Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Physicochemical Characteristics of Soils
2.2. DNA Extraction and High-Throughput Sequencing
2.3. Microbial Culture and Isolation of Actinobacteria
2.4. The 16S rRNA Gene Sequencing and Sequencing Data Processing
2.5. Strain Characteristics
3. Results
3.1. Physicochemical and Bacteriological Characteristics of Flaming Mountain Soils
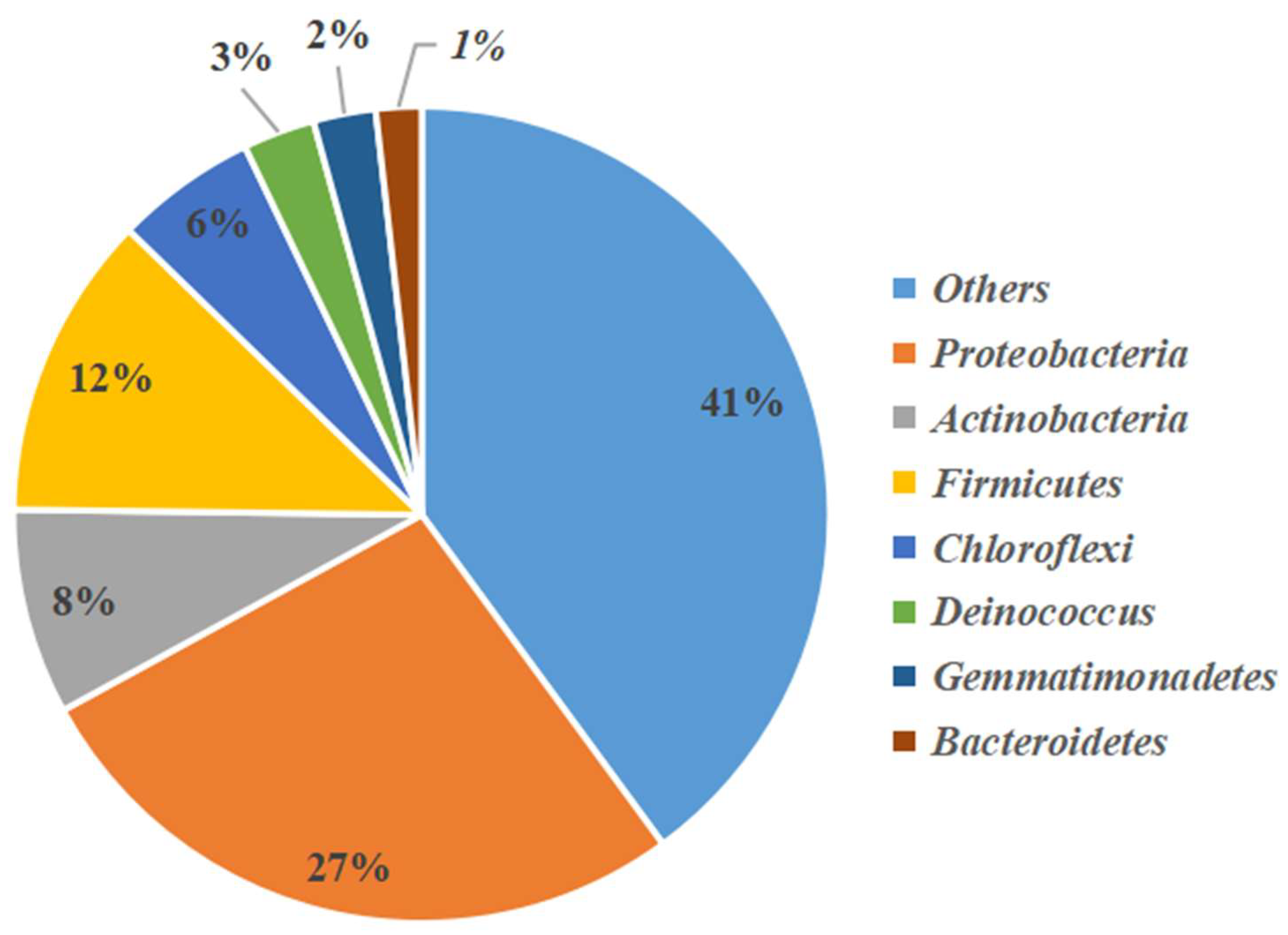
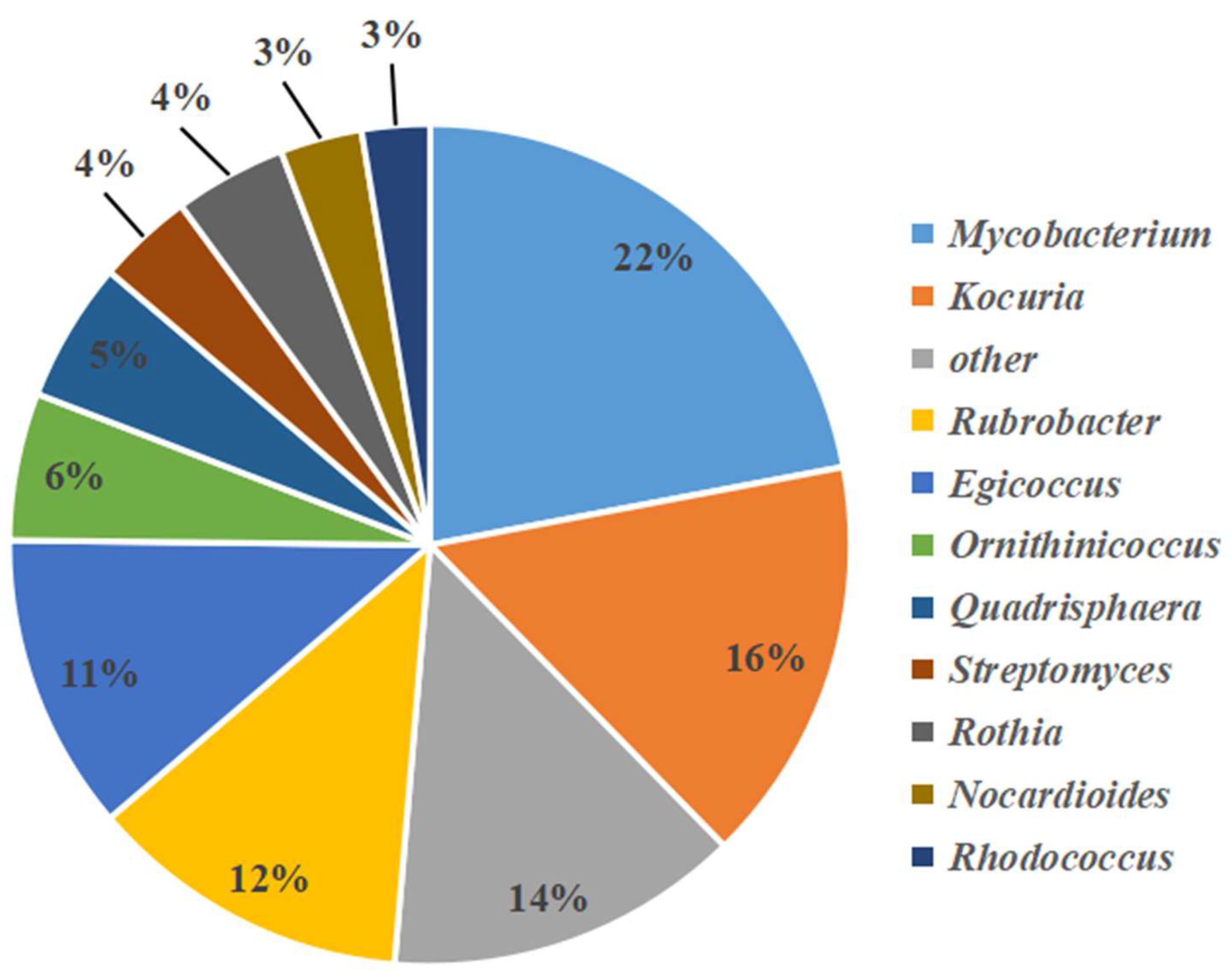
3.2. Analysis of Bacterial and Actinobacterial Community Structures by High-Throughput Sequencing
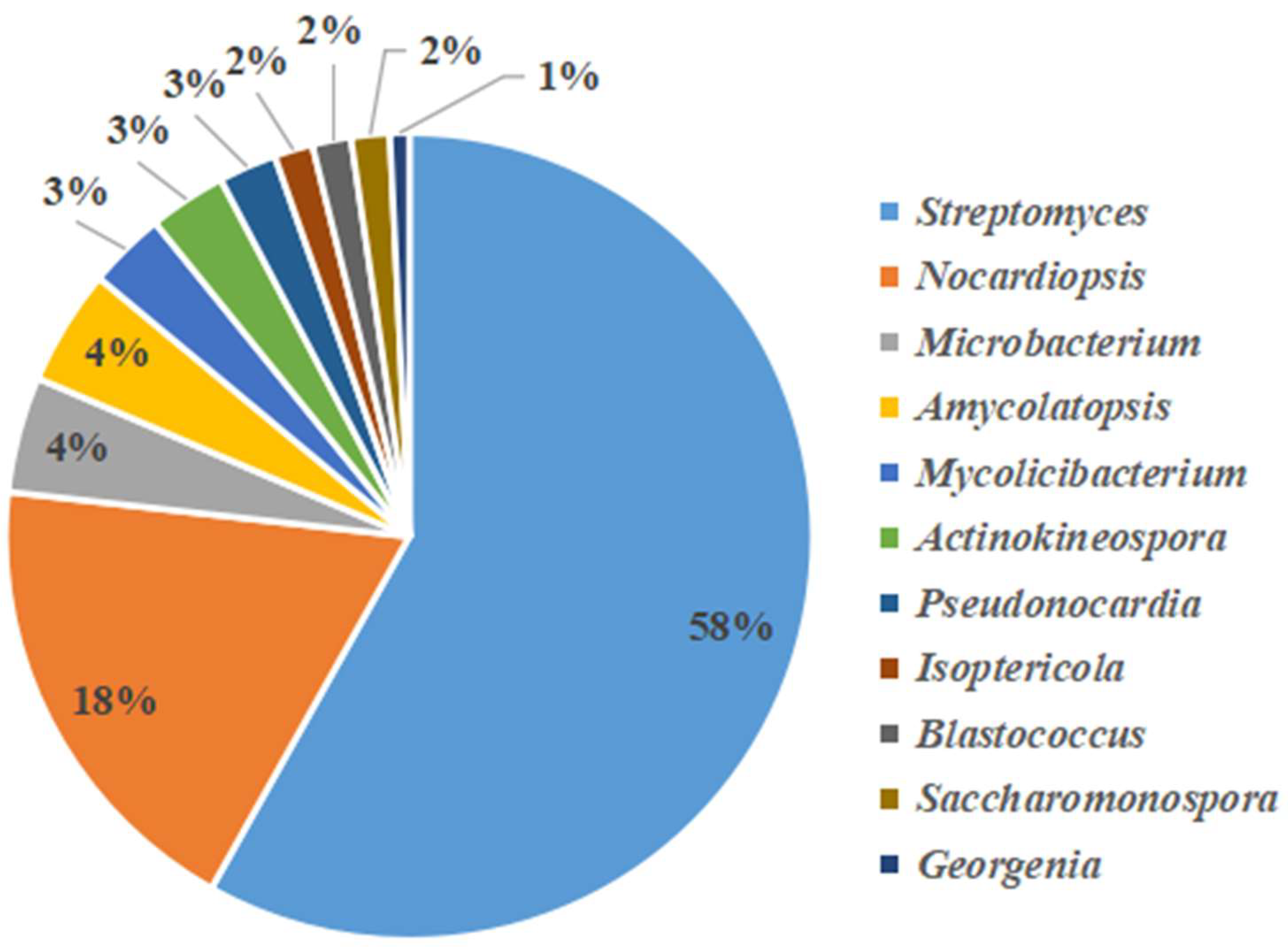
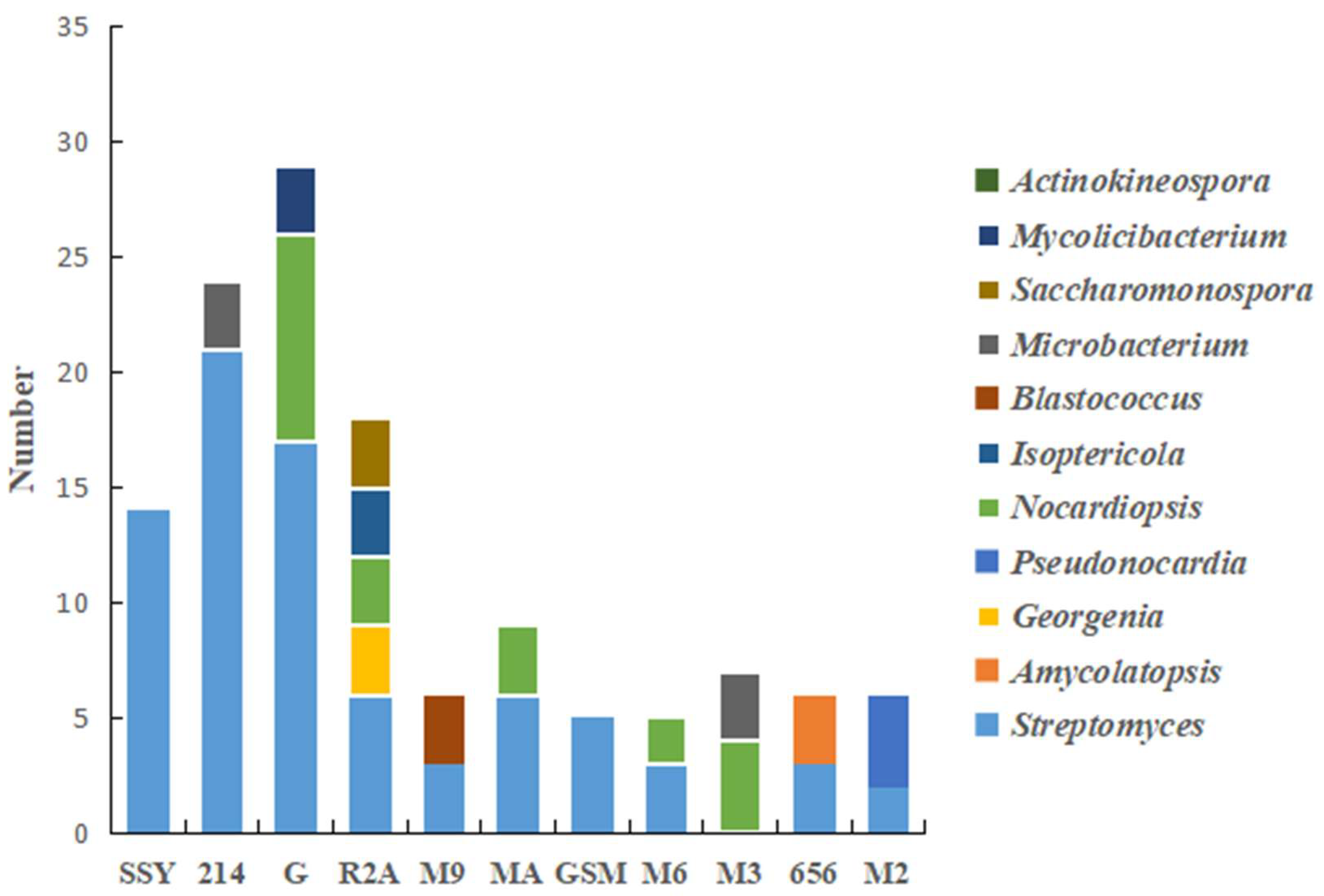
3.3. Analysis of Culturable Actinobacteria Diversity
3.3.1. Screening and Identification of Culturable Actinobacteria
3.3.2. Effects of Different Media on the Screening of Actinobacteria
3.3.3. Potential New Species of Actinobacteria
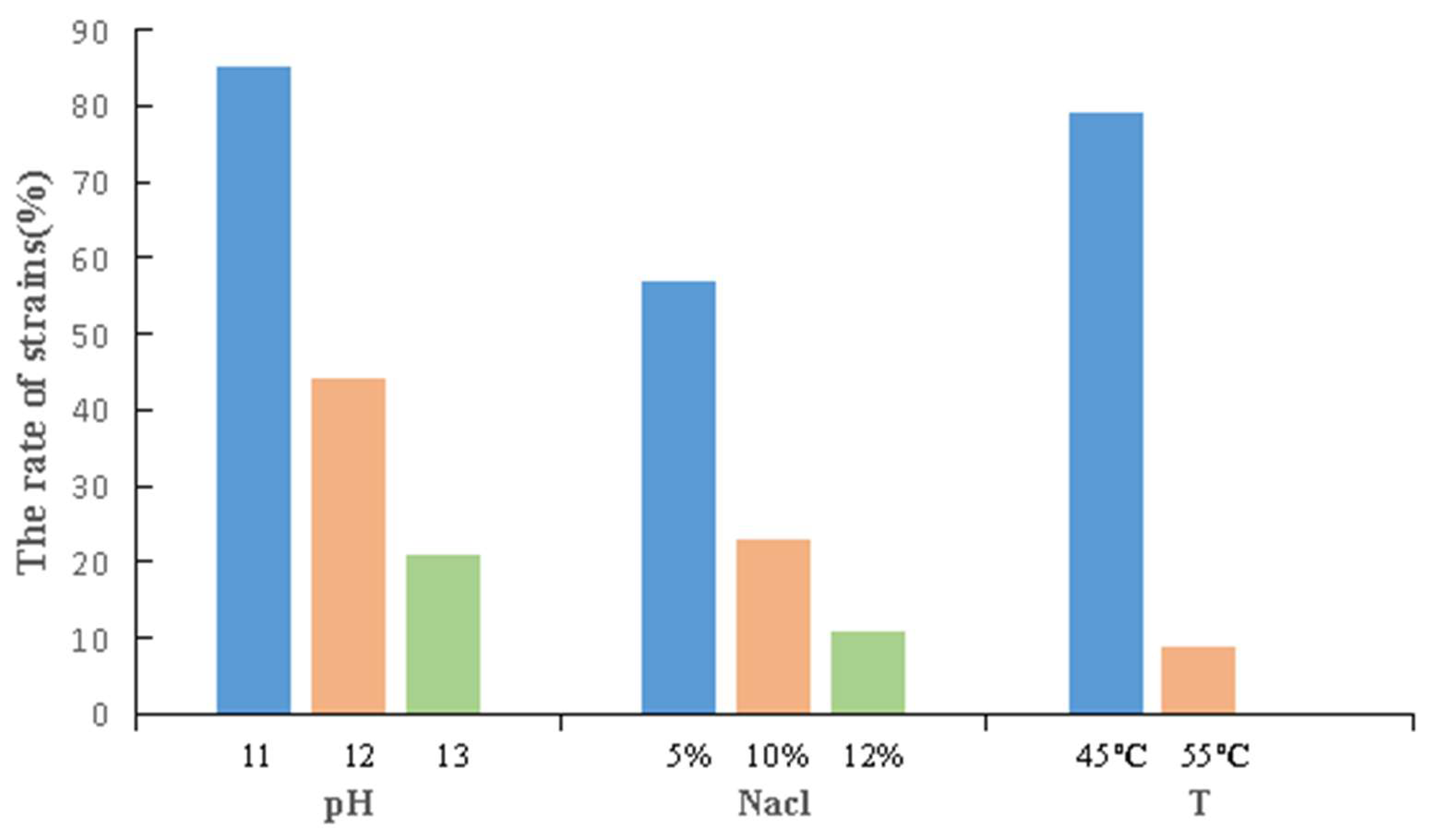
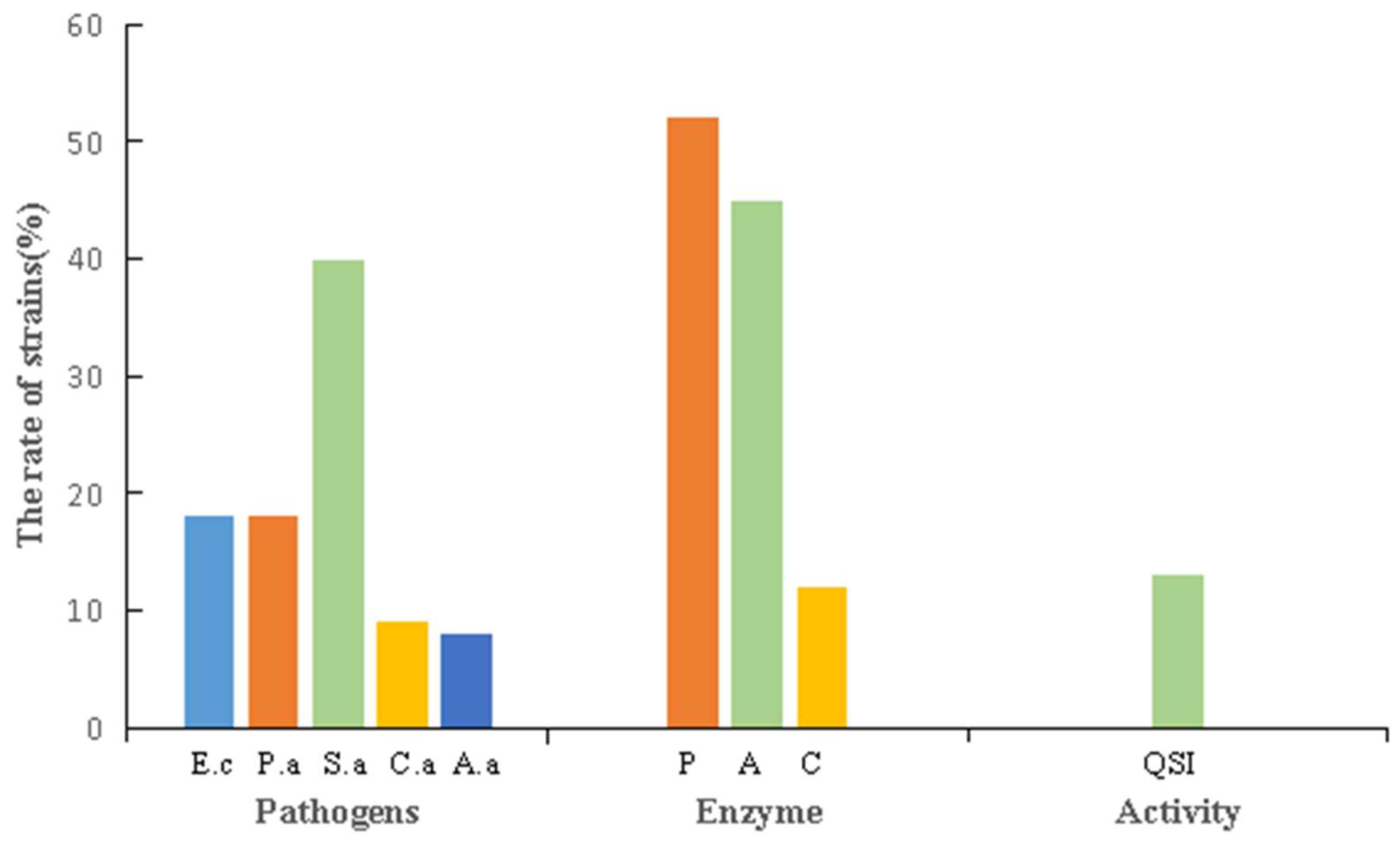
3.4. Stress Tolerance and Biological Activity of the Strains
3.4.1. Stress Tolerance of the Strains
3.4.2. Biological Activity of the Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Almuhayawi, M.S.; Hassan AH, A.; Al Jaouni, S.K.; Alkhalifah, D.H.M.; Hozzein, W.N.; Selim, S.; Khamis, G. Influence of elevated CO2 on nutritive value and health-promoting prospective of three genotypes of Alfalfa sprouts (Medicago Sativa). Food Chem. 2021, 15, 340. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Carro, L.; García López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare taxa and dark microbial matter: Novel bioactive actinobacteria abound in Atacama Desert soils. Antonie van Leeuwenhoek 2018, 111, 1315–1332. [Google Scholar] [CrossRef]
- Idris, H.; Goodfellow, M.; Sanderson, R.; Asenjo, J.A.; Bull, A.T. Actinobacterial Rare Biospheres and Dark Matter Revealed in Habitats of the Chilean Atacama Desert. Sci. Rep. 2017, 7, 8373. [Google Scholar] [CrossRef]
