Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Cohort
2.2. Sample Collection
2.3. 16S rRNA High-Throughput Sequencing
2.4. Microbiome Analyses
2.5. Functional Analyses
2.6. Network Analyses
3. Results
3.1. Bacteriome Taxonomic Diversity and Structure
3.2. Bacteriome Functional Diversity
3.3. Bacteriome Network Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Busse, W.W.; Banks-Schlegel, S.; Wenzel, S.E. Pathophysiology of severe asthma. J. Allergy Clin. Immunol. 2000, 106, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- King, G.G.; James, A.; Harkness, L.; Wark, P.A.B. Pathophysiology of severe asthma: We’ve only just started. Respirology 2018, 23, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of Asthma in Children and Adults. Front. Pediatr. 2019, 7, 246. [Google Scholar] [CrossRef]
- Mims, J.W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. 2015, 5 (Suppl. S1), S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Licari, A.; Brambilla, I.; Marseglia, A.; De Filippo, M.; Paganelli, V.; Marseglia, G.L. Difficult vs. Severe Asthma: Definition and Limits of Asthma Control in the Pediatric Population. Front. Pediatr. 2018, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Serebrisky, D.; Wiznia, A. Pediatric Asthma: A Global Epidemic. Ann. Glob. Health 2019, 85, 6. [Google Scholar] [CrossRef]
- Dierick, B.J.H.; van der Molen, T.; Flokstra-de Blok, B.M.J.; Muraro, A.; Postma, M.J.; Kocks, J.W.H.; van Boven, J.F.M. Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy. Expert Rev. Pharm. Outcomes Res. 2020, 20, 437–453. [Google Scholar] [CrossRef]
- GBD Diseases Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Asher, M.I.; Rutter, C.E.; Bissell, K.; Chiang, C.Y.; El Sony, A.; Ellwood, E.; Ellwood, P.; Garcia-Marcos, L.; Marks, G.B.; Morales, E.; et al. Worldwide trends in the burden of asthma symptoms in school-aged children: Global Asthma Network Phase I cross-sectional study. Lancet 2021, 398, 1569–1580. [Google Scholar] [CrossRef]
- GBD Disease Injury Incidence Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- The Global Asthma Report. Auckland, New Zealand: Global Asthma Network. 2018. Available online: http://globalasthmareport.org/2018/index.html (accessed on 6 February 2023).
- Muc, M.; Mota-Pinto, A.; Padez, C. Prevalence of asthma and rhinitis symptoms among children living in Coimbra, Portugal. Rev. Port Pneumol. 2014, 20, 208–210. [Google Scholar] [CrossRef]
- Sa-Sousa, A.; Morais-Almeida, M.; Azevedo, L.F.; Carvalho, R.; Jacinto, T.; Todo-Bom, A.; Loureiro, C.; Bugalho-Almeida, A.; Bousquet, J.; Fonseca, J.A. Prevalence of asthma in Portugal—The Portuguese National Asthma Survey. Clin. Transl. Allergy 2012, 2, 15. [Google Scholar] [CrossRef]
- Ferreira-Magalhaes, M.; Sa-Sousa, A.; Morais-Almeida, M.; Pite, H.; Azevedo, L.F.; Azevedo, M.I.; Bugalho-Almeida, A.; Fonseca, J.A. Asthma-like symptoms, diagnostic tests, and asthma medication use in children and adolescents: A population-based nationwide survey. J. Asthma Off. J. Assoc. Care Asthma 2016, 53, 269–276. [Google Scholar] [CrossRef]
- Meltzer, E.O.; Bukstein, D.A. The economic impact of allergic rhinitis and current guidelines for treatment. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2011, 106, S12–S16. [Google Scholar] [CrossRef]
