Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era
Abstract
1. Overview of Human Coronaviruses and Their Public Health Impacts
1.1. Human Coronaviruses Associated with the Common Cold
1.2. Emergent Coronaviruses—SARS-CoV, MERS-CoV, and SARS-CoV-2
2. Coronavirus Biology
2.1. Classification
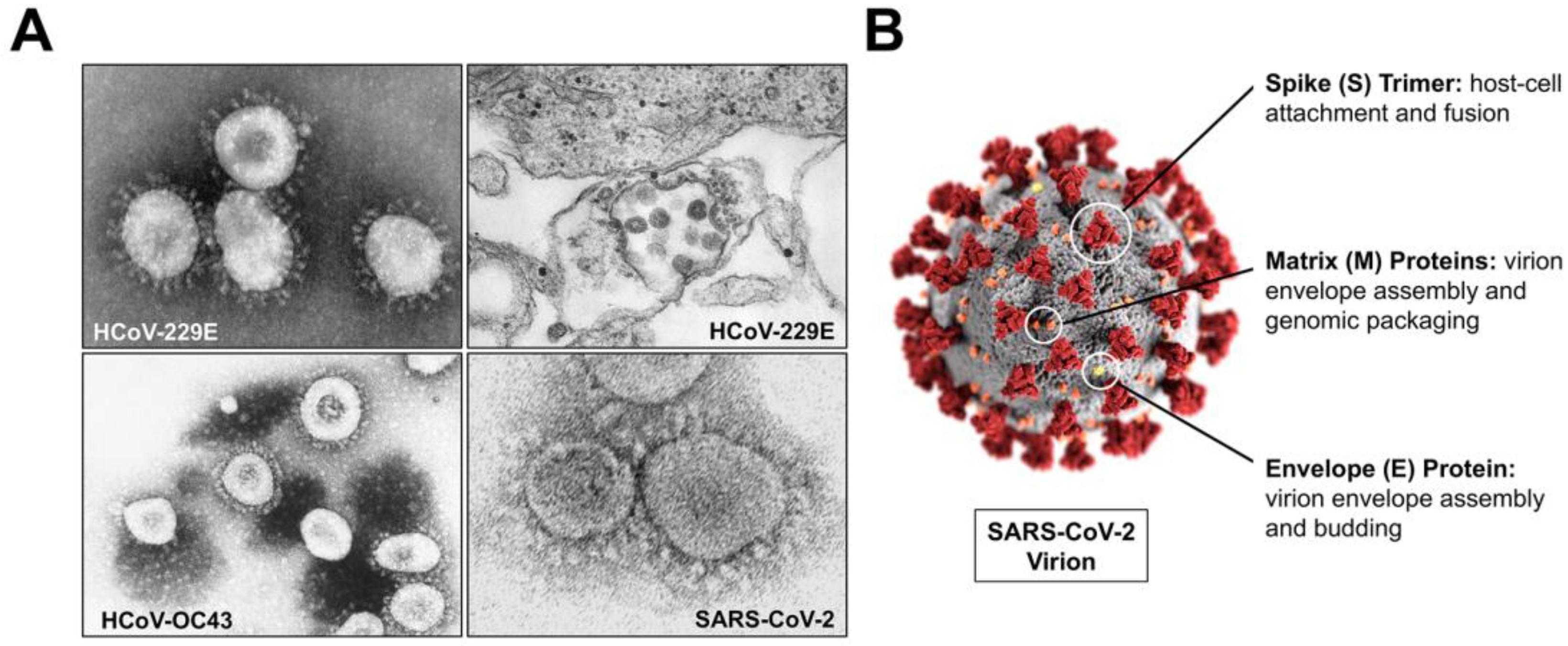
2.2. Structure
2.3. Genomic Organization and Phylogeny of Human Coronavirus Common Cold Strains and SARS-CoV-2
2.4. Replication
3. Coronavirus Pathogenesis and Disease
3.1. Human Coronavirus Diseases
3.2. Transmission and Cellular Infection
3.3. Modulation of Innate Immunity to Coronavirus Infection
3.4. Immunopathology and Modulation of Adaptive Immunity to Coronavirus Infections
3.5. Seasonality and Endemicity of Human Coronaviruses
4. Emergence and Evolution of Common Cold Coronaviruses and SARS-CoV-2
4.1. Emergence and Evolution of Human Coronaviruses Associated with the Common Cold
4.2. Emergence and Evolution of SARS-CoV-2
4.3. Zoonotic Potential for Novel Human Coronaviruses
5. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encycl. Virol. 2021, 2, 428–440. [Google Scholar] [CrossRef]
- Tyrrell, D.A.J.; Bynoe, M.L. Cultivation of a Novel Type of Common-Cold Virus in Organ Cultures. Br. Med. J. 1965, 1, 1467–1470. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic Origins of Human Coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rasool, S.; Fielding, B.C. Understanding Human Coronavirus HCoV-NL63. Open Virol. J. 2010, 4, 76–84. [Google Scholar] [CrossRef] [PubMed]
- van der Hoek, L.; Sure, K.; Ihorst, G.; Stang, A.; Pyrc, K.; Jebbink, M.F.; Petersen, G.; Forster, J.; Berkhout, B.; Überla, K. Croup Is Associated with the Novel Coronavirus NL63. PLoS Med. 2005, 2, e240. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.; Chan, K.; Tsoi, H.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef]
- Talbot, H.K.; Shepherd, B.E.; Crowe, J.E.; Griffin, M.R.; Edwards, K.M.; Podsiad, A.B.; Tollefson, S.J.; Wright, P.F.; Williams, J.V. The Pediatric Burden of Human Coronaviruses Evaluated for Twenty Years. Pediatr. Infect. Dis. J. 2009, 28, 682–687. [Google Scholar] [CrossRef]
- Huang, S.-H.; Su, M.-C.; Tien, N.; Huang, C.-J.; Lan, Y.-C.; Lin, C.-S.; Chen, C.-H.; Lin, C.-W. Epidemiology of Human Coronavirus NL63 Infection among Hospitalized Patients with Pneumonia in Taiwan. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan Ran Za Zhi 2017, 50, 763–770. [Google Scholar] [CrossRef]
- Xu, R.-H.; He, J.-F.; Evans, M.R.; Peng, G.-W.; Field, H.E.; Yu, D.-W.; Lee, C.-K.; Luo, H.-M.; Lin, W.-S.; Lin, P.; et al. Epidemiologic Clues to SARS Origin in China. Emerg. Infect. Dis. 2004, 10, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gargan, S.; Lu, Y.; Stevenson, N.J. An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity. Viruses 2021, 13, 560. [Google Scholar] [CrossRef]
- Bell, D.M. Public Health Interventions and SARS Spread, 2003. Emerg. Infect. Dis. 2004, 10, 1900–1906. [Google Scholar] [CrossRef]
