Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequences
2.2. Identification and Analysis of the Thermophilin 13 Biosynthetic Gene Cluster (BGC)
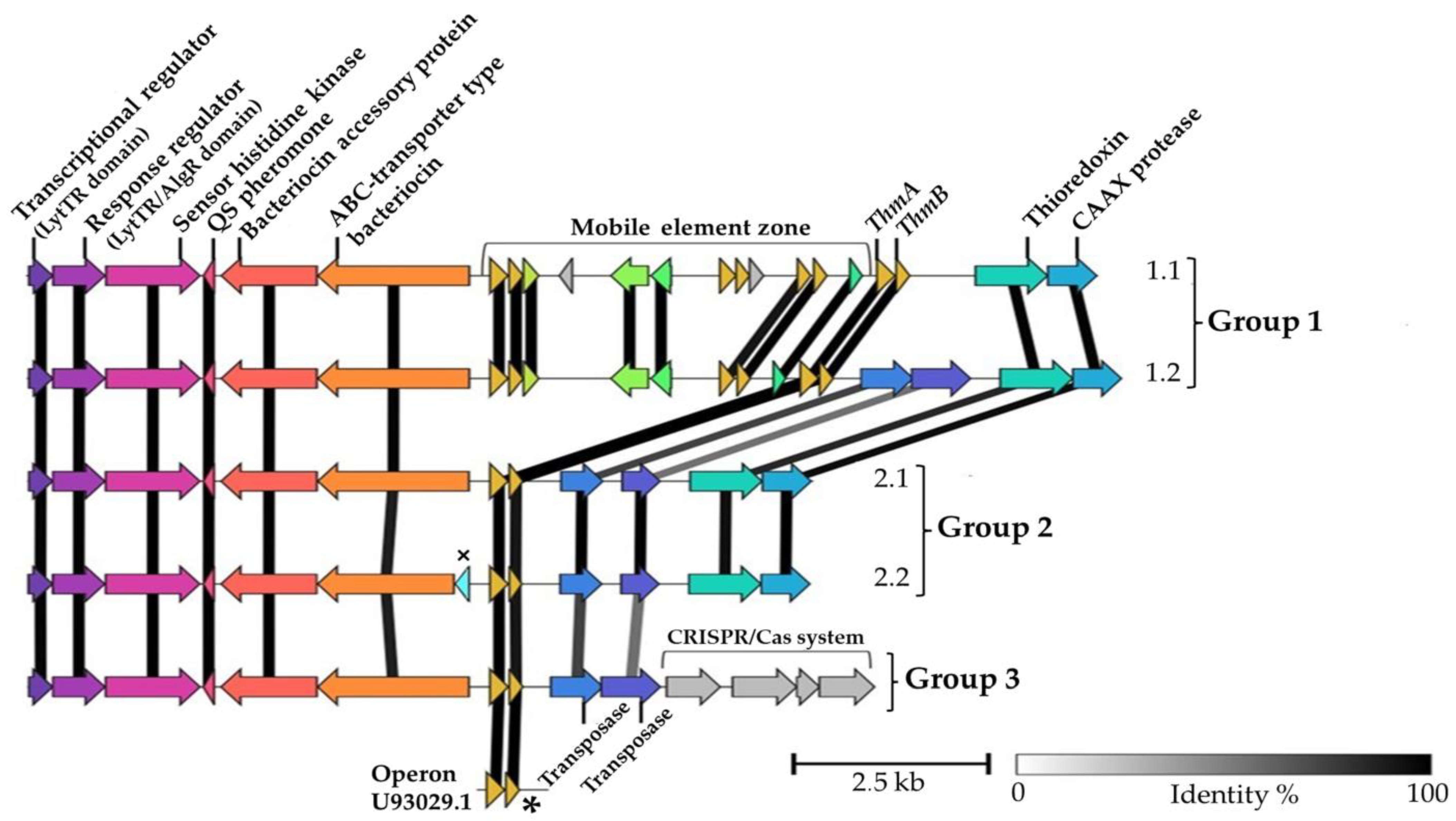
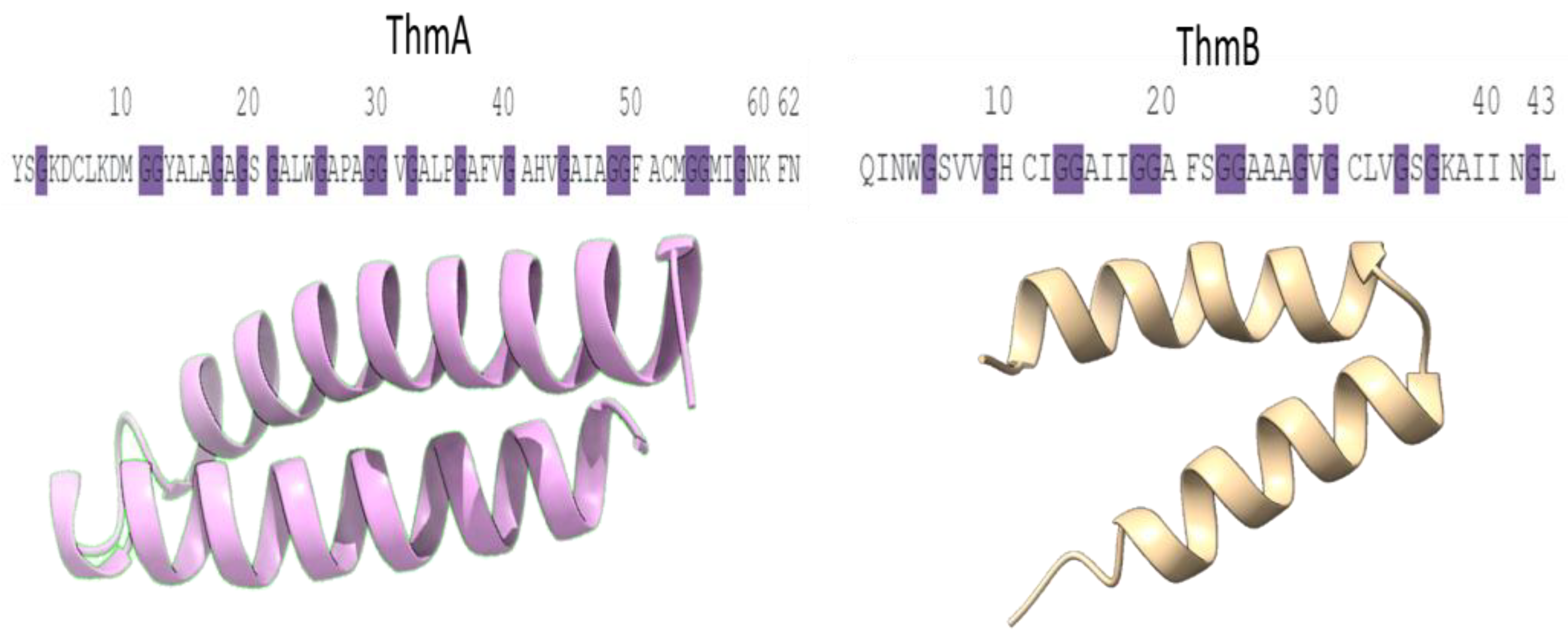
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Delorme, C.; Legravet, N.; Jamet, E.; Hoarau, C.; Alexandre, B.; El-Sharoud, W.M.; Darwish, M.S.; Renault, P. Study of Streptococcus thermophilus Population on a World-Wide and Historical Collection by a New MLST Scheme. Int. J. Food Microbiol. 2017, 242, 70–81. [Google Scholar] [CrossRef]
- Rossi, F.; Marzotto, M.; Cremonese, S.; Rizzotti, L.; Torriani, S. Diversity of Streptococcus thermophilus in Bacteriocin Production; Inhibitory Spectrum and Occurrence of Thermophilin Genes. Food Microbiol. 2013, 35, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.J.; Somkuti, G.A. Characterization of a bacteriocin produced by Streptococcus thermophilus ST134. Appl. Microbiol. Biotechnol. 1995, 42, 330–335. [Google Scholar] [CrossRef]
- Anastasios, A.; George, K. Purification and Characterization of Thermophilin ST-1, a Novel Bacteriocin Produced by Streptococcus thermophilus ACA-DC 0001. Le Lait 2003, 83, 365–378. [Google Scholar] [CrossRef]
- Gilbreth, S.E.; Somkuti, G.A. Thermophilin 110: A Bacteriocin of Streptococcus thermophilus ST110. Curr. Microbiol. 2005, 51, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Kabuki, T.; Uenishi, H.; Watanabe, M.; Seto, Y.; Nakajima, H. Characterization of a Bacteriocin, Thermophilin 1277, Produced by Streptococcus thermophilus SBT1277. J. Appl. Microbiol. 2007, 102, 971–980. [Google Scholar] [CrossRef]
- Ivanova, I.; Miteva, V.; Stefanova, T.; Pantev, A.; Budakov, I.; Danova, S.; Moncheva, P.; Nikolova, I.; Dousset, X.; Boyaval, P. Characterization of a Bacteriocin Produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 1998, 42, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Mathot, A.G.; Beliard, E.; Thuault, D. Streptococcus thermophilus 580 Produces a Bacteriocin Potentially Suitable for Inhibition of Clostridium Tyrobutyricum in Hard Cheese. J. Dairy Sci. 2003, 86, 3068–3074. [Google Scholar] [CrossRef]