- Savoure, M.; Bousquet, J.; Jaakkola, J.J.K.; Jaakkola, M.S.; Jacquemin, B.; Nadif, R. Worldwide prevalence of rhinitis in adults: A review of definitions and temporal evolution. Clin. Transl. Allergy 2022, 12, e12130. [Google Scholar] [CrossRef]
- Steelant, B.; Farre, R.; Wawrzyniak, P.; Belmans, J.; Dekimpe, E.; Vanheel, H.; Van Gerven, L.; Kortekaas Krohn, I.; Bullens, D.M.A.; Ceuppens, J.L.; et al. Impaired barrier function in patients with house dust mite-induced allergic rhinitis is accompanied by decreased occludin and zonula occludens-1 expression. J. Allergy Clin. Immunol. 2016, 137, 1043–1053. [Google Scholar] [CrossRef]
- Steelant, B.; Seys, S.F.; Van Gerven, L.; Van Woensel, M.; Farre, R.; Wawrzyniak, P.; Kortekaas Krohn, I.; Bullens, D.M.; Talavera, K.; Raap, U.; et al. Histamine and T helper cytokine-driven epithelial barrier dysfunction in allergic rhinitis. J. Allergy Clin. Immunol. 2018, 141, 951–963.e958. [Google Scholar] [CrossRef]
- Acevedo-Prado, A.; Seoane-Pillado, T.; Lopez-Silvarrey-Varela, A.; Salgado, F.J.; Cruz, M.J.; Faraldo-Garcia, A.; Nieto-Fontarigo, J.J.; Pertega-Diaz, S.; Sanchez-Lastres, J.; San-Jose-Gonzalez, M.A.; et al. Association of rhinitis with asthma prevalence and severity. Sci. Rep. 2022, 12, 6389. [Google Scholar] [CrossRef]
- Nur Husna, S.M.; Tan, H.T.; Md Shukri, N.; Mohd Ashari, N.S.; Wong, K.K. Allergic Rhinitis: A Clinical and Pathophysiological Overview. Front. Med. 2022, 9, 874114. [Google Scholar] [CrossRef]
- Falcão, H.; Ramos, E.; Marques, A.; Barros, H. Prevalence of asthma and rhinitis in 13 year old adolescents in Porto, Portugal. Rev. Port Pneumol. 2008, 14, 747–768. [Google Scholar] [CrossRef]
- Todo-Bom, A.; Loureiro, C.; Almeida, M.M.; Nunes, C.; Delgado, L.; Castel-Branco, G.; Bousquet, J. Epidemiology of rhinitis in Portugal: Evaluation of the intermittent and the persistent types. Allergy 2007, 62, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Compalati, E.; Ridolo, E.; Passalacqua, G.; Braido, F.; Villa, E.; Canonica, G.W. The link between allergic rhinitis and asthma: The united airways disease. Expert Rev. Clin. Immunol. 2010, 6, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, J.; Hellings, P.W.; Agache, I.; Amat, F.; Annesi-Maesano, I.; Ansotegui, I.J.; Anto, J.M.; Bachert, C.; Bateman, E.D.; Bedbrook, A.; et al. Allergic Rhinitis and its Impact on Asthma (ARIA) Phase 4 (2018): Change management in allergic rhinitis and asthma multimorbidity using mobile technology. J. Allergy Clin. Immunol. 2019, 143, 864–879. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Magalhaes, M.; Pereira, A.M.; Sa-Sousa, A.; Morais-Almeida, M.; Azevedo, I.; Azevedo, L.F.; Fonseca, J.A. Asthma control in children is associated with nasal symptoms, obesity, and health insurance: A nationwide survey. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2015, 26, 466–473. [Google Scholar] [CrossRef]
- Pite, H.; Pereira, A.M.; Morais-Almeida, M.; Nunes, C.; Bousquet, J.; Fonseca, J.A. Prevalence of asthma and its association with rhinitis in the elderly. Respir. Med. 2014, 108, 1117–1126. [Google Scholar] [CrossRef]
- Bergeron, C.; Hamid, Q. Relationship between Asthma and Rhinitis: Epidemiologic, Pathophysiologic, and Therapeutic Aspects. Allergy Asthma Clin. Immunol. 2005, 1, 81–87. [Google Scholar] [CrossRef]