- Pillaiyar, T. Middle East Respiratory Syndrome-Coronavirus (MERS-CoV): An Updated Overview and Pharmacotherapeutics. Med. Chem. 2015, 5, 8. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Singh, R.; Dar, Z.; Bijarnia, R.K.; Dhingra, N.; Kaur, T. Genetic Comparison among Various Coronavirus Strains for the Identification of Potential Vaccine Targets of SARS-CoV2. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2021, 89, 104490. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.A.; Verkleij, A.; Rottier, P.J.M.; Koster, A.J.; Bosch, B.J. Cryo-Electron Tomography of Mouse Hepatitis Virus: Insights into the Structure of the Coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef]
- Neuman, B.W.; Adair, B.D.; Yoshioka, C.; Quispe, J.D.; Orca, G.; Kuhn, P.; Milligan, R.A.; Yeager, M.; Buchmeier, M.J. Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. J. Virol. 2006, 80, 7918–7928. [Google Scholar] [CrossRef]
- Tyrrell, D.A.; Almeida, J.D.; Cunningham, C.H.; Dowdle, W.R.; Hofstad, M.S.; McIntosh, K.; Tajima, M.; Zakstelskaya, L.Y.; Easterday, B.C.; Kapikian, A.; et al. Coronaviridae. Intervirology 1975, 5, 76–82. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Dediego, M.L.; Alvarez, E.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular Location and Topology of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus Envelope Protein: Current Knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.M.; et al. The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-like Particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar] [CrossRef]
- Mortola, E.; Roy, P. Efficient Assembly and Release of SARS Coronavirus-like Particles by a Heterologous Expression System. FEBS Lett. 2004, 576, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A Structural Analysis of M Protein in Coronavirus Assembly and Morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Ruch, T.R.; Machamer, C.E. The Coronavirus E Protein: Assembly and Beyond. Viruses 2012, 4, 363–382. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Verheije, M.H.; Hagemeijer, M.C.; Ulasli, M.; Reggiori, F.; Rottier, P.J.M.; Masters, P.S.; de Haan, C.A.M. The Coronavirus Nucleocapsid Protein Is Dynamically Associated with the Replication-Transcription Complexes. J. Virol. 2010, 84, 11575–11579. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Beniac, D.R.; Andonov, A.; Grudeski, E.; Booth, T.F. Architecture of the SARS Coronavirus Prefusion Spike. Nat. Struct. Mol. Biol. 2006, 13, 751–752. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Berardi, M.; Li, W.; Farzan, M.; Dormitzer, P.R.; Harrison, S.C. Conformational States of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Ectodomain. J. Virol. 2006, 80, 6794–6800. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Tortorici, M.A.; Bosch, B.-J.; Frenz, B.; Rottier, P.J.M.; DiMaio, F.; Rey, F.A.; Veesler, D. Cryo-Electron Microscopy Structure of a Coronavirus Spike Glycoprotein Trimer. Nature 2016, 531, 114–117. [Google Scholar] [CrossRef]
- Cueno, M.E.; Imai, K. Structural Comparison of the SARS CoV 2 Spike Protein Relative to Other Human-Infecting Coronaviruses. Front. Med. 2021, 7, 594439. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Che, X.-Y.; Qiu, L.-W.; Liao, Z.-Y.; Wang, Y.; Wen, K.; Pan, Y.-X.; Hao, W.; Mei, Y.-B.; Cheng, V.C.C.; Yuen, K.-Y. Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome-Associated Coronavirus and Human Coronaviruses 229E and OC43. J. Infect. Dis. 2005, 191, 2033–2037. [Google Scholar] [CrossRef]