- Zhao, K.; Yang, D.; Zeng, P.; Huang, Z.; Wu, W.; Li, B.; Teng, T. Effect of water content on the failure pattern and acoustic emission characteristics of red sandstone. Int. J. Rock Mech. Min. Sci. 2021, 142, 104709. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.G.; Wang, H.F.; Yang, L.L.; Zhou, X.K.; Zhi, X.Y.; Duan, Y.Q.; Li, W.J. Egibacter rhizosphaerae gen. nov.sp. nov.an obligately halophilic, facultatively alkaliphilic actinobacterium and proposal of Egibaceraceae fam. nov. and Egibacterales ord. nov. Microbiol. Soc. 2016, 66, 283–289. [Google Scholar] [CrossRef]
- Gulijamila, A.; Dilinuer, A. Climate change in Turpan city in recent 57 years. J. Guangzhou Univ. 2012, 11, 83–87. [Google Scholar]
- Pan, H.X.; Cheng, Z.M.; Wang, L.X. Microbial resources in extreme environment in Turpan Basin of Xinjiang. J. Arid. Land Resour. Environ. 2002, 4, 91–94. [Google Scholar]
- Xia, Z.F. Polyphasic Identification and Antifungal Metabolite Analysis of Extreme Actinomycetes Isolated in Xinjian. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2015. [Google Scholar]
- Oberhardt, M.A.; Zarecki, R.; Gronow, S.; Lang, E.; Klenk, H.-P.; Gophna, U.; Ruppin, E. Harnessing the landscape of microbial culture media to predict new organism–media pairings. Nat. Commun. 2015, 6, 8493. [Google Scholar] [CrossRef]
- Campos-M, M.; Campos-C, R. Applications of quartering method in soils and foods. Int. J. Eng. Res. Appl. 2017, 7, 35–39. [Google Scholar] [CrossRef]