- Marciset, O.; Jeronimus-Stratingh, M.C.; Mollet, B.; Poolman, B. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 1997, 272, 14277–14284. [Google Scholar] [CrossRef]
- Oppegård, C.; Rogne, P.; Emanuelsen, L.; Kristiansen, P.E.; Fimland, G.; Nissen-Meyer, J. The Two-Peptide Class II Bacteriocins: Structure, Production, and Mode of Action. J. Mol. Microbiol. Biotechnol. 2007, 13, 210–219. [Google Scholar] [CrossRef]
- Dimov, S.; Ivanova, P.; Harizanova, N. Genetics of Bacteriocins Biosynthesis by Lactic Acid Bacteria. Biotechnol. Biotechnol. Equip. 2005, 19, 4–10. [Google Scholar] [CrossRef]
- Ditu, L.-M.; Chifiriuc, M.; Pelinescu, D.; Avram, I.; Pircalabioru, G.; Mihaescu, G. Class I and II Bacteriocins: Structure, Biosynthesis and Drug Delivery Systems for the Improvement of Their Antimicrobial Activity. Curr. Proteom. 2016, 11, 121–127. [Google Scholar] [CrossRef]
- Zouhir, A.; Hammami, R.; Fliss, I.; Hamida, J. Ben A New Structure-Based Classification of Gram-Positive Bacteriocins. Protein J. 2010, 29, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A More Efficient Report with Usability Improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef]
- Rangwala, S.H.; Kuznetsov, A.; Ananiev, V.; Asztalos, A.; Borodin, E.; Evgeniev, V.; Joukov, V.; Lotov, V.; Pannu, R.; Rudnev, D.; et al. Accessing NCBI Data Using the NCBI Sequence Viewer and Genome Data Viewer (GDV). Genome Res. 2021, 31, 159–169. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST : A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Genome Analysis Operon-Mapper: A Web Server for Precise Operon Identification in Bacterial and Archaeal Genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Booth, T.J.; van Wersch, B.; van Grieken, L.; Medema, M.H.; Chooi, Y.