- Kim, H.; Bouchard, J.; Renzi, P.M. The link between allergic rhinitis and asthma: A role for antileukotrienes? Can. Respir J. 2008, 15, 91–98. [Google Scholar] [CrossRef]
- Valero, A.; Pereira, C.; Loureiro, C.; Martinez-Cocera, C.; Murio, C.; Rico, P.; Palomino, R.; Davila, I. Interrelationship between skin sensitization, rhinitis, and asthma in patients with allergic rhinitis: A study of Spain and Portugal. J. Investig. Allergol. Clin. Immunol. 2009, 19, 167–172. [Google Scholar]
- Huang, Y.J.; Boushey, H.A. The microbiome and asthma. Ann. Am. Thorac. Soc. 2014, 11 (Suppl. S1), S48–S51. [Google Scholar] [CrossRef]
- Huang, Y.J.; Boushey, H.A. The microbiome in asthma. J. Allergy Clin. Immunol. 2015, 135, 25–30. [Google Scholar] [CrossRef]
- Brar, T.; Nagaraj, S.; Mohapatra, S. Microbes and asthma: The missing cellular and molecular links. Curr. Opin. Pulm. Med. 2012, 18, 14–22. [Google Scholar] [CrossRef]
- Dickson, R.P.; Huffnagle, G.B. The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLoS Pathog. 2015, 11, e1004923. [Google Scholar] [CrossRef]
- Castro-Nallar, E.; Shen, Y.; Freishtat, R.J.; Pérez-Losada, M.; Manimaran, S.; Liu, G.; Spira, A.; Johnson, W.E.; Crandall, K.A. Integrating metagenomics and host gene expression to characterize asthma-associated microbial communities. BMC Med. Genom. 2015, 8, 50. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef]
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Alamri, L.; Crandall, K.A.; Freishtat, R.J. Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma. PLoS ONE 2017, 12, e0170543. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Authelet, K.J.; Hoptay, C.E.; Kwak, C.; Crandall, K.A.; Freishtat, R.J. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome 2018, 6, 179. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Castro-Nallar, E.; Bendall, M.L.; Freishtat, R.J.; Crandall, K.A. Dual Transcriptomic Profiling of Host and Microbiota during Health and Disease in Pediatric Asthma. PLoS ONE 2015, 10, e0131819. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Crandall, K.A.; Freishtat, R.J. Comparison of two commercial DNA extraction kits for the analysis of nasopharyngeal bacterial communities. AIMS Microbiol. 2016, 2, 108–119. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Crandall, K.A.; Freishtat, R.J. Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome 2016, 4, 25. [Google Scholar] [CrossRef]
- Raita, Y.; Perez-Losada, M.; Freishtat, R.J.; Harmon, B.; Mansbach, J.M.; Piedra, P.A.; Zhu, Z.; Camargo, C.A.; Hasegawa, K. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat. Commun. 2021, 12, 3601. [Google Scholar] [CrossRef] [PubMed]
- Dinwiddie, D.L.; Denson, J.L.; Kennedy, J.L. Role of the Airway Microbiome in Respiratory Infections and Asthma in Children. Pediatr. Allergy Immunol. Pulmonol. 2018, 31, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, K.; Pali-Scholl, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Losol, P.; Choi, J.P.; Kim, S.H.; Chang, Y.S. The Role of Upper Airway Microbiome in the Development of Adult Asthma. Immune Netw. 2021, 21, e19. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut(-)Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef]