- Li, S.; Lei, X.; Qin, L.; Sun, X.; Wang, L.; Zhao, S.; Wang, M.; Chen, S. Fe(III) reduction due to low pe+pH contributes to reducing Cd transfer within a soil-rice system. J. Hazard. Mater. 2021, 415, 125668. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.Y.; Chen, T.; Zhang, G.; Wu, F.; Zhang, W.; Liu, G.; Wu, Y.; Xu, Y.; Tian, M. The diversity of culturable bacteria in the Eastern Edge of Kumtag Desert and primary study of their Coupling Characteristic of Radiation-Resistance and Anti-Oxidation. Environ. Sci. China 2021, 12, 5921–5932. [Google Scholar] [CrossRef]
- Parlapani, F.; Meziti, A.; Kormas, K.; Boziaris, I. Indigenous and spoilage microbiota of farmed sea bream stored in ice identified by phenotypic and 16S rRNA gene analysis. Food Microbiol. 2013, 104. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Andreevskaya, M.; Johansson, P.; Laine, P.; Smolander, O.P.; Sonck, M.; Rahkila, R.; Jääskeläinen, E.; Paulin, L.; Auvinen, P.; Björkroth, J. Genome sequence and transcriptome analysis of meat-spoilage-associated lactic acid bacterium Lactococcus piscium MKFS47. Appl. Environ. Microbiol. 2015, 105, 6178–6186. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2013, 32, 1792–1797. [Google Scholar] [CrossRef]
- Li, L. Isolation and identification cultivable microbes from the Antarctic Krill (Euphausia superba) and other Antarctic samples. Int. J. Ecol. 2018, 7, 29–36. [Google Scholar] [CrossRef]
- Fang, B.-Z.; Han, M.-X.; Jiao, J.-Y.; Xie, Y.-G.; Zhang, X.-T.; Liu, L.; Zhang, Z.-T.; Xiao, M.; Li, W.-J. Streptomyces cavernae sp. nov., a novel actinobacterium isolated from a karst cave sediment sample. Int. J. Syst. Evol. Microbiol. 2020, 70, 120–125. [Google Scholar] [CrossRef]
- Salina, E.G.; Ekins, S.; Makarov, V.A. A rapid method for estimation of the efficacy of potential antimicrobials in humans and animals by agar diffusion assay. Chem. Biol. Drug Des. 2019, 93, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Finstad, K.; Pfeiffer, M.; Amundson, R. Hyperarid Soils and the Soil Taxonomy. Soil Sci. Soc. Am. J. 2014, 78, 1845–1851. [Google Scholar] [CrossRef]