-H. Cblaster: A Remote Search Tool for Rapid Identification and Visualization of Homologous Gene Clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Comelli, E.M.; Guggenheim, B.; Stingele, F.; Neeser, J.R. Selection of Dairy Bacterial Strains as Probiotics for Oral Health. Eur. J. Oral Sci. 2002, 110, 218–224. [Google Scholar] [CrossRef]
- Comelli, E.; Guggenheim, B.; Neeser, J.; Stingele, F.; Cocconcelli, P.S. Treatment fo Actinomtces Naeslundii-Related Diseases with Exogenous Lactic Acid Bacteria Strains. U.S. Patent US 7491386 B2, 17 February 2009. [Google Scholar]
- Renye, J.A.; Somkuti, G.A.; Steinberg, D.H. Thermophilin 109 Is a Naturally Produced Broad Spectrum Bacteriocin Encoded within the Blp Gene Cluster of Streptococcus thermophilus. Biotechnol. Lett. 2019, 41, 283–292. [Google Scholar] [CrossRef]
- Alexandraki, V.; Kazou, M.; Blom, J.; Pot, B.; Papadimitriou, K.; Tsakalidou, E. Comparative Genomics of Streptococcus thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species. Front. Microbiol. 2019, 10, 2916. [Google Scholar] [CrossRef]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative Genomics of the Lactic Acid Bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef]
- Treu, L.; de Diego-Díaz, B.; Papadimitriou, K.; Tsakalidou, E.; Giacomini, A.; Corich, V. Whole-Genome Sequences of Three Streptococcus macedonicus Strains Isolated from Italian Cheeses in the Veneto Region. Genome Announc. 2017, 5, e01358-17. [Google Scholar] [CrossRef]
- Roux, E.; Nicolas, A.; Valence, F.; Siekaniec, G.; Chuat, V.; Nicolas, J.; Le Loir, Y.; Guédon, E. The Genomic Basis of the Streptococcus Thermophilus Health-Promoting Properties. BMC Genom. 2022, 23, 210. [Google Scholar] [CrossRef] [PubMed]
- CIRM-BIA: International Centre for Microbial Resources-Food-Associated Bacteria, STLO UMR 1253 INRAE-Agrocampus 65 rue de saint Brieuc, F-35042 Rennes Cedex, France. CIRM Associated Bacteria. Available online: https://collection-cirmbia.fr/page/Cite%20CIRM-BIA (accessed on 20 November 2022).