- Bender, M.E.; Read, T.D.; Edwards, T.S.; Hargita, M.; Cutler, A.J.; Wissel, E.F.; Wise, S.K. A Comparison of the Bacterial Nasal Microbiome in Allergic Rhinitis Patients Before and After Immunotherapy. Laryngoscope 2020, 130, E882–E888. [Google Scholar] [CrossRef]
- Chen, M.; He, S.; Miles, P.; Li, C.; Ge, Y.; Yu, X.; Wang, L.; Huang, W.; Kong, X.; Ma, S.; et al. Nasal Bacterial Microbiome Differs Between Healthy Controls and Those With Asthma and Allergic Rhinitis. Front. Cell Infect Microbiol. 2022, 12, 841995. [Google Scholar] [CrossRef]
- Gan, W.; Yang, F.; Meng, J.; Liu, F.; Liu, S.; Xian, J. Comparing the nasal bacterial microbiome diversity of allergic rhinitis, chronic rhinosinusitis and control subjects. Eur. Arch. Otorhinolaryngol. 2021, 278, 711–718. [Google Scholar] [CrossRef]
- Lal, D.; Keim, P.; Delisle, J.; Barker, B.; Rank, M.A.; Chia, N.; Schupp, J.M.; Gillece, J.D.; Cope, E.K. Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int. Forum Allergy Rhinol 2017, 7, 561–569. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Han, S.; Kim, W. Compositional alteration of the nasal microbiome and Staphylococcus aureus-characterized dysbiosis in the nasal mucosa of patients with allergic rhinitis. Clin. Exp. Otorhinolaryngol. 2022, 15, 335–345. [Google Scholar] [CrossRef]
- Azevedo, A.C.; Hilario, S.; Goncalves, M.F.M. Microbiome in Nasal Mucosa of Children and Adolescents with Allergic Rhinitis: A Systematic Review. Children 2023, 10, 226. [Google Scholar] [CrossRef]
- Garcia-Rodriguez, J.A.; Fresnadillo Martinez, M.J. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 2002, 50 (Suppl. S2), 59–73. [Google Scholar] [CrossRef]
- Biesbroek, G.; Tsivtsivadze, E.; Sanders, E.A.; Montijn, R.; Veenhoven, R.H.; Keijser, B.J.; Bogaert, D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014, 190, 1283–1292. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med. 2013, 7, 245–257. [Google Scholar] [CrossRef]
- Prevaes, S.M.; de Winter-de Groot, K.M.; Janssens, H.M.; de Steenhuijsen Piters, W.A.; Tramper-Stranders, G.A.; Wyllie, A.L.; Hasrat, R.; Tiddens, H.A.; van Westreenen, M.; van der Ent, C.K.; et al. Development of the Nasopharyngeal Microbiota in Infants with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2016, 193, 504–515. [Google Scholar] [CrossRef]
- Huang, Y.J. Nasopharyngeal Microbiota: Gatekeepers or Fortune Tellers of Susceptibility to Respiratory Tract Infections? Am. J. Respir. Crit. Care Med. 2017, 196, 1504–1505. [Google Scholar] [CrossRef]
- Esposito, S.; Principi, N. Impact of nasopharyngeal microbiota on the development of respiratory tract diseases. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 1–7. [Google Scholar] [CrossRef]
- Koppen, I.J.N.; Bosch, A.; Sanders, E.A.M.; van Houten, M.A.; Bogaert, D. The respiratory microbiota during health and disease: A paediatric perspective. Pneumonia 2015, 6, 90–100. [Google Scholar] [CrossRef]
- Man, W.H.; De Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Cao, W.; Sun, Y.; Zhao, N.; Song, J.; Zhang, N.; Liu, L.; Liu, Q. Characteristics of the bacterial microbiota in the upper respiratory tract of children. Eur. Arch. Otorhinolaryngol. 2022, 279, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Mahdavinia, M. The nasal microbiome: Opening new clinical research avenues for allergic disease. Expert Rev. Clin. Immunol. 2018, 14, 645–647. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Leong, L.E.X.; Choo, J.M.; Wesselingh, S.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Jenkins, C.; et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J. Allergy Clin. Immunol. 2018, 141, 94–103.e115. [Google Scholar] [CrossRef]