- Bull, A. Actinobacteria of the extremobiosphere. In Extremophiles Handbook; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1203–1240. [Google Scholar] [CrossRef]
- Acosta-Martinez, V.; Moore-Kucera, J.; Cotton, J.; Gardner, T.; Wester, D. Soil enzyme activities during the 2011 Texas record drought/heat wave and implications to biogeochemical cycling and organic matter dynamics. Appl. Soil Ecol. 2014, 75, 43–51. [Google Scholar] [CrossRef]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 2009, 95, 2. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Asenjo, J.A. Microbiology of hyper-arid environments: Recent insights from the Atacama Desert, Chile. Van Leeuw J. Microb. 2013, 103, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shi, Y.-L.; Wang, H.; Zhang, T.; Yu, L.-Y.; Sun, H.; Zhang, Y.-Q. Diversity of Bacteria and the Characteristics of Actinobacteria Community Structure in Badain Jaran Desert and Tengger Desert of China. Front. Microbiol. 2018, 9, 1068. [Google Scholar] [CrossRef]
- León-Sobrino, C.; Ramond, J.B.; Maggs-Kölling, G.; Cowan, D.A. Nutrient acquisition, rather than stress response over diel cycles, drives microbial transcription in a hyper-arid Namib Desert soil. Front. Microbiol. 2019, 10, 1054. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Pan, H. Study on extreme environmental microbial resources in Turpan Basin of Xinjiang-Ecological distribution of soil microorganisms in Turpan Basin. Arid. Zone Res. 1991, 43–46. [Google Scholar] [CrossRef]
- Chen, D.; Li, K.; Wu, Q.; He, Y.; Yin, X. Comparison of two extraction methods in extraction of total DNA from soil microbes in the rhizosphere of pitaya. J. Trop. Biol. 2019, 10. [Google Scholar] [CrossRef]
