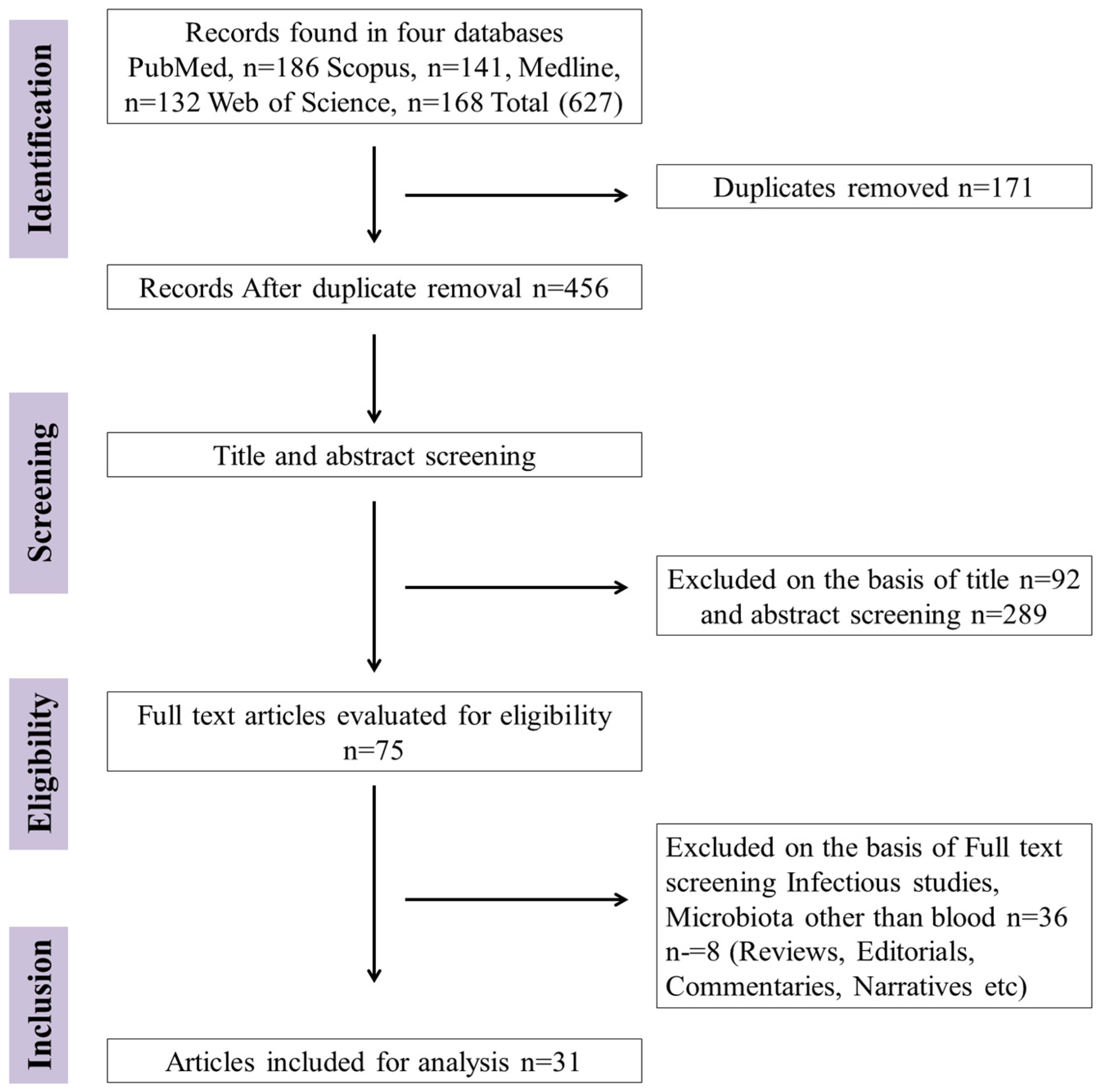
3.1. Study Selection and Exclusion
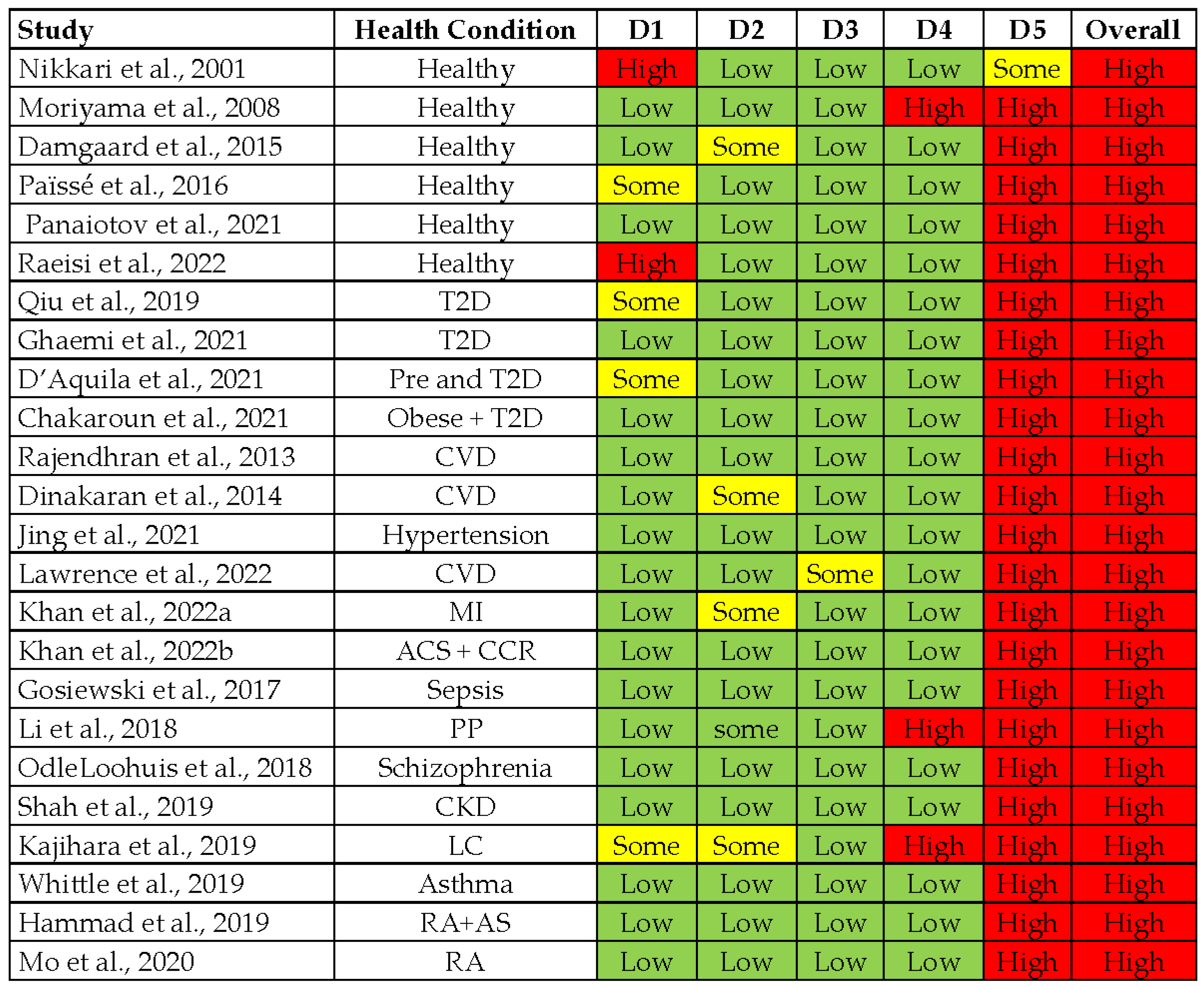
3.2. Quality Assessment of Included Studies
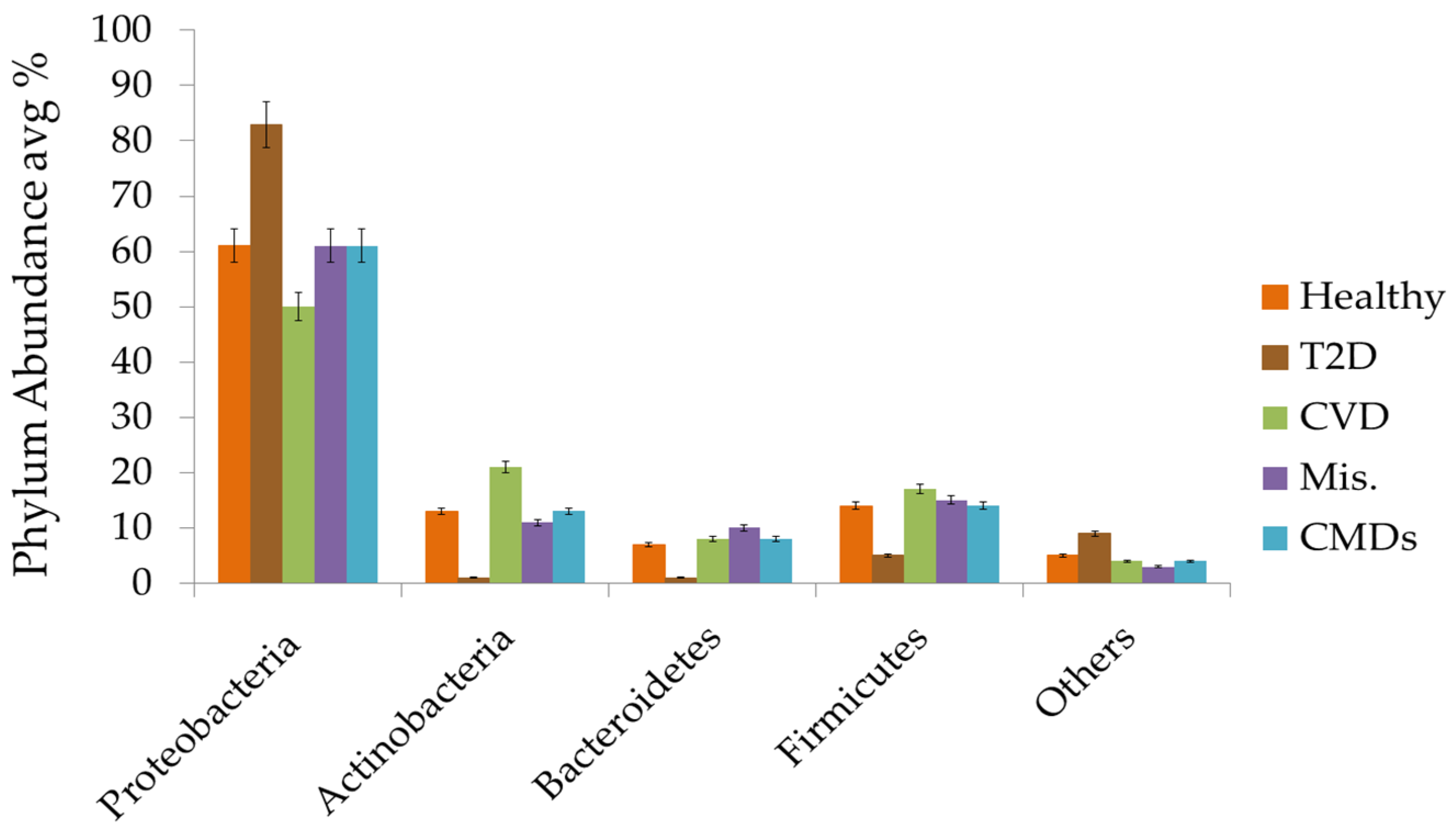
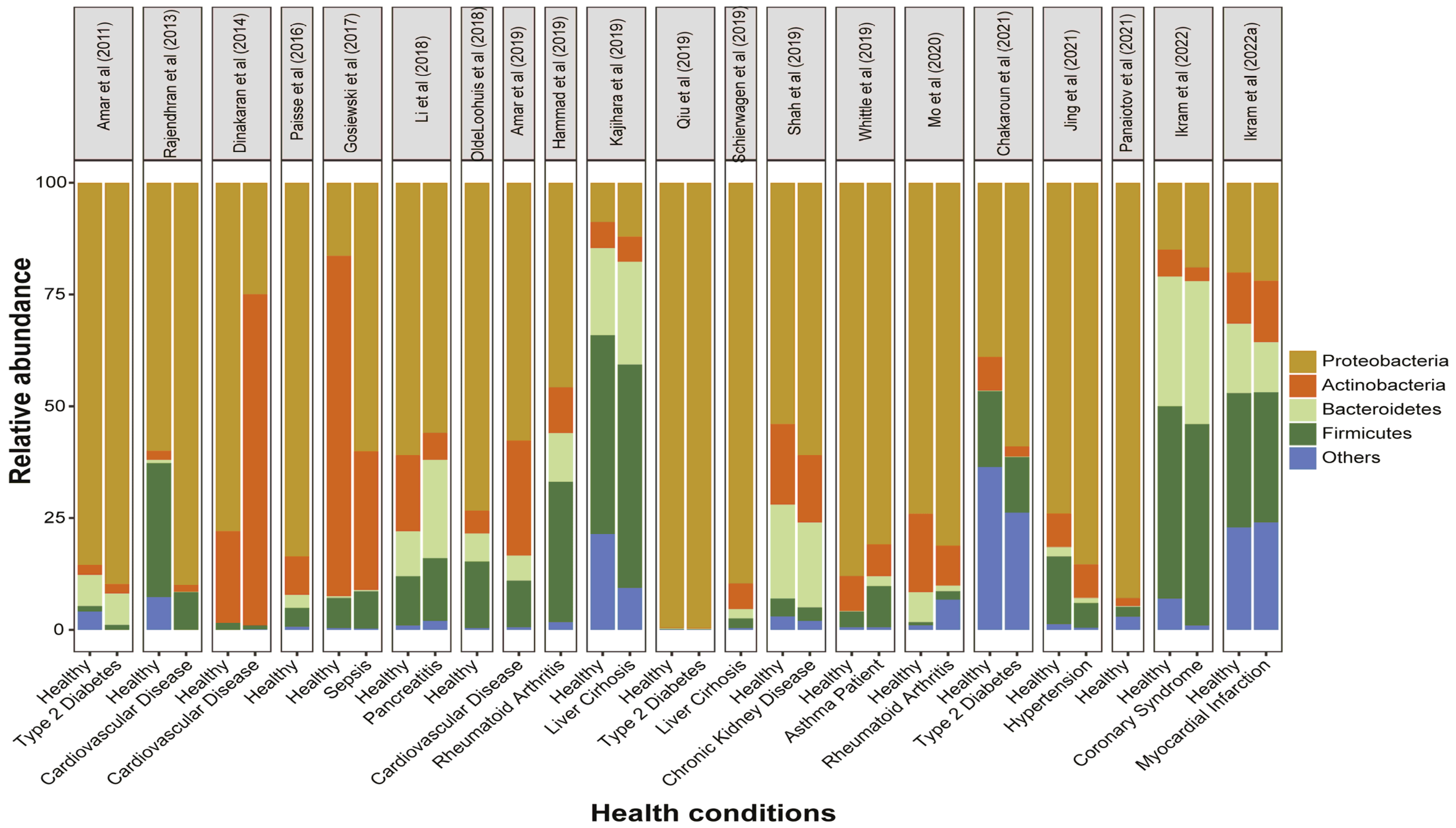
3.3. Circulating Bacterial Signatures in Healthy Individuals
3.4. Circulating Bacterial Dysbiosis and Diabetes
3.5. Circulating Bacterial Dysbiosis and Cardiovascular Diseases
3.6. Circulating Bacterial Dysbiosis and Miscellaneous Non-Infectious Diseases
4. Discussion
4.1. Study Strengths
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heidenreich, P.A.; Trogdon, J.G.; Khavjou, O.A.; Butler, J.; Dracup, K.; Ezekowitz, M.D.; Finkelstein, E.A.; Hong, Y.; Johnston, S.C.; Khera, A. Forecasting the future of cardiovascular disease in the United States: A policy statement from the american heart association. Circulation 2011, 123, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Arena, R.; Guazzi, M.; Lianov, L.; Whitsel, L.; Berra, K.; Lavie, C.J.; Kaminsky, L.; Williams, M.; Hivert, M.-F.; Cherie Franklin, N. Healthy lifestyle interventions to combat noncommunicable disease—A novel nonhierarchical connectivity model for key stakeholders: A policy statement from the american heart association, european society of cardiology, european association for cardiovascular prevention and rehabilitation, and american college of preventive medicine. Eur. Heart J. 2015, 36, 2097–2109. [Google Scholar]
- Miranda, J.J.; Barrientos-Gutiérrez, T.; Corvalan, C.; Hyder, A.A.; Lazo-Porras, M.; Oni, T.; Wells, J.C.K. Understanding the rise of cardiometabolic diseases in low- and middle-income countries. Nat. Med. 2019, 25, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Kopp, W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2221–2236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Nakayama, T. Inflammation, a link between obesity and cardiovascular disease. Mediat. Inflamm. 2010, 2010, 535918. [Google Scholar] [CrossRef]
- Piché, M.-E.; Tchernof, A.; Després, J.-P. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ. Res. 2020, 126, 1477–1500. [Google Scholar] [CrossRef]
- De Rooij, S.R.; Nijpels, G.; Nilsson, P.M.; Nolan, J.J.; Gabriel, R.; Bobbioni-Harsch, E.; Mingrone, G.; Dekker, J.M.; Sensitivity, R.B.I.; Care, C.D.I.J.D. Low-grade chronic inflammation in the relationship between insulin sensitivity and cardiovascular disease (risc) population: Associations with insulin resistance and cardiometabolic risk profile. Diabetes Care 2009, 32, 1295–1301. [Google Scholar] [CrossRef]
- Pereira, S.S.; Alvarez-Leite, J.I. Low-grade inflammation, obesity, and diabetes. Curr. Obes. Rep. 2014, 3, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Yaribeygi, H.; Atkin, S.L.; Sahebkar, A. Mitochondrial dysfunction in diabetes and the regulatory roles of antidiabetic agents on the mitochondrial function. J. Cell. Physiol. 2019, 234, 8402–8410. [Google Scholar] [CrossRef]
- Ferrante, A.W., Jr. Obesity-induced inflammation: A metabolic dialogue in the language of inflammation. J. Intern. Med. 2007, 262, 408–414. [Google Scholar] [CrossRef]
- Schindler, R. Causes and therapy of microinflammation in renal failure. Nephrol. Dial. Transplant. 2004, 19, v34–v40. [Google Scholar] [CrossRef] [PubMed]
- Akchurin, O.M.; Kaskel, F. Update on inflammation in chronic kidney disease. Blood Purif. 2015, 39, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Schertzer, J.D. Immunometabolism of obesity and diabetes: Microbiota link compartmentalized immunity in the gut to metabolic tissue inflammation. Clin. Sci. 2015, 129, 1083–1096. [Google Scholar] [CrossRef]