- Dijkman, R.; Jebbink, M.F.; El Idrissi, N.B.; Pyrc, K.; Müller, M.A.; Kuijpers, T.W.; Zaaijer, H.L.; van der Hoek, L. Human Coronavirus NL63 and 229E Seroconversion in Children. J. Clin. Microbiol. 2008, 46, 2368–2373. [Google Scholar] [CrossRef]
- Patrick, D.M.; Petric, M.; Skowronski, D.M.; Guasparini, R.; Booth, T.F.; Krajden, M.; McGeer, P.; Bastien, N.; Gustafson, L.; Dubord, J.; et al. An Outbreak of Human Coronavirus OC43 Infection and Serological Cross-Reactivity with SARS Coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006, 17, 330–336. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the Largest RNA Virus Genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef]
- Brierley, I.; Digard, P.; Inglis, S.C. Characterization of an Efficient Coronavirus Ribosomal Frameshifting Signal: Requirement for an RNA Pseudoknot. Cell 1989, 57, 537–547. [Google Scholar] [CrossRef]
- Lu, Y.; Lu, X.; Denison, M.R. Identification and Characterization of a Serine-like Proteinase of the Murine Coronavirus MHV-A59. J. Virol. 1995, 69, 3554–3559. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses Post-SARS: Update on Replication and Pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Eckerle, L.D.; Lu, X.; Sperry, S.M.; Choi, L.; Denison, M.R. High Fidelity of Murine Hepatitis Virus Replication Is Decreased in Nsp14 Exoribonuclease Mutants. J. Virol. 2007, 81, 12135–12144. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef] [PubMed]
- Mielech, A.M.; Chen, Y.; Mesecar, A.D.; Baker, S.C. Nidovirus Papain-like Proteases: Multifunctional Enzymes with Protease, Deubiquitinating and DeISGylating Activities. Virus Res. 2014, 194, 184–190. [Google Scholar] [CrossRef]
- Nakagawa, K.; Narayanan, K.; Wada, M.; Popov, V.L.; Cajimat, M.; Baric, R.S.; Makino, S. The Endonucleolytic RNA Cleavage Function of Nsp1 of Middle East Respiratory Syndrome Coronavirus Promotes the Production of Infectious Virus Particles in Specific Human Cell Lines. J. Virol. 2018, 92, e01157-18. [Google Scholar] [CrossRef]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Pyrc, K.; Dijkman, R.; Deng, L.; Jebbink, M.F.; Ross, H.A.; Berkhout, B.; van der Hoek, L. Mosaic Structure of Human Coronavirus NL63, One Thousand Years of Evolution. J. Mol. Biol. 2006, 364, 964–973. [Google Scholar] [CrossRef]
- Dijkman, R.; van der Hoek, L. Human Coronaviruses 229E and NL63: Close Yet Still So Far. J. Formos. Med. Assoc. 2009, 108, 270–279. [Google Scholar] [CrossRef]
- Vabret, A.; Dina, J.; Gouarin, S.; Petitjean, J.; Corbet, S.; Freymuth, F. Detection of the New Human Coronavirus HKU1: A Report of 6 Cases. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006, 42, 634–639. [Google Scholar] [CrossRef]
- Stobart, C.C.; Sexton, N.R.; Munjal, H.; Lu, X.; Molland, K.L.; Tomar, S.; Mesecar, A.D.; Denison, M.R. Chimeric Exchange of Coronavirus Nsp5 Proteases (3CLpro) Identifies Common and Divergent Regulatory Determinants of Protease Activity. J. Virol. 2013, 87, 12611–12618. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Yuen, K.-Y. More and More Coronaviruses: Human Coronavirus HKU1. Viruses 2009, 1, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Wong, B.H.L.