- Hu, T.; Cui, Y.; Zhang, Y.; Qu, X.; Zhao, C. Genome Analysis and Physiological Characterization of Four Streptococcus thermophilus Strains Isolated From Chinese Traditional Fermented Milk. Front. Microbiol. 2020, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-S.; Huang, Y.-Y.; Kuang, J.-H.; Yu, J.-J.; Zhou, Q.-Y.; Liu, D.-M. Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea. Microorganisms 2020, 8, 1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Q.; Lu, B.; Shen, H.; Liu, S.; Shi, Y.; Leptihn, S.; Li, H.; Wei, J.; Liu, C.; et al. Whole-Genome Analysis of Probiotic Product Isolates Reveals the Presence of Genes Related to Antimicrobial Resistance, Virulence Factors, and Toxic Metabolites, Posing Potential Health Risks. BMC Genom. 2021, 22, 210. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Park, K.; Kim, K. Genome Analysis of Streptococcus salivarius Subsp. Thermophilus Type Strain ATCC 19258 and Its Comparison to Equivalent Strain NCTC 12958. Arch. Microbiol. 2021, 203, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Upton, M.; Renault, P.; Wirawan, R.E.; Power, D.; Burton, J.P.; Chilcott, C.N.; Tagg, J.R. Salivaricin 9, a New Lantibiotic Produced by Streptococcus salivarius. Microbiology 2011, 157, 1290–1299. [Google Scholar] [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Ekblad, B.; Kristiansen, P.E. NMR Structures and Mutational Analysis of the Two Peptides Constituting the Bacteriocin Plantaricin S. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Russell, A.H.; Truman, A.W. Genome Mining Strategies for Ribosomally Synthesised and Post-Translationally Modified Peptides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [Google Scholar] [CrossRef]
- Oppegård, C.; Schmidt, J.; Kristiansen, P.E.; Nissen-Meyer, J. Mutational Analysis of Putative Helix-Helix Interacting GxxxG-Motifs and Tryptophan Residues in the Two-Peptide Bacteriocin Lactococcin G. Biochemistry 2008, 47, 5242–5249. [Google Scholar] [CrossRef]
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Nikolskaya, A.N.; Galperin, M.Y. A Novel Type of Conserved DNA-Binding Domain in the Transcriptional Regulators of the AlgR/AgrA/LytR Family. Nucleic Acids Res. 2002, 30, 2453–2459. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Qin, H.; Brenner, A.E.; Raghavan, R.; Millar, J.A.; Gu, Q.; Xie, Z.; Kreth, J.; Merritt, J. LytTR Regulatory Systems: A Potential New Class of Prokaryotic Sensory System. PLoS Genet. 2018, 14, e1007709. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, S.E.; Oyston, P.C.F. Structure and Function of the LysR-Type Transcriptional Regulator (LTTR) Family Proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Jia, F.F.; Pang, X.H.; Zhu, D.Q.; Zhu, Z.T.; Sun, S.R.; Meng, X.C. Role of the LuxS Gene in Bacteriocin Biosynthesis by Lactobacillus Plantarum KLDS1.0391: A Proteomic Analysis. Sci. Rep. 2017, 7, 13871. [Google Scholar] [CrossRef]
- Hühne, K.; Axelsson, L.; Holck, A.; Kröckel, L. Analysis of the Sakacin P Gene Cluster from Lactobacillus Sake Lb674 and Its Expression in Sakacin-Negative Lb. Sake Strains. Microbiology 1996, 142, 1437–1448. [Google Scholar] [CrossRef]
- Maldonado-Barragán, A.; West, S.A. The Cost and Benefit of Quorum Sensing-Controlled Bacteriocin Production in Lactobacillus plantarum. J. Evol. Biol. 2020, 33, 101–111. [Google Scholar] [CrossRef]