- Nishida, K.; Otsu, K. Inflammation and metabolic cardiomyopathy. Cardiovasc. Res. 2017, 113, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Attaye, I.; Warmbrunn, M.V.; Boot, A.N.A.F.; van der Wolk, S.C.; Hutten, B.A.; Daams, J.G.; Herrema, H.; Nieuwdorp, M. A systematic review and meta-analysis of dietary interventions modulating gut microbiota and cardiometabolic diseases—Striving for new standards in microbiome studies. Gastroenterology 2022, 162, 1911–1932. [Google Scholar] [CrossRef]
- Chakaroun, R.; Massier, L.; Musat, N.; Kovacs, P. New paradigms for familiar diseases: Lessons learned on circulatory bacterial signatures in cardiometabolic diseases. Exp. Clin. Endocrinol. Diabetes 2022, 130, 313–326. [Google Scholar] [CrossRef]
- Sumida, K.; Han, Z.; Chiu, C.-Y.; Mims, T.S.; Bajwa, A.; Demmer, R.T.; Datta, S.; Kovesdy, C.P.; Pierre, J.F. Circulating microbiota in cardiometabolic disease. Front. Cell. Infect. Microbiol. 2022, 12, 892232. [Google Scholar] [CrossRef]
- Goraya, M.U.; Li, R.; Mannan, A.; Gu, L.; Deng, H.; Wang, G. Human circulating bacteria and dysbiosis in non-infectious diseases. Front. Cell. Infect. Microbiol. 2022, 12, 932702. [Google Scholar] [CrossRef]
- Anhê, F.F.; Jensen, B.A.H.; Varin, T.V.; Servant, F.; Van Blerk, S.; Richard, D.; Marceau, S.; Surette, M.; Biertho, L.; Lelouvier, B.; et al. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2020, 2, 233–242. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Sumida, K.; Kovesdy, C. The gut-kidney-heart axis in chronic kidney disease. Physiol. Int. 2019, 106, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Schierwagen, R.; Alvarez-Silva, C.; Madsen, M.S.A.; Kolbe, C.C.; Meyer, C.; Thomas, D.; Uschner, F.E.; Magdaleno, F.; Jansen, C.; Pohlmann, A.; et al. Circulating microbiome in blood of different circulatory compartments. Gut 2019, 68, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Wiedermann, C.J.; Kiechl, S.; Dunzendorfer, S.; Schratzberger, P.; Egger, G.; Oberhollenzer, F.; Willeit, J. Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: Prospective results from the bruneck study. J. Am. Coll. Cardiol. 1999, 34, 1975–1981. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, G.; Dinakaran, V.; Rajendhran, J.; Swaminathan, K. Blood microbiota and circulating microbial metabolites in diabetes and cardiovascular disease. Trends Endocrinol. 2020, 31, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, P.B.; Brennan, M.T.; Sasser, H.C.; Fox, P.C.; Paster, B.J.; Bahrani-Mougeot, F.K. Bacteremia associated with toothbrushing and dental extraction. Circulation 2008, 117, 3118–3125. [Google Scholar] [CrossRef]
- Mougeot, F.K.; Saunders, S.E.; Brennan, M.T.; Lockhart, P.B. Associations between bacteremia from oral sources and distant-site infections: Tooth brushing versus single tooth extraction. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 119, 430–435. [Google Scholar] [CrossRef]
- Lelouvier, B.; Servant, F.; Païssé, S.; Brunet, A.-C.; Benyahya, S.; Serino, M.; Valle, C.; Ortiz, M.R.; Puig, J.; Courtney, M.; et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology 2016, 64, 2015–2027. [Google Scholar] [CrossRef]
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut dysbiosis and detection of “live gut bacteria” in blood of japanese patients with type 2 diabetes. Diabetes Care 2014, 37, 2343–2350. [Google Scholar] [CrossRef]
- Païssé, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive description of blood microbiome from healthy donors assessed by 16s targeted metagenomic sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef]
- Nikkari, S.; McLaughlin, I.J.; Bi, W.; Dodge, D.E.; Relman, D.A. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 2001, 39, 1956–1959. [Google Scholar] [CrossRef] [PubMed]