- Martin, J.; Chrystala, C.; Tristan, C.; Charles, W.; Mark, J. High-throughput sequencing of 16S rRNA gene amplicons: Effects of extraction procedure, primer length and annealing temperature. PLoS ONE 2017, 7. [Google Scholar] [CrossRef]
- Saker, R.; Meklat, A.; Bouras, N.; Zitouni, A.; Mathieu, F.; Spröer, C.; Klenk, H.-P.; Sabaou, N. Diversity and antagonistic properties of culturable halophilic actinobacteria in soils of two arid regions of septentrional Sahara: M’zab and Zibans. Ann. Microbiol. 2015, 65, 2241–2253. [Google Scholar] [CrossRef]
- Fatima, A.; Aftab, U.; Shaaban, K.A.; Thorson, J.S.; Sajid, I. Spore forming Actinobacterial diversity of Cholistan Desert Pakistan: Polyphasic taxonomy, antimicrobial potential and chemical profiling. BMC Microbiol. 2019, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, R.; Moll, G. Ein neues knospendes Bakterium aus der Ostsee. Arch. Microbiol. 1970, 70, 243–265. [Google Scholar] [CrossRef]
- Fan, G.N.; Fu, S.Z.; Cai, H.Y. Overview of studies on microorganisms in extreme environments. Fujian Hot Work. Sci. Technol. 2000, 12–15. [Google Scholar]
- Connon, S.; Giovannoni, S. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 2002, 68. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S. The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 2013, 16, 636–642. [Google Scholar] [CrossRef]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Genet. 2015, 13, 217–229. [Google Scholar] [CrossRef]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef]
- Martiny, A.C. High proportions of bacteria are culturable across major biomes. ISME J. 2019, 13, 2125–2128. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Qin, Y.; Ou, X.L.; Cui, Z.L.; Wang, A.Y.; Zhu, J.B. Screening of thermostable protease-producing bacterium and its enzyme-producing condition. China Brew. 2012, 31, 98–101. [Google Scholar]