- Renye, J.A.; Somkuti, G.A. BlpC-Regulated Bacteriocin Production in Streptococcus thermophilus. Biotechnol. Lett. 2013, 35, 407–412. [Google Scholar] [CrossRef]
- Smits, S.H.J.; Schmitt, L.; Beis, K. Self-Immunity to Antibacterial Peptides by ABC Transporters. FEBS Lett. 2020, 594, 3920–3942. [Google Scholar] [CrossRef]
- Venema, K.; Kok, J.; Marugg, J.D.; Toonen, M.Y.; Ledeboer, A.M.; Venema, G.; Chikindas, M.L. Functional Analysis of the Pediocin Operon of Pediococcus Acidilactici PAC1.0: PedB Is the Immunity Protein and PedD Is the Precursor Processing Enzyme. Mol. Microbiol. 1995, 17, 515–522. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Y.; Li, L.; Jiang, X.; Chen, Z.; Zhao, F.; Yi, Y. Biosynthesis and Production of Class II Bacteriocins of Food-Associated Lactic Acid Bacteria. Fermentation 2022, 8, 217. [Google Scholar] [CrossRef]
- Franke, C.M.; Tiemersma, J.; Venema, G.; Kok, J. Membrane Topology of the Lactococcal Bacteriocin ATP-Binding Cassette Transporter Protein LcnC. Involvement of LcnC in Lactococcin A Maturation. J. Biol. Chem. 1999, 274, 8484–8490. [Google Scholar] [CrossRef] [PubMed]
- Beis, K. Structural Basis for the Mechanism of ABC Transporters. Biochem. Soc. Trans. 2015, 43, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Kanonenberg, K.; Schwarz, C.K.W.; Schmitt, L. Type I Secretion Systems—A Story of Appendices. Res. Microbiol. 2013, 164, 596–604. [Google Scholar] [CrossRef]
- Beis, K.; Rebuffat, S. Multifaceted ABC Transporters Associated to Microcin and Bacteriocin Export. Res. Microbiol. 2019, 170, 399–406. [Google Scholar] [CrossRef]
- Wu, K.H.; Tai, P.C. Cys32 and His105 Ate the Critical Residues for the Calcium-Dependent Cysteine Proteolytic Activity of CvaB, an ATP-Binding Cassette Transporter. J. Biol. Chem. 2004, 279, 901–909. [Google Scholar] [CrossRef]
- Mesa-Pereira, B.; O’Connor, P.M.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Controlled Functional Expression of the Bacteriocins Pediocin PA-1 and Bactofencin A in Escherichia Coli. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- García-Curiel, L.; del Rocío López-Cuellar, M.; Rodríguez-Hernández, A.I.; Chavarría-Hernández, N. Toward Understanding the Signals of Bacteriocin Production by Streptococcus Spp. and Their Importance in Current Applications. World J. Microbiol. Biotechnol. 2021, 37, 1–14. [Google Scholar] [CrossRef]
- Duan, Y.; Niu, W.; Pang, L.; Bian, X.; Zhang, Y.; Zhong, G. Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. Int. J. Mol. Sci. 2022, 23, 7231. [Google Scholar] [CrossRef]
- Davey, L.; Halperin, S.A.; Lee, S.F. Thiol-Disulfide Exchange in Gram-Positive Firmicutes. Trends Microbiol. 2016, 24, 902–915. [Google Scholar] [CrossRef]
- Kjos, M.; Snipen, L.; Salehian, Z.; Nes, I.F.; Diep, D.B. The Abi Proteins and Their Involvement in Bacteriocin Self-Immunity. J. Bacteriol. 2010, 192, 2068–2076. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.L.; Abrudan, M.I.; Roberts, I.S.; Rozen, D.E. Diverse Ecological Strategies Are Encoded by Streptococcus pneumoniae Bacteriocin-Like Peptides. Genome Biol. Evol. 2016, 8, 1072–1090. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Mitchell, D.A.; Dixon, J.E.; Grishin, N.V. Expansion of Type II CAAX Proteases Reveals Evolutionary Origin of γ-Secretase Subunit APH-1. J. Mol. Biol. 2011, 410, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, L.; Boutry, C.; Guédon, E.; Guillot, A.; Ibrahim, M.; Grossiord, B.; Hols, P. Quorum-Sensing Regulation of the Production of Blp Bacteriocins in Streptococcus thermophilus. J. Bacteriol. 2007, 189, 7195–7205. [Google Scholar] [CrossRef] [PubMed]