- Whittle, E.; Leonard, M.O.; Harrison, R.; Gant, T.W.; Tonge, D.P. Multi-method characterization of the human circulating microbiome. Front. Microbiol. 2019, 9, 3266. [Google Scholar] [CrossRef] [PubMed]
- Rajendhran, J.; Shankar, M.; Dinakaran, V.; Rathinavel, A.; Gunasekaran, P. Contrasting circulating microbiome in cardiovascular disease patients and healthy individuals. Int. J. Cardiol. 2013, 168, 5118–5120. [Google Scholar] [CrossRef] [PubMed]
- Herrema, H.; Niess, J.H. Intestinal microbial metabolites in human metabolism and type 2 diabetes. Diabetologia 2020, 63, 2533–2547. [Google Scholar] [CrossRef]
- Dinakaran, V.; Rathinavel, A.; Pushpanathan, M.; Sivakumar, R.; Gunasekaran, P.; Rajendhran, J. Elevated levels of circulating DNA in cardiovascular disease patients: Metagenomic profiling of microbiome in the circulation. PLoS ONE 2014, 9, e105221. [Google Scholar] [CrossRef]
- Amar, J.; Lange, C.; Payros, G.; Garret, C.; Chabo, C.; Lantieri, O.; Courtney, M.; Marre, M.; Charles, M.A.; Balkau, B.; et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: The DESIR Study. PLoS ONE 2013, 8, e54461. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The prisma statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Altman, D.G.; Gøtzsche, P.C.; Jüni, P.; Moher, D.; Oxman, A.D.; Savović, J.; Schulz, K.F.; Weeks, L.; Sterne, J.A.C. The cochrane collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011, 343, d5928. [Google Scholar] [CrossRef]
- Moskalewicz, A.; Oremus, M. No clear choice between newcastle–ottawa scale and appraisal tool for cross-sectional studies to assess methodological quality in cross-sectional studies of health-related quality of life and breast cancer. J. Clin. Epidemiol. 2020, 120, 94–103. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef]
- Moriyama, K.; Ando, C.; Tashiro, K.; Kuhara, S.; Okamura, S.; Nakano, S.; Takagi, Y.; Miki, T.; Nakashima, Y.; Hirakawa, H. Polymerase chain reaction detection of bacterial 16s rrna gene in human blood. Microbiol. Immunol. 2008, 52, 375–382. [Google Scholar] [CrossRef]
- Damgaard, C.; Magnussen, K.; Enevold, C.; Nilsson, M.; Tolker-Nielsen, T.; Holmstrup, P.; Nielsen, C.H. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS ONE 2015, 10, e0120826. [Google Scholar] [CrossRef] [PubMed]
- Raeisi, J.; Oloomi, M.; Zolfaghari, M.; Siadat, S.D.; Zargar, M.; Pourramezan, Z. Bacterial DNA detection in the blood of healthy subjects. Iran. Biomed. J. 2022, 26, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Panaiotov, S.; Hodzhev, Y.; Tsafarova, B.; Tolchkov, V.; Kalfin, R. Culturable and Non-Culturable Blood Microbiota of Healthy Individuals. Microorganisms 2021, 9, 1464. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.W.; Vali, H.; Lau, P.C.; Palfree, R.G.; De Ciccio, A.; Sirois, M.; Ahmad, D.; Villemur, R.; Desrosiers, M.; Chan, E.C. Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J. Clin. Microbiol. 2002, 40, 4771–4775. [Google Scholar] [CrossRef]
- Amar, J.; Serino, M.; Lange, C.; Chabo, C.; Iacovoni, J.; Mondot, S.; Lepage, P.; Klopp, C.; Mariette, J.; Bouchez, O.; et al. Involvement of tissue bacteria in the onset of diabetes in humans: Evidence for a concept. Diabetologia 2011, 54, 3055–3061. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, H.; Jing, Y.; Dong, C. Association between blood microbiome and type 2 diabetes mellitus: A nested case-control study. J. Clin. Lab. Anal. 2019, 33, e22842. [Google Scholar] [CrossRef]
- Massier, L.; Chakaroun, R.; Tabei, S.; Crane, A.; Didt, K.D.; Fallmann, J.; von Bergen, M.; Haange, S.B.; Heyne, H.; Stumvoll, M.; et al. Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes. Gut 2020, 69, 1796–1806. [Google Scholar] [CrossRef]