- Nguyen, H.T.; Pokhrel, A.R.; Nguyen, C.T.; Pham, V.T.T.; Dhakal, D.; Lim, H.N.; Jung, H.J.; Kim, T.-S.; Yamaguchi, T.; Sohng, J.K. Streptomyces sp. VN1, a producer of diverse metabolites including non-natural furan-type anticancer compound. Sci. Rep. 2020, 10, 1756. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.; Houssen, W.; Harrison, W.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, S.; Hu, X.; You, X.; Sun, C. Studies on secondary metabolites from the Taklimakan Desert-derived Streptomyces sp. 90. Chin. J. Antibiot. 2022, 47, 654–660. [Google Scholar] [CrossRef]
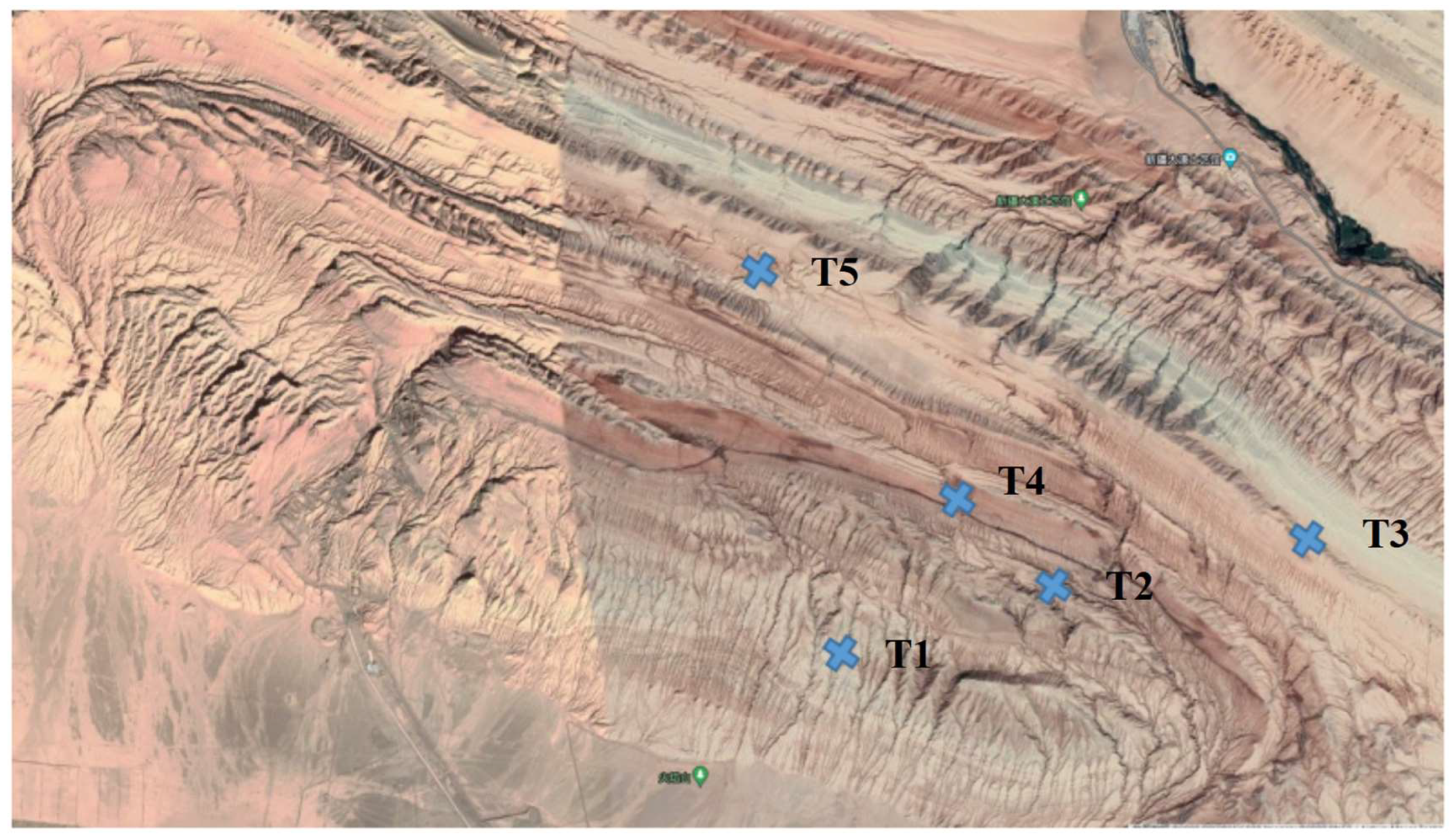
| Sample Name | Location (Latitude, Longitude) | Soil Moisture (%) | pH | Organic Matter (g/kg) | Soluble Salt (g/kg) | Total Nitrogen (g/kg) |
|---|---|---|---|---|---|---|
| T1 | 42°92′98″ N, 89°52′54″ E | 0.2 | 8.84 | 9.8 | 343.7 | 0.26 |
| T2 | 42°93′40″ N, 89°55′40″ E | 0.22 | 8.97 | 6.5 | 551 | 0.1 |
| T3 | 42°93′64″ N, 89°53′12″ E | 0.27 | 9.08 | 7.9 | 668.4 | 0.13 |
| T4 | 42°93′47″ N, 89°53′62″ E | 0.33 | 8.94 | 1.1 | 219.6 | 0.12 |
| T5 | 42°94′51″ N, 89°52′12″ E | 0.26 | 8.74 | 10 | 283.8 | 0.22 |
| Samples | CFU/g | CFU/g (Actinobacteria) | Actinobacteria Ratio (%) |
|---|---|---|---|
| T1 | 3.58 × 103 | 2.54 × 103 | 71 |
| T2 | 2.1 × 103 | 1.72 × 103 | 82 |
| T3 | 7.88 × 103 | 6.14 × 103 | 78 |
| T4 | 2.47 × 103 | 1.95 × 103 | 79 |
| T5 | 5.32 × 103 | 4.25 × 103 | 80 |
| Samples | Raw Number | Clean Number | Effective Number | OTU Number |
|---|---|---|---|---|
| T1 | 239,111 | 230,725 | 188,077 | 1690 |
| T2 | 257,611 | 252,630 | 189,403 | 1157 |
| T3 | 274,307 | 268,002 | 195,385 | 2762 |
| T4 | 254,272 | 248,614 | 190,818 | 1611 |
| T5 | 255,370 | 254,253 | 199,780 | 1226 |
| Total | 1,280,671 | 1,254,224 | 963,463 | 2994 |
| NO. | Family | Genus | Species | Number | Ratio (%) |