- Benton, A.S.; Wang, Z.; Lerner, J.; Foerster, M.; Teach, S.J.; Freishtat, R.J. Overcoming heterogeneity in pediatric asthma: Tobacco smoke and asthma characteristics within phenotypic clusters in an African American cohort. J. Asthma Off. J. Assoc. Care Asthma 2010, 47, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, P.J.; Burbach, G.; Heinzerling, L.M.; Edenharter, G.; Bachert, C.; Bindslev-Jensen, C.; Bonini, S.; Bousquet-Rouanet, L.; Demoly, P.; Bresciani, M.; et al. GA2LEN skin test study III: Minimum battery of test inhalent allergens needed in epidemiological studies in patients. Allergy 2009, 64, 1656–1662. [Google Scholar] [CrossRef]
- Pereira, C.; Valero, A.; Loureiro, C.; Davila, I.; Martinez-Cocera, C.; Murio, C.; Rico, P.; Palomino, R. Iberian study of aeroallergens sensitisation in allergic rhinitis. Eur. Ann. Allergy Clin. Immunol. 2006, 38, 186–194. [Google Scholar]
- Silva, D.; Severo, M.; Paciencia, I.; Rufo, J.; Martins, C.; Moreira, P.; Padrao, P.; Delgado, L.; Moreira, A. Setting definitions of childhood asthma in epidemiologic studies. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2019, 30, 708–715. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Lee, T.D.; Andrade, J.; Oguntuyo, K.; Chun, Y.; Grishina, G.; Grishin, A.; Bunyavanich, S. The nasal microbiome in asthma. J. Allergy Clin. Immunol. 2018, 142, 834–843.e832. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vazquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cook, R.D. Detection of Influential Observation in Linear Regression. Technometrics 1977, 19, 15–18. [Google Scholar] [CrossRef]
- Team, R.D.C. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- RStudio, R.T. Integrated Development for R; RStudio Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Muller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Peschel, S.; Muller, C.L.; von Mutius, E.; Boulesteix, A.L.; Depner, M. NetCoMi: Network construction and comparison for microbiome data in R. Brief. Bioinform. 2021, 22, bbaa290. [Google Scholar] [CrossRef]
- Backhed, F.; Fraser, C.M.; Ringel, Y.; Sanders, M.E.; Sartor, R.B.; Sherman, P.M.; Versalovic, J.; Young, V.; Finlay, B.B. Defining a Healthy Human Gut Microbiome: Current Concepts, Future Directions, and Clinical Applications. Cell Host Microbe 2012, 12, 611–622. [Google Scholar] [CrossRef]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef]
- Losol, P.; Park, H.S.; Song, W.J.; Hwang, Y.K.; Kim, S.H.; Holloway, J.W.; Chang, Y.S. Association of upper airway bacterial microbiota and asthma: Systematic review. Asia Pac. Allergy 2022, 12, e32. [Google Scholar] [CrossRef]
- Sherret, J.; Gajjar, B.; Ibrahim, L.; Mohamed Ahmed, A.; Panta, U.R. Dolosigranulum pigrum: Predicting Severity of Infection. Cureus 2020, 12, e9770. [Google Scholar] [CrossRef]
- Gaugris, S.; Sazonov-Kocevar, V.; Thomas, M. Burden of concomitant allergic rhinitis in adults with asthma. J. Asthma Off. J. Assoc. Care Asthma 2006, 43, 1–7. [Google Scholar] [CrossRef]