- Ghaemi, F.; Fateh, A.; Sepahy, A.A.; Zangeneh, M.; Ghanei, M.; Siadat, S.D. Blood microbiota composition in iranian pre-diabetic and type 2 diabetic patients. Hum. Antibodies 2021, 29, 243–248. [Google Scholar] [CrossRef]
- D’Aquila, P.; Giacconi, R.; Malavolta, M.; Piacenza, F.; Bürkle, A.; Villanueva, M.M.; Dollé, M.E.T.; Jansen, E.; Grune, T.; Gonos, E.S.; et al. Microbiome in blood samples from the general population recruited in the mark-age project: A pilot study. Front. Cell. Infect. Microbiol. 2021, 12, 707515. [Google Scholar] [CrossRef]
- Chakaroun, R.M.; Massier, L.; Heintz-Buschart, A.; Said, N.; Fallmann, J.; Crane, A.; Schütz, T.; Dietrich, A.; Blüher, M.; Stumvoll, M.; et al. Circulating bacterial signature is linked to metabolic disease and shifts with metabolic alleviation after bariatric surgery. Genome Med. 2021, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.; Lelouvier, B.; Servant, F.; Lluch, J.; Burcelin, R.; Bongard, V.; Elbaz, M. Blood microbiota modification after myocardial infarction depends upon low-density lipoprotein cholesterol levels. J. Am. Heart Assoc. 2019, 8, e011797. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Zhou, H.; Lu, H.; Chen, X.; Zhou, L.; Zhang, J.; Wu, J.; Dong, C. Associations between peripheral blood microbiome and the risk of hypertension. Am. J. Hypertens. 2021, 34, 1064–1070. [Google Scholar] [CrossRef]
- Lawrence, G.; Midtervoll, I.; Samuelsen, S.O.; Kristoffersen, A.K.; Enersen, M.; Håheim, L.L. The blood microbiome and its association to cardiovascular disease mortality: Case-cohort study. BMC Cardiovasc. Disord. 2022, 22, 344. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Khan, I.; Kakakhel, M.A.; Xiaowei, Z.; Ting, M.; Ali, I.; Fei, Y.; Jianye, Z.; Zhiqiang, L.; Lizhe, A. Comparison of microbial populations in the blood of patients with myocardial infarction and healthy individuals. Front. Microbiol. 2022, 13, 845038. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Khan, I.; Usman, M.; Jianye, Z.; Wei, Z.X.; Ping, X.; Zhiqiang, L.; Lizhe, A. Analysis of the blood bacterial composition of patients with acute coronary syndrome and chronic coronary syndrome. Front. Cell. Infect. Microbiol. 2022, 12, 943808. [Google Scholar] [CrossRef]
- Gosiewski, T.; Ludwig-Galezowska, A.H.; Huminska, K.; Sroka-Oleksiak, A.; Radkowski, P.; Salamon, D.; Wojciechowicz, J.; Kus-Slowinska, M.; Bulanda, M.; Wolkow, P.P. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method—The observation of dnaemia. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 329–336. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Tang, C.; Zhao, X.; He, Q.; Li, J. Identification and characterization of blood and neutrophil-associated microbiomes in patients with severe acute pancreatitis using next-generation sequencing. Front. Cell. Infect. Microbiol. 2018, 8, 5. [Google Scholar] [CrossRef]
- Olde Loohuis, L.M.; Mangul, S.; Ori, A.P.S.; Jospin, G.; Koslicki, D.; Yang, H.T.; Wu, T.; Boks, M.P.; Lomen-Hoerth, C.; Wiedau-Pazos, M.; et al. Transcriptome analysis in whole blood reveals increased microbial diversity in schizophrenia. Transl. Psychiatry 2018, 8, 96. [Google Scholar] [CrossRef]
- Shah, N.B.; Allegretti, A.S.; Nigwekar, S.U.; Kalim, S.; Zhao, S.; Lelouvier, B.; Servant, F.; Serena, G.; Thadhani, R.I.; Raj, D.S.; et al. Blood microbiome profile in ckd: A pilot study. Clin. J. Am. Soc. Nephrol. CJASN 2019, 14, 692–701. [Google Scholar] [CrossRef]
- Kajihara, M.; Koido, S.; Kanai, T.; Ito, Z.; Matsumoto, Y.; Takakura, K.; Saruta, M.; Kato, K.; Odamaki, T.; Xiao, J.-z.; et al. Characterisation of blood microbiota in patients with liver cirrhosis. Eur. J. Gastroenterol. Hepatol. 2019, 31, 1577–1583. [Google Scholar] [CrossRef] [PubMed]