|---|---|---|---|---|---|
| 1 | Streptomycetaceae | Streptomyces | S.thermolilacinus | 15 | 11.36% |
| S.werraensis | 2 | 1.51% | |||
| S.glomeratus | 4 | 3.03% | |||
| S.euryhalinus | 3 | 2.27% | |||
| S.pini | 2 | 1.51% | |||
| S.mutabilis | 1 | 0.75% | |||
| S.indiaensis | 6 | 4.54% | |||
| S.qinglanensis | 7 | 5.30% | |||
| S. nanhaiensis | 3 | 2.27% | |||
| S.rochei | 4 | 3.03% | |||
| S.radiopugnans | 12 | 9.09% | |||
| S.heliomycini | 6 | 4.54% | |||
| S.mangrovicola | 3 | 2.27% | |||
| S.lavendulocolor | 6 | 4.54% | |||
| S.kanasensis | 3 | 2.27% | |||
| 2 | Mycobacteriaceae | Mycobacterium | M.arcueilense | 4 | 3.03% |
| 3 | Promicromonosporaceae | Isoptericola | I.halotolerans | 2 | 1.51% |
| 4 | Microbacteriaceae | Microbacterium | M.algeriense | 4 | 3.03% |
| M. oryzae | 2 | 1.51% | |||
| 5 | Nocardiopsaceae | Nocardiopsis | N.dassonvillei | 16 | 12.12% |
| N.alborubida | 8 | 6.06% | |||
| 6 | Pseudonocardiaceae | Saccharomonospora | S.azurea | 2 | 1.51% |
| Pseudonocardia | P.cypriaca | 3 | 2.27% | ||
| Amycolatopsis | A.marina | 7 | 5.30% | ||
| Actinokineospora | A.fastidiosa | 4 | 3.03% | ||
| 7 | Geodermatophilaceae | Blastococcus | B.aggregatus | 2 | 1.51% |
| 8 | Micrococcales | Georgenia | G.satyanarayanai | 1 | 0.75% |
| NO. | Similar Strain | Accession | Similarity |
|---|---|---|---|
| G24 | Streptomyces thermolilacinus | NR 125444.1 | 98.35% |
| G36 | Saccharomonospora azurea | NR 029371.1 | 98.07% |
| G85 | Streptomyces qinglanensis | NR 109303.1 | 97.95% |
| G86 | Nocardiopsis dassonvillei | NR 074635.1 | 98.32% |
| GH22 | Streptomyces pini | NR 108264.1 | 98.08% |
| G150 | Georgenia satyanarayanai | NR 117051.1 | 96.72% |
| Name | Similar Strain | pH 12 | pH 13 | 5% NaCl | 10% NaCl | 12% NaCl | 45 °C | 55 °C |
|---|---|---|---|---|---|---|---|---|
| G131 | Streptomyces indiaensis | + | + | + | − | − | + | + |