- Kritikos, V.; Price, D.; Papi, A.; Infantino, A.; Stallberg, B.; Ryan, D.; Lavorini, F.; Chrystyn, H.; Haughney, J.; Lisspers, K.; et al. The Burden of Self-Reported Rhinitis and Associated Risk for Exacerbations with Moderate-Severe Asthma in Primary Care Patients. J. Asthma Allergy 2020, 13, 415–428. [Google Scholar] [CrossRef]
- Kang, H.Y.; Park, C.S.; Bang, H.R.; Sazonov, V.; Kim, C.J. Effect of allergic rhinitis on the use and cost of health services by children with asthma. Yonsei Med. J. 2008, 49, 521–529. [Google Scholar] [CrossRef]
- Sanders, D.J.; Inniss, S.; Sebepos-Rogers, G.; Rahman, F.Z.; Smith, A.M. The role of the microbiome in gastrointestinal inflammation. Biosci. Rep. 2021, 41, 543–556. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal. Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef]
- Lira-Lucio, J.A.; Falfan-Valencia, R.; Ramirez-Venegas, A.; Buendia-Roldan, I.; Rojas-Serrano, J.; Mejia, M.; Perez-Rubio, G. Lung Microbiome Participation in Local Immune Response Regulation in Respiratory Diseases. Microorganisms 2020, 8, 1059. [Google Scholar] [CrossRef]
- Lynch, S.V. The Lung Microbiome and Airway Disease. Ann. Am. Thorac. Soc. 2016, 13 (Suppl. S2), S462–S465. [Google Scholar] [CrossRef]
- Paudel, K.R.; Dharwal, V.; Patel, V.K.; Galvao, I.; Wadhwa, R.; Malyla, V.; Shen, S.S.; Budden, K.F.; Hansbro, N.G.; Vaughan, A.; et al. Role of Lung Microbiome in Innate Immune Response Associated With Chronic Lung Diseases. Front. Med. 2020, 7, 554. [Google Scholar] [CrossRef]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef]
- Brugger, S.D.; Bomar, L.; Lemon, K.P. Commensal-Pathogen Interactions along the Human Nasal Passages. PLoS Pathog. 2016, 12, e1005633. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Liang, J.Q. Nasal Microbiome and Its Interaction with the Host in Childhood Asthma. Cells 2022, 11, 3155. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Pamp, S.J.; Fukuyama, J.; Hwang, P.H.; Cho, D.Y.; Holmes, S.; Relman, D.A. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 2013, 14, 631–640. [Google Scholar] [CrossRef]
- Chung, K.F. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J. Allergy Clin. Immunol. 2017, 139, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Dimitri-Pinheiro, S.; Soares, R.; Barata, P. The Microbiome of the Nose-Friend or Foe? Allergy Rhinol. 2020, 11, 2152656720911605. [Google Scholar] [CrossRef]
- Chiang, T.Y.; Yang, Y.R.; Zhuo, M.Y.; Yang, F.; Zhang, Y.F.; Fu, C.H.; Lee, T.J.; Chung, W.H.; Chen, L.; Chang, C.J. Microbiome profiling of nasal extracellular vesicles in patients with allergic rhinitis. World Allergy Organ J. 2022, 15, 100674. [Google Scholar] [CrossRef]
- Espuela-Ortiz, A.; Lorenzo-Diaz, F.; Baez-Ortega, A.; Eng, C.; Hernandez-Pacheco, N.; Oh, S.S.; Lenoir, M.; Burchard, E.G.; Flores, C.; Pino-Yanes, M. Bacterial salivary microbiome associates with asthma among african american children and young adults. Pediatr. Pulmonol. 2019, 54, 1948–1956. [Google Scholar] [CrossRef]
- Huang, Y.J.; Nelson, C.E.; Brodie, E.L.; Desantis, T.Z.; Baek, M.S.; Liu, J.; Woyke, T.; Allgaier, M.; Bristow, J.; Wiener-Kronish, J.P.; et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011, 127, 372–381.e3. [Google Scholar] [CrossRef]