- Hammad, D.B.M.; Hider, S.L.; Liyanapathirana, V.C.; Tonge, D.P. Molecular characterization of circulating microbiome signatures in rheumatoid arthritis. Front. Cell. Infect. Microbiol. 2019, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.-B.; Dong, C.-Y.; He, P.; Wu, L.-F.; Lu, X.; Zhang, Y.-H.; Deng, H.-W.; Deng, F.-Y.; Lei, S.-F. Alteration of circulating microbiome and its associated regulation role in rheumatoid arthritis: Evidence from integration of multiomics data. Clin. Transl. Med. 2020, 10, e229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Lederman, M.M.; Hunt, P.; Sieg, S.F.; Haley, K.; Rodriguez, B.; Landay, A.; Martin, J.; Sinclair, E.; Asher, A.I.; et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated hiv infection. J. Infect. Dis. 2009, 199, 1177–1185. [Google Scholar] [CrossRef]
- Markova, N.D. L-form bacteria cohabitants in human blood: Significance for health and diseases. Discov. Med. 2017, 23, 305–313. [Google Scholar] [PubMed]
- Suparan, K.; Sriwichaiin, S.; Chattipakorn, N.; Chattipakorn, S.C. Human blood bacteriome: Eubiotic and dysbiotic states in health and diseases. Cells 2022, 11, 2015. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell. Infect. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef]
- Emery, D.C.; Cerajewska, T.L.; Seong, J.; Davies, M.; Paterson, A.; Allen-Birt, S.J.; West, N.X. Comparison of blood bacterial communities in periodontal health and periodontal disease. Front. Cell. Infect. Microbiol. 2020, 10, 577485. [Google Scholar] [CrossRef]
- Chang, C.J.; Zhang, J.; Tsai, Y.L.; Chen, C.B.; Lu, C.W.; Huo, Y.P.; Liou, H.M.; Ji, C.; Chung, W.H. Compositional features of distinct microbiota base on serum extracellular vesicle metagenomics analysis in moderate to severe psoriasis patients. Cells 2021, 10, 2349. [Google Scholar] [CrossRef]
- Buford, T.W.; Carter, C.S.; VanDerPol, W.J.; Chen, D.; Lefkowitz, E.J.; Eipers, P.; Morrow, C.D.; Bamman, M.M. Composition and richness of the serum microbiome differ by age and link to systemic inflammation. GeroScience 2018, 40, 257–268. [Google Scholar] [CrossRef]
- Leibovici, L.; Samra, Z.; Konisberger, H.; Kalter-Leibovici, O.; Pitlik, S.D.; Drucker, M. Bacteremia in adult diabetic patients. Diabetes Care 1991, 14, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Karusheva, Y.; Koessler, T.; Strassburger, K.; Markgraf, D.; Mastrototaro, L.; Jelenik, T.; Simon, M.-C.; Pesta, D.; Zaharia, O.-P.; Bódis, K.; et al. Short-term dietary reduction of branched-chain amino acids reduces meal-induced insulin secretion and modifies microbiome composition in type 2 diabetes: A randomized controlled crossover trial. Am. J. Clin. Nutr. 2019, 110, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P.J. Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe 2019, 26, 265–272.e264. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4592–4598. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, R.; Zhang, Y.; Pan, D.; Zhu, Y.; Zhang, X.; Yang, R.; Jiang, R.; Xu, Y.; Qin, H. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol. Genom. 2018, 50, 893–903. [Google Scholar] [CrossRef]
- Hoyles, L.; Fernández-Real, J.-M.; Federici, M.; Serino, M.; Abbott, J.; Charpentier, J.; Heymes, C.; Luque, J.L.; Anthony, E.; Barton, R.H.; et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018, 24, 1070–1080. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Moschen, A.R. Multiple parallel hits hypothesis in nonalcoholic fatty liver disease: Revisited after a decade. Hepatology 2021, 73, 833–842. [Google Scholar] [CrossRef]
- Vanholder, R.; Schepers, E.; Pletinck, A.; Nagler, E.V.; Glorieux, G. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: A systematic review. J. Am. Soc. Nephrol. 2014, 25, 1897. [Google Scholar] [CrossRef]