- Hols, P.; Hancy, F.; Fontaine, L.; Grossiord, B.; Prozzi, D.; Leblond-Bourget, N.; Decaris, B.; Bolotin, A.; Delorme, C.; Duskoehrlich, S. New Insights in the Molecular Biology and Physiology of Streptococcus thermophilus Revealed by Comparative Genomics. FEMS Microbiol. Rev. 2005, 29, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Renye, J.R.; Wales, N.; Somkuti, G.A. Bacteriocin with Novel Activity. US 2016/0368953 A1, 22 December 2016. [Google Scholar]
- Blanchard, A.E.; Liao, C.; Lu, T. An Ecological Understanding of Quorum Sensing-Controlled Bacteriocin Synthesis. Cell. Mol. Bioeng. 2016, 9, 443–454. [Google Scholar] [CrossRef]
- Meng, F.; Zhao, H.; Nie, T.; Lu, F.; Zhang, C.; Lu, Y.; Lu, Z. Acetate Activates Lactobacillus Bacteriocin Synthesis by Controlling Quorum Sensing. Appl. Environ. Microbiol. 2021, 87, e007202. [Google Scholar] [CrossRef]
- Rohde, B.H.; Quadri, L.E.N. Functional Characterization of a Three-Component Regulatory System Involved in Quorum Sensing-Based Regulation of Peptide Antibiotic Production in Carnobacterium maltaromaticum. BMC Microbiol. 2006, 6, 93. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D. In Silico Identification of Bacteriocin Gene Clusters in the Gastrointestinal Tract, Based on the Human Microbiome Project’s Reference Genome Database. BMC Microbiol. 2015, 15, 183. [Google Scholar] [CrossRef]

| Strains | GenBank Code | Isolation/Source | Reference |
|---|---|---|---|
| Streptococcus thermophilus B59671 | CP022547.1 | Milk | [28] |
| Streptococcus thermophilus KLDS 3.1003 | CP016877.1 | Lactic starter (for yoghurt production) | [29] |
| Streptococcus thermophilus LMD-9 | CP086001.1 | Lactic starter (for yoghurt and mozzarella production) | [30] |
| Streptococcus macedonicus 19AS | PEBN00000000.1 *** | Cheese | [31] |
| Streptococcus thermophilus STH_CIRM_1049 | LR822034.1 | Lactic starter (for yoghurt production) | [32,33] ** |
| Streptococcus thermophilus STH_CIRM_1048 | LR822033.1 | Lactic starter (for yoghurt production) | [32,33] ** |
| Streptococcus thermophilus CS9 | CP030927.1 | Fermented milk | [34] |
| Streptococcus thermophilus DMST-H2 | CP063275.1 | Probiotic products | [35] |
| Streptococcus thermophilus TK-P3A | CP045596.1 | Pasteurised milk | [36] |
| Streptococcus thermophilus ATCC 19258 * | CP038020.1 | Milk | [37] |
| Streptococcus thermophilus NCTC 12958 * | LS483339.1 | Milk | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salini, F.; Iacumin, L.; Comi, G.; Dicks, L.M.T. Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production. Microorganisms 2023, 11, 611. https://doi.org/10.3390/microorganisms11030611
Salini F, Iacumin L, Comi G, Dicks LMT. Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production. Microorganisms. 2023; 11(3):611. https://doi.org/10.3390/microorganisms11030611
Chicago/Turabian StyleSalini, Francesco, Lucilla Iacumin, Giuseppe Comi, and Leon M. T. Dicks. 2023. "Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production" Microorganisms 11, no. 3: 611. https://doi.org/10.3390/microorganisms11030611
APA StyleSalini, F., Iacumin, L., Comi, G., & Dicks, L. M. T. (2023). Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production. Microorganisms, 11(3), 611. https://doi.org/10.3390/microorganisms11030611