- Choi, C.H.; Poroyko, V.; Watanabe, S.; Jiang, D.; Lane, J.; deTineo, M.; Baroody, F.M.; Naclerio, R.M.; Pinto, J.M. Seasonal allergic rhinitis affects sinonasal microbiota. Am. J. Rhinol. Allergy 2014, 28, 281–286. [Google Scholar] [CrossRef]
- Depner, M.; Ege, M.J.; Cox, M.J.; Dwyer, S.; Walker, A.W.; Birzele, L.T.; Genuneit, J.; Horak, E.; Braun-Fahrlander, C.; Danielewicz, H.; et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J. Allergy Clin. Immunol. 2017, 139, 826–834.e813. [Google Scholar] [CrossRef]
- Bisgaard, H.; Hermansen, M.N.; Buchvald, F.; Loland, L.; Halkjaer, L.B.; Bonnelykke, K.; Brasholt, M.; Heltberg, A.; Vissing, N.H.; Thorsen, S.V.; et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007, 357, 1487–1495. [Google Scholar] [CrossRef]
- Ma, Z.S. Testing the Anna Karenina Principle in Human Microbiome-Associated Diseases. iScience 2020, 23, 101007. [Google Scholar] [CrossRef]
- Ma, Z.S.; Li, L.; Gotelli, N.J. Diversity-disease relationships and shared species analyses for human microbiome-associated diseases. ISME J. 2019, 13, 1911–1919. [Google Scholar] [CrossRef]
- Hoffman, D.J.; Campos-Ponce, M.; Taddei, C.R.; Doak, C.M. Microbiome, growth retardation and metabolism: Are they related? Ann. Hum. Biol. 2017, 44, 201–207. [Google Scholar] [CrossRef]
- Ta, L.D.H.; Yap, G.C.; Tay, C.J.X.; Lim, A.S.M.; Huang, C.H.; Chu, C.W.; De Sessions, P.F.; Shek, L.P.; Goh, A.; Van Bever, H.P.S.; et al. Establishment of the nasal microbiota in the first 18 months of life: Correlation with early-onset rhinitis and wheezing. J. Allergy Clin. Immunol. 2018, 142, 86–95. [Google Scholar] [CrossRef]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef]
- Hu, Q.; Jin, L.; Zeng, J.; Wang, J.; Zhong, S.; Fan, W.; Liao, W. Tryptophan metabolite-regulated Treg responses contribute to attenuation of airway inflammation during specific immunotherapy in a mouse asthma model. Hum. Vaccin. Immunother. 2020, 16, 1891–1899. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Cheng, M.L.; Chiang, M.H.; Wang, C.J.; Tsai, M.H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887. [Google Scholar] [CrossRef]
- Samra, M.S.; Lim, D.H.; Han, M.Y.; Jee, H.M.; Kim, Y.K.; Kim, J.H. Bacterial Microbiota-derived Extracellular Vesicles in Children With Allergic Airway Diseases: Compositional and Functional Features. Allergy Asthma Immunol. Res. 2021, 13, 56–74. [Google Scholar] [CrossRef]
- Li, K.J.; Chen, Z.L.; Huang, Y.; Zhang, R.; Luan, X.Q.; Lei, T.T.; Chen, L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019, 20, 272. [Google Scholar] [CrossRef]
- Sharma, A.; Laxman, B.; Naureckas, E.T.; Hogarth, D.K.; Sperling, A.I.; Solway, J.; Ober, C.; Gilbert, J.A.; White, S.R. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J. Allergy Clin. Immunol. 2019, 144, 1214–1227.e1217. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yu, Y.; Du, W.; Liu, Y.; Dai, R.; Tang, W.; Wang, P.; Zhang, C.; Shi, G. Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin. Transl. Allergy 2020, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Xia, Y. Further analysis reveals new gut microbiome markers of type 2 diabetes mellitus. Antonie Van Leeuwenhoek 2017, 110, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Kong, X.; Liu, J.; Cetinbas, M.; Sadreyev, R.; Koh, M.; Huang, H.; Adeseye, A.; He, P.; Zhu, J.; Russell, H.; et al. New and Preliminary Evidence on Altered Oral and Gut Microbiota in Individuals with Autism Spectrum Disorder (ASD): Implications for ASD Diagnosis and Subtyping Based on Microbial Biomarkers. Nutrients 2019, 11, 2128. [Google Scholar] [CrossRef]