| Study | Selection (4*) | Comparability (2*) | Outcome (3*) | NOS Score |
|---|---|---|---|---|
| McLaughlin et al., 2002 | * * | * | 3/9 | |
| Amar et al., 2011 | * * * | * | * | 5/9 |
| Massier et al., 2020 | * * | * | * | 4/9 |
| Amar et al., 2013 | * * * | * * | 5/9 | |
| Amar et al., 2019 | * * * | * * | 5/9 | |
| Lelouvier et al., 2016 | * * | * * | 4/9 | |
| Schierwagen et al., 2019 | * * * | * * | 5/9 |
| Authors | Study Population | Sample | Study Design | Country | Methodology | Changes in Circulating Microbiota | Ref. |
|---|---|---|---|---|---|---|---|
| Healthy individuals | |||||||
| Nikkari et al., 2001 | 4 HI | Whole Blood | Cohort Study | USA | qPCR and rRNA targeting the conserved region of 16S rDNA by fluorescent probe | Bacteria from 5 divisions and 7 distinct phylogenetic groups detected in the blood. | [31] |
| McLaughlin et al., 2002 | 25 HI | Whole blood | Cohort Study | Canada | Characterization by 16S rRNA and gyrB genes Dark-field microscopy and FISH. | Pleomorphic antibiotic susceptible bacteria existing in healthy blood with limited growth a (possibly Pseudomonas). | [45] |
| Moriyama et al., 2008 | 2 HI | Whole Blood | Preliminary Study | Japan | 16S rRNA PCR and Sanger sequencing. | Aquabacterium, Budvicia, Stenotrophomonas, Serratia, Bacillus and Flavobacteria identified only in clones. | [41] |
| Damgaard et al., 2015 | 60 HI | Blood plasma and RBCs | Cross Sectional Study | Denmark | Blood samples incubated on TSA or blue lactose plates, 16S rRNA gene sequencing and colony PCR | Bacterial growth observed in 35% of RBC fractions and 53% of plasma fractions. Staphylococci, Propionibacterium, Micrococcus and Bacillus most frequently found. | [42] |
| Païssé et al., 2016 | 30 HI | Buffy Coat, Plasma and RBCs | Cohort Study | France | 16S rRNA gene qPCR and 16S targeted metagenomic sequencing (Illumina MiSeq) | All blood fractions were positive for bacterial DNA. Most prevalent Phyla were Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes | [30] |
| Panaiotov et al., 2021 | 28 HI | Whole blood, | Cohort Study | Bulgaria | 16S rRNA genes and ITS2 targeted sequencing on Illumina MiSeq and TEM. | Cultural and molecular characterization of healthy blood microbiota (Proteobacteria and Frimicutes were prominent) | [44] |
| Raeisi et al., 2022 | 50 HI | Whole blood | Cohort Study | Iran | 16S rRNA gene PCR pyro-sequencing | Cultures positive (12%) PCR positive (12%) Staphylococcus, Bacilli were main findings Direct blood PCR-sequencing: Bulkholderia | [43] |
| Diabetic vs. healthy Individuals | |||||||
| Amar et al., 2011 | 3280T2D | Leukocytes | Longitudinal cohort study | France | 16S rDNA quantitative PCR and Pooled pyrosequencing (V1-V2) | High 16S rDNA levels caused diabetes despite risk factors. Proteobacteria phylum represented the highest relative abundance (~90%). High prevalence of Ralstonia spp. in diabetes patients. | [46] |
| Qiu et al., 2019 | 150 (50 T2D 100 HI) | Blood plasma | Nested case-control study | China | 16S rRNA amplicon sequencing by Illumina MiSeq (V5-V6) | Aquabacterium, Pseudonocardia, and Xanthomonas genera and Bacteroides spp. showed an inverse while Alishewanella, Actinotalea, Pseudoclavibacter, Sediminibacterium spp. showed a positive correlation with diabetes. | [47] |
| Massier et al., 2020 | 75, (42 obese and 33 T2D) | Blood | Comparative Cohort study | Germany | 16S rRNA gene sequencing (V4-V5), CARD-FISH | Genus Lactobacillus, Acinetobacter and Lactococcus decreased in T2D while Tahibacter increased in T2D | [48] |
| Ghaemi et al., 2021 | 90(30 T2D, 30 Pre-D and 30 HI) | Buffy Coat | Cohort Study | Iran | Real-time PCR using genus-specific 16srRNA primers | Akkermansia, and Faecalibacterium were higher in HI compared to pre-diabetic and T2D | [49] |