| G86 | Nocardiopsis. sp. | + | − | + | + | + | + | − |
| D119 | Streptomyces euryhalinus | + | + | + | + | + | − | − |
| G24 | Streptomyces. sp. | + | − | + | − | − | + | − |
| G114 | Streptomyces heliomycini | + | + | + | + | − | + | + |
| D70 | Streptomyces mutabilis | + | − | + | − | − | − | − |
| GH22 | Streptomyces. sp. | + | − | + | − | − | + | − |
| NO. | Protease | Amylase | Cellulase | E. coli | P. aeruginosa | S. aureus | C. albicans | A. alternata | QSI Activity |
|---|---|---|---|---|---|---|---|---|---|
| G131 | + | − | − | + | − | + | + | − | − |
| G86 | + | + | − | − | + | + | − | − | − |
| D119 | + | + | + | − | + | + | − | + | − |
| G24 | − | + | − | − | + | − | + | − | + |
| G114 | + | + | − | − | − | + | − | + | − |
| D70 | − | + | − | + | − | − | + | − | − |
| GH22 | + | + | − | − | − | + | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Wang, Y.; Bai, X.; Chu, M.; Yi, Y.; Zhu, J.; Gu, M.; Jiang, L.; Zhang, Z. Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China. Microorganisms 2023, 11, 489. https://doi.org/10.3390/microorganisms11020489
He Z, Wang Y, Bai X, Chu M, Yi Y, Zhu J, Gu M, Jiang L, Zhang Z. Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China. Microorganisms. 2023; 11(2):489. https://doi.org/10.3390/microorganisms11020489
Chicago/Turabian StyleHe, Zixuan, Yuxian Wang, Xiaoyu Bai, Min Chu, Yuanyang Yi, Jing Zhu, Meiying Gu, Ling Jiang, and Zhidong Zhang. 2023. "Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China" Microorganisms 11, no. 2: 489. https://doi.org/10.3390/microorganisms11020489
APA StyleHe, Z., Wang, Y., Bai, X., Chu, M., Yi, Y., Zhu, J., Gu, M., Jiang, L., & Zhang, Z. (2023). Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China. Microorganisms, 11(2), 489. https://doi.org/10.3390/microorganisms11020489