- Kim, Y.H.; Jang, H.; Kim, S.Y.; Jung, J.H.; Kim, G.E.; Park, M.R.; Hong, J.Y.; Kim, M.N.; Kim, E.G.; Kim, M.J.; et al. Gram-negative microbiota is related to acute exacerbation in children with asthma. Clin. Transl. Allergy 2021, 11, e12069. [Google Scholar] [CrossRef]
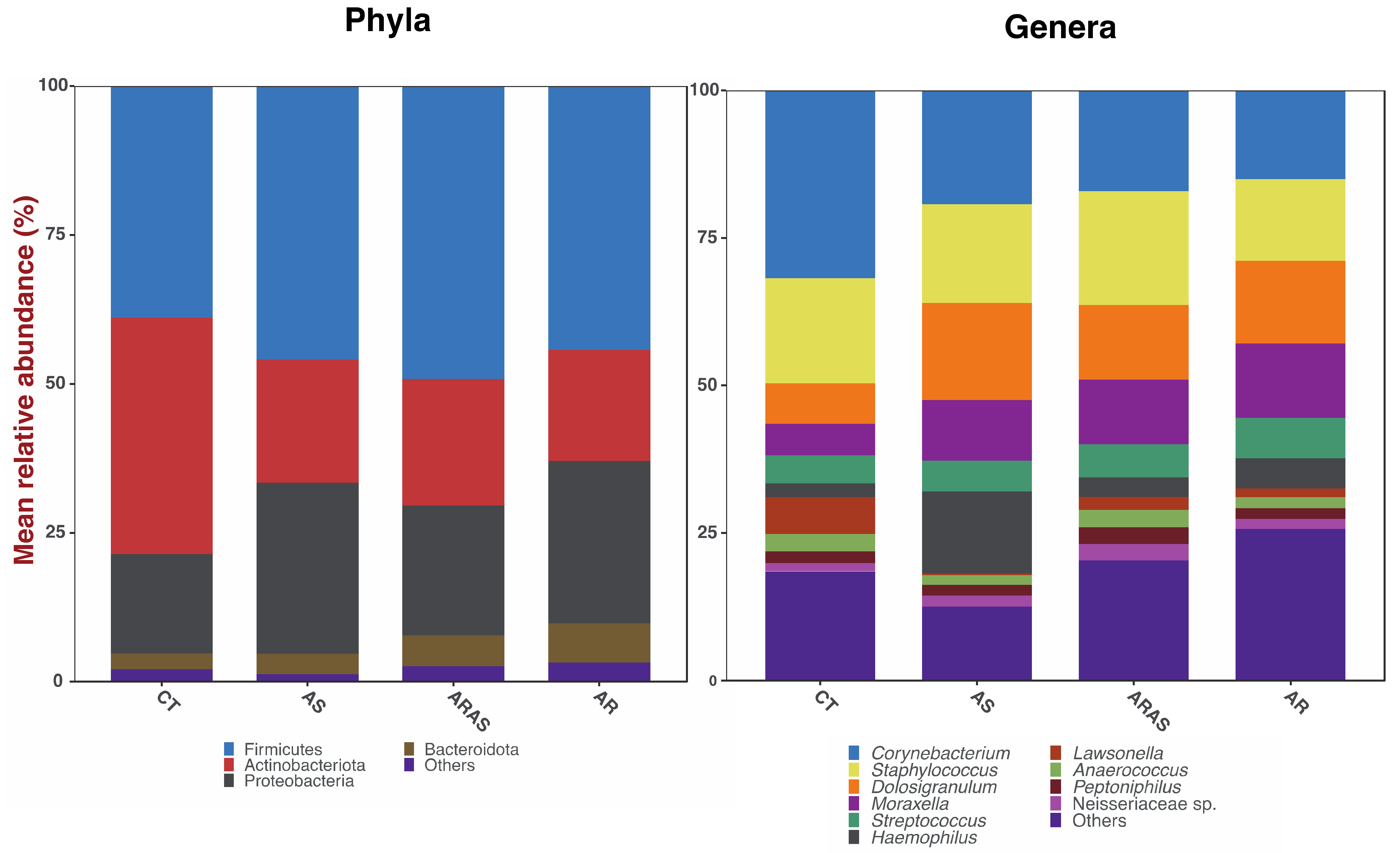
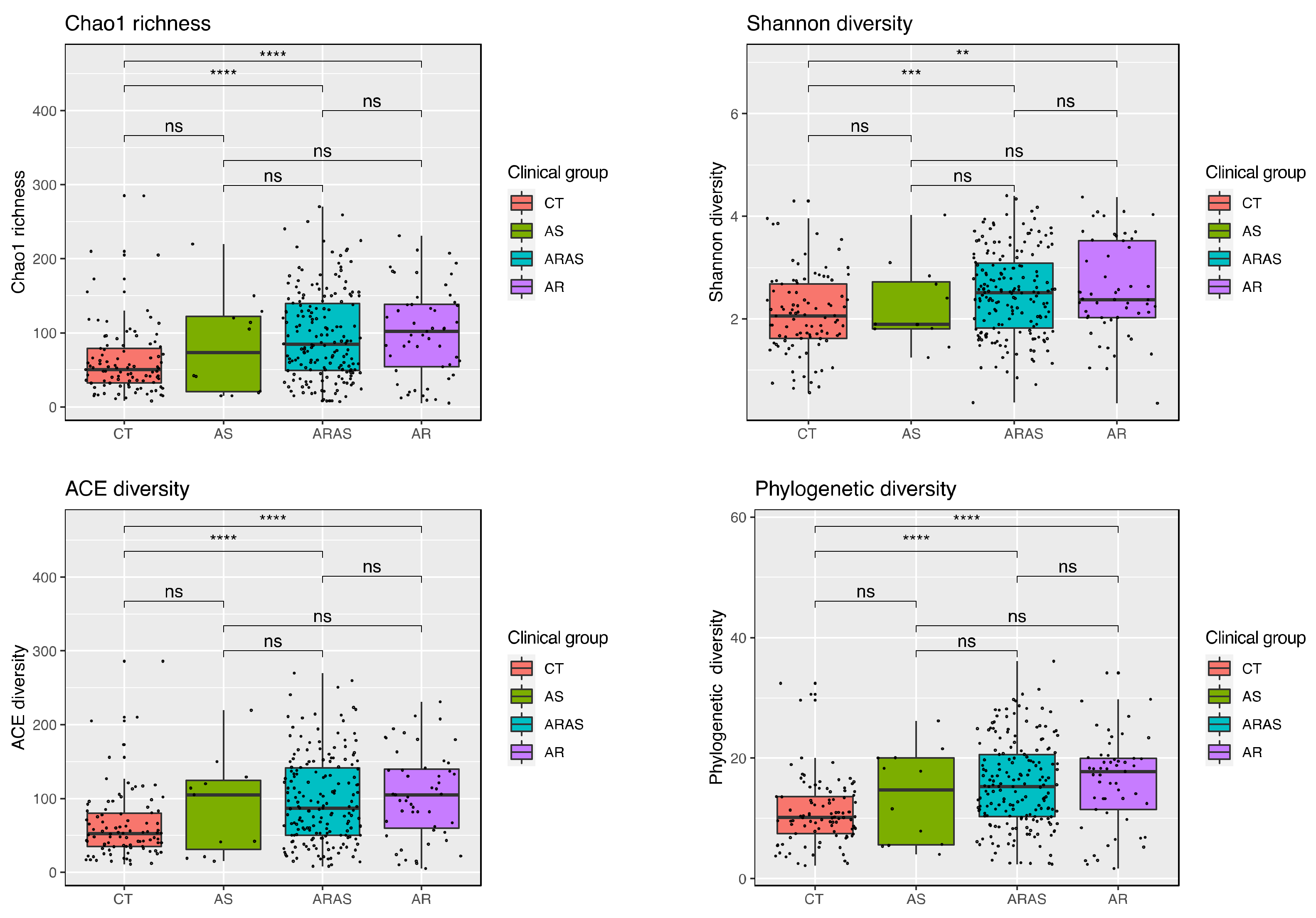
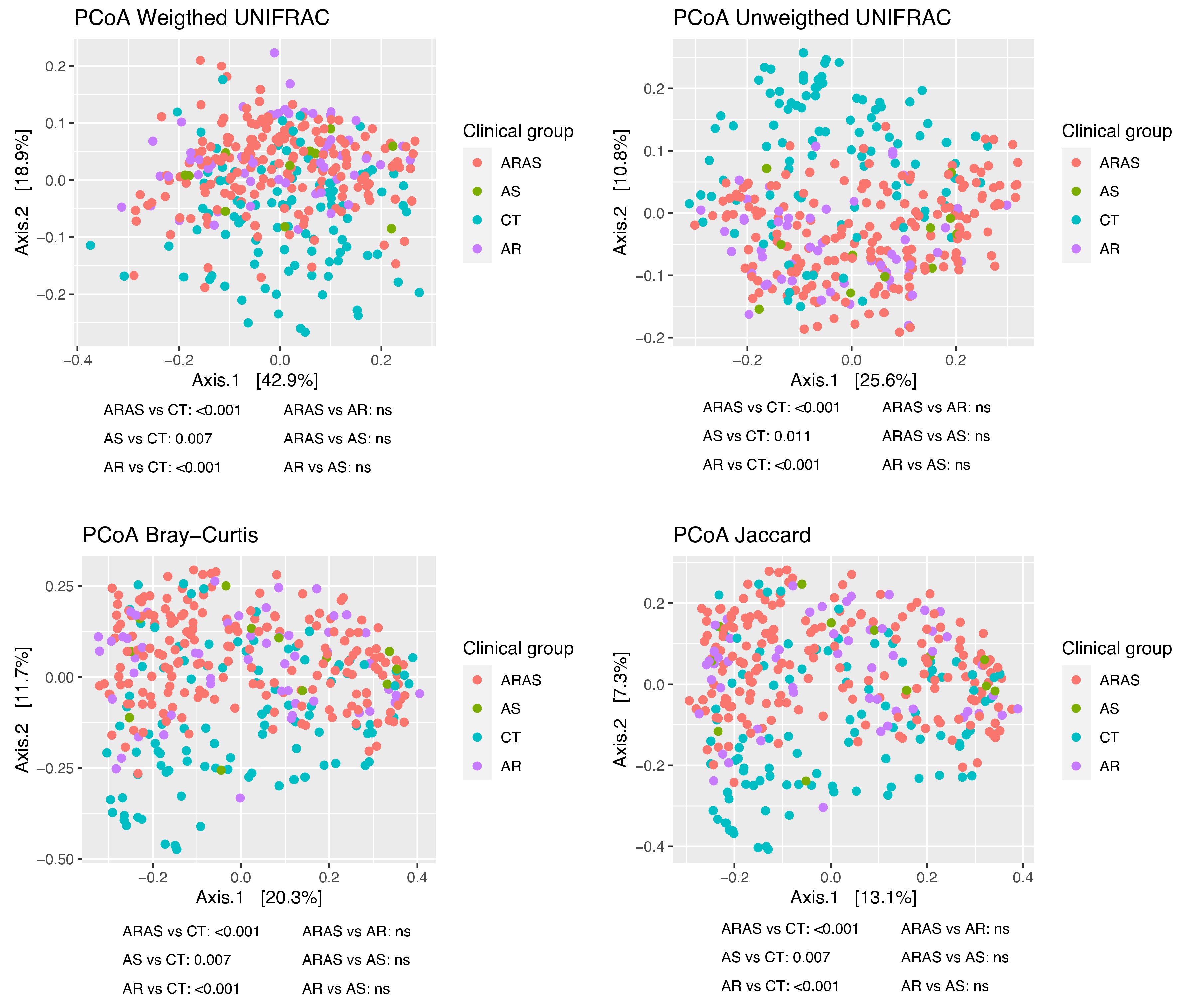
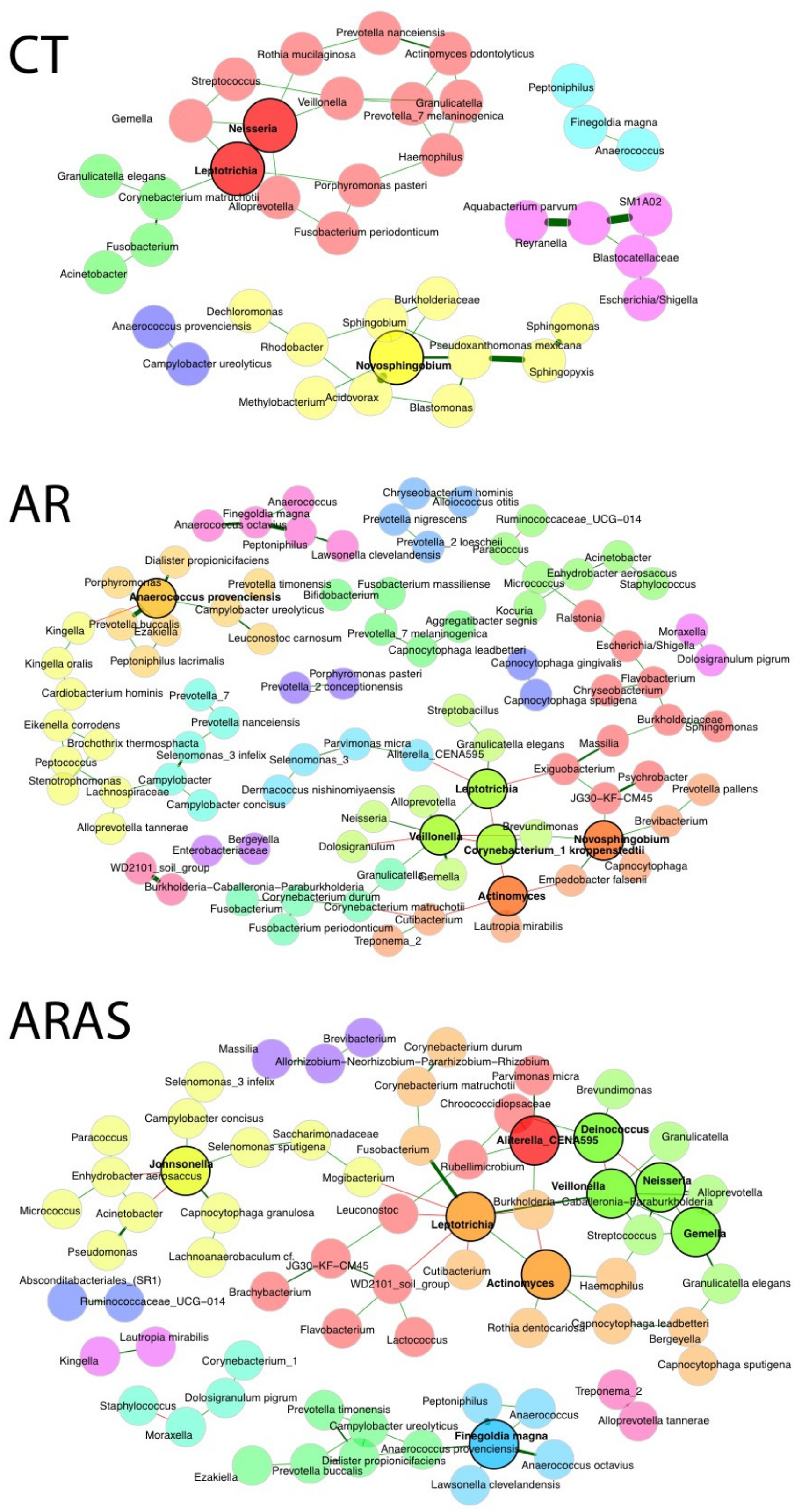
| Mean Relative Proportions (%) | Wilcoxon Test Significance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CT | AS | ARAS | AR | AS-AR | AS-ARAS | ARAS-AR | AS-CT | ARAS-CT | AR-CT | |
| Phylum | ||||||||||
| Actinobacteriota | 42.0 | 21.1 | 21.1 | 18.1 | ns | ns | ns | 0.021 | <0.001 | <0.001 |
| Bacteroidota | 2.3 | 4.1 | 5.3 | 6.5 | ns | ns | ns | ns | <0.001 | <0.001 |
| Firmicutes | 38.4 | 49.8 | 50.4 | 43.7 | ns | ns | 0.036 | ns | <0.001 | ns |
| Proteobacteria | 15.4 | 23.6 | 20.6 | 28.7 | ns | ns | ns | ns | ns | <0.001 |
| Others | 1.9 | 1.4 | 2.5 | 3.1 | - | - | - | - | - | - |
| Genus | ||||||||||
| Anaerococcus | 2.9 | 2.2 | 3.1 | 2.1 | ns | ns | 0.031 | ns | ns | ns |
| Corynebacterium | 33.2 | 19.3 | 16.6 | 14.4 | ns | ns | ns | ns | <0.001 | <0.001 |
| Dolosigranulum | 7.5 | 15.3 | 12.2 | 13.0 | ns | ns | ns | 0.021 | <0.001 | <0.001 |
| Haemophilus | 1.6 | 10.0 | 2.7 | 4.2 | ns | ns | ns | 0.002 | <0.001 | <0.001 |
| Lawsonella | 7.3 | 0.5 | 2.5 | 1.4 | ns | ns | ns | 0.001 | <0.001 | <0.001 |
| Moraxella | 5.2 | 8.6 | 9.3 | 15.3 | ns | ns | ns | 0.002 | <0.001 | <0.001 |
| Neisseriaceae sp. | 2.8 | 2.1 | 3.9 | 2.3 | ns | ns | ns | 0.018 | <0.001 | <0.001 |
| Peptoniphilus | 1.9 | 2.7 | 2.8 | 1.9 | ns | ns | ns | ns | ns | ns |
| Staphylococcus | 18.0 | 19.0 | 20.4 | 13.0 | ns | ns | 0.013 | ns | ns | ns |
| Streptococcus | 3.8 | 6.0 | 5.9 | 7.2 | ns | ns | ns | ns | <0.001 | <0.001 |
| Others | 15.8 | 14.4 | 20.8 | 25.2 | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Losada, M.; Castro-Nallar, E.; Laerte Boechat, J.; Delgado, L.; Azenha Rama, T.; Berrios-Farías, V.; Oliveira, M. Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions. Microorganisms 2023, 11, 683. https://doi.org/10.3390/microorganisms11030683
Pérez-Losada M, Castro-Nallar E, Laerte Boechat J, Delgado L, Azenha Rama T, Berrios-Farías V, Oliveira M. Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions. Microorganisms. 2023; 11(3):683. https://doi.org/10.3390/microorganisms11030683
Chicago/Turabian StylePérez-Losada, Marcos, Eduardo Castro-Nallar, José Laerte Boechat, Luis Delgado, Tiago Azenha Rama, Valentín Berrios-Farías, and Manuela Oliveira. 2023. "Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions" Microorganisms 11, no. 3: 683. https://doi.org/10.3390/microorganisms11030683
APA StylePérez-Losada, M., Castro-Nallar, E., Laerte Boechat, J., Delgado, L., Azenha Rama, T., Berrios-Farías, V., & Oliveira, M. (2023). Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions. Microorganisms, 11(3), 683. https://doi.org/10.3390/microorganisms11030683