| D’Aquila et al., 2021 | 1285 MARK-AGE Study | Whole blood | Cross-sectional Study | Selected European Countries | Quantification of 16S rRNA by Real-time qPCR (V3-V4) | High level of Bacterial DNA were associated with higher level of Insulin and glucose | [50] |
| Chakaroun et al., 2021 | 112(64 BO 24 T2D and 24 HI) | Whole Blood | Cohort Study | Germany | 16s rRNA sequencing (V4-V5) Bacteria were visualized by CARD-FISH | Loss of Bacillaceae and Bukholderiaceae in T2D and Anoxybacillus, Duganella, Acidibacter, Chryseomicrobium, Sphingomonas were decreased | [51] |
| Cardiovascular vs. Healthy Individuals | |||||||
| Amar et al., 2013 | 3936 CVD | Leukocytes | Longitudinal Study | France | Eubacteria and Proteobacteria 16S rDNA by qPCR | There was a positive correlation of Proteobacteria, and vice versa of Eubacteria, with cardiovascular events. | [36] |
| Rajendhran et al., 2013 | 41 (31 CVD 10 HI) | Whole blood | Comparative Case Study | India | Amplicon sequencing of 16S rDNA V3 region (Ion Torrent PGM) | Increase of Proteobacteria (Pseudomonadaceae and Gammaproteobacteria), decrease in Firmicutes (Staphylococcaceae) and Bacillales, in CVDs | [33] |
| Dinakaran et al., 2014 | 120(80 CVD 40 HI) | Blood plasma | Comparative quantitative study | India | 16S rDNA and β-globin gene concentrations by qRT-PCR. Shotgun and amplicon sequencing V3 region (Ion Torrent PGM) | The 16S rRNA/β-globin gene ratio was higher in CVDs patients than in controls Actinobacteria and Bacteriophages were dominant in CVDs patients whereas Proteobacteria and eukaryotic viruses were dominant in controls. | [35] |
| Amar et al., 2019 | 202 (99 CVD with MI 103 HRCVD) | Whole Blood | Case Control Study | France | 16S rRNA sequencing for V3–V4 regions | Norcardiaceae and Aerococcaceae families and Propionibacterium, Gordonia, Chryseobacterium, and Rhodococcus genera (cholesterol-degrading bacteria) were lower in patients with (vs. without) myocardial infarction. An increase in 16S rRNA gene concentration in CVDs. | [52] |
| Jing et al., 2021 | 300 (150 Hypertension and 150 HI) | Blood Plasma | Case Control Study | China | 16S rRNA gene (V6-V7) Miseq Illumina sequencing | Staphylococcus might be a protective factor for hypertension while either Acinetobacter and Sphingomonas might be are risk factor for hypertension | [53] |
| Lawrence et al., 2022 | 405 (CVD227, HI 178) | Whole Blood | Case Cohort Study | Norway | 16S rRNA gene sequencing (V3-V5) | Higher abundance of Staphylococcus, Kocuria, Enhydrobacter and low levels of Paracoccus were associated with CVDs. While Streptococcus, Paracoccus, Veillonella, and Bacteroides were in abundance in HI. | [54] |
| Khan et al., 2022a | 58 (MI 29 and 29 HI) | Whole Blood | Comparative Case control study | China | 16S rRNA genes (V3-V4) Miseq Illumina sequencing | Decreased alpha diversity in MI patients. Abundance of Bifidobacterium and Bacteroides was increased in MI patients and HI respectively. | [55] |
| Khan et al., 2022b | 210 (ACS, CCR and HI 70 each) | Whole Blood | Comparative Case control study | China | 16S rRNA genes (V3-V4) Illumina sequencing | Proteobacteria and Desulfabacterota abundance was higher in ACS and Actinobacteria in HI | [56] |
| Miscellaneous diseases and Healthy | |||||||
| Lelouvier et al., 2016 | 108 (NAFLD 22 with 86 without fibrosis) | Buffy Coat | cross-sectional study | France | 16S rDNA PCR and (V-3V4) Miseq | Increased 16S rDNA and proportions of Proteobacteria (specifically Sphingomonas, Variovorax, Bosea taxa) in NAFLD | [28] |
| Gosiewski et al., 2017 | 85 (62 sepsis 23 HI) | Whole Blood | Comparative Cohort Study | Poland | 16S rRNA gene targeted metagenomic Miseq Illumina (V3-V4) and Cultures | Healthy samples presented higher diversity than sepsis patients. Proteobacteria were lower in healthy individuals, while Actinobacteria decreased in sepsis patients. HI had predominance of anaerobic bacteria of order Bifidobacteriales | [57] |
| Li et al., 2018 | 62 (50 PP and 12 HI) | Whole blood and Neutrophils | Cohort Study | China | 16S rDNA, qPCR and targeted metagenomic sequencing V3 region (Ion Torrent PGM) | 16S rDNA gene copies were higher in patients. Bacteroidetes were high and Actinobacteria lower in patients. | [58] |
| OdleLoohuis et al., 2018 | 192 (48 SCZ, ALS 47, BPD 48, 49 HI) | Whole Blood | Cohort Study | USA | High Quality unmapped RNA sequencing | Proteobacteria, Firmicutes and Cyanobacteria were most prevalent phyla while Schizophrenia patients have diverse microbes. Bacteria and CD8+ memory T cells are inversely related. | [59] |
| Shah et al., 2019 | 40(20CKD and 20 HI) | Buffy Coat | Cross sectional Pilot Study | France | 16S rRNA (V3–V4) regions gene sequencing | Enterobacteriaceae and Pseudomonadaceae significantly higher in CKD than HC (10% versus 7%; and 23% versus 18%, respectively. | [60] |
| Schierwagen et al., 2019 | 7 LC | Venous Blood | Cohort Study | Germany | 16s rRNA Gene sequencing, Anaerobic Cultivation | Positive cultivation of Staphylococcus and Acinetobacter, Inflammatory cytokine positively correlated with abundance of blood microbiome | [23] |
| Kajihara et al., 2019 | 80 (66 LC and 14 HI) | Peripheral Blood | Cohort Study | Japan | 16s rRNA Gene sequencing, (V3-V4) | Enterobacteriaceae was higher in LC. On the contrary, the abundance of Akkermansia, Rikenellaceae and Erysipelotrichales were lower in LC | [61] |
| Whittle et al., 2019 | 10 (5 Asthma and 5 H women) | Plasma fractions | Cohort Study | United Kingdom | Cultures an16S rRNA gene sequencing | Most abundant phyla were Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes. Achromobacter and Pseudomonas were decreased in Asthma patients | [32] |
| Hammad et al., 2019 | 32 (20RA, 4AS, 4 PA, 4HI) | Serum | Cohort Study | United Kingdom | 16S rDNA sequencing V4 region | Blood dysbiosis in RA characterized by an increase in genera Halomonas, Anaerococcus, Shewanella, and members of Lachnospiraceae, while decrease in Corynebacterium 1 and Streptococcus | [62] |
| Mo et al., 2020 | 43 (28 RA and 15 HI) | Mononuclear cells | Cohort Study | China | 16S rDNA sequencing | Bacteroidetes had less abundance in RA while Candidatus Saccharibacteria was increased. | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah Goraya, M.; Li, R.; Gu, L.; Deng, H.; Wang, G. Blood Stream Microbiota Dysbiosis Establishing New Research Standards in Cardio-Metabolic Diseases, A Meta-Analysis Study. Microorganisms 2023, 11, 777. https://doi.org/10.3390/microorganisms11030777
Ullah Goraya M, Li R, Gu L, Deng H, Wang G. Blood Stream Microbiota Dysbiosis Establishing New Research Standards in Cardio-Metabolic Diseases, A Meta-Analysis Study. Microorganisms. 2023; 11(3):777. https://doi.org/10.3390/microorganisms11030777
Chicago/Turabian StyleUllah Goraya, Mohsan, Rui Li, Liming Gu, Huixiong Deng, and Gefei Wang. 2023. "Blood Stream Microbiota Dysbiosis Establishing New Research Standards in Cardio-Metabolic Diseases, A Meta-Analysis Study" Microorganisms 11, no. 3: 777. https://doi.org/10.3390/microorganisms11030777
APA StyleUllah Goraya, M., Li, R., Gu, L., Deng, H., & Wang, G. (2023). Blood Stream Microbiota Dysbiosis Establishing New Research Standards in Cardio-Metabolic Diseases, A Meta-Analysis Study. Microorganisms, 11(3), 777. https://doi.org/10.3390/microorganisms11030777