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Cytosine Deamination and Selection of CpG Suppressed Clones Are the Two Major Independent Biological Forces That Shape Codon Usage Bias in Coronaviruses. Virology 2007, 369, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, S.R.; Sims, G.E.; Wentworth, D.E.; Halpin, R.A.; Robinson, C.C.; Town, C.D.; Holmes, K.V. Genomic Analysis of 16 Colorado Human NL63 Coronaviruses Identifies a New Genotype, High Sequence Diversity in the N-Terminal Domain of the Spike Gene and Evidence of Recombination. J. Gen. Virol. 2012, 93, 2387–2398. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Lee, P.; Tsang, A.K.L.; Yip, C.C.Y.; Tse, H.; Lee, R.A.; So, L.-Y.; Lau, Y.-L.; Chan, K.-H.; Woo, P.C.Y.; et al. Molecular Epidemiology of Human Coronavirus OC43 Reveals Evolution of Different Genotypes over Time and Recent Emergence of a Novel Genotype Due to Natural Recombination. J. Virol. 2011, 85, 11325–11337. [Google Scholar] [CrossRef]
- Shao, N.; Zhang, C.; Dong, J.; Sun, L.; Chen, X.; Xie, Z.; Xu, B.; An, S.; Zhang, T.; Yang, F. Molecular Evolution of Human Coronavirus-NL63, -229E, -HKU1 and -OC43 in Hospitalized Children in China. Front. Microbiol. 2022, 13, 1023847. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Tsoi, H.-W.; Chan, K.-H.; Yuen, K.-Y. Comparative Analysis of 22 Coronavirus HKU1 Genomes Reveals a Novel Genotype and Evidence of Natural Recombination in Coronavirus HKU1. J. Virol. 2006, 80, 7136–7145. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [PubMed]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and Bovine Coronaviruses Recognize Sialic Acid-Containing Receptors Similar to Those of Influenza C Viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Zmora, P.; Gierer, S.; Heurich, A.; Pöhlmann, S. Proteolytic Activation of the SARS-Coronavirus Spike Protein: Cutting Enzymes at the Cutting Edge of Antiviral Research. Antiviral Res. 2013, 100, 605–614. [Google Scholar] [CrossRef]
- Baranov, P.V.; Henderson, C.M.; Anderson, C.B.; Gesteland, R.F.; Atkins, J.F.; Howard, M.T. Programmed Ribosomal Frameshifting in Decoding the SARS-CoV Genome. Virology 2005, 332, 498–510. [Google Scholar] [CrossRef]
- Grum-Tokars, V.; Ratia, K.; Begaye, A.; Baker, S.C.; Mesecar, A.D. Evaluating the 3C-like Protease Activity of SARS-Coronavirus: Recommendations for Standardized Assays for Drug Discovery. Virus Res. 2008, 133, 63–73. [Google Scholar] [CrossRef]
- Ziebuhr, J.; Snijder, E.J.; Gorbalenya, A.E. Virus-Encoded Proteinases and Proteolytic Processing in the Nidovirales. J. Gen. Virol. 2000, 81, 853–879. [Google Scholar] [CrossRef]
- Anderson-Daniels, J.; Gribble, J.; Denison, M. Proteolytic Processing of the Coronavirus Replicase Nonstructural Protein 14 Exonuclease Is Not Required for Virus Replication but Alters RNA Synthesis and Viral Fitness. J. Virol. 2022, 96, e0084122. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Oostra, M.; Hagemeijer, M.C.; van Gent, M.; Bekker, C.P.J.; te Lintelo, E.G.; Rottier, P.J.M.; de Haan, C.A.M. Topology and Membrane Anchoring of the Coronavirus Replication Complex: Not All Hydrophobic Domains of Nsp3 and Nsp6 Are Membrane Spanning. J. Virol. 2008, 82, 12392–12405. [Google Scholar] [CrossRef]
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4, e00524-13. [Google Scholar] [CrossRef] [PubMed]
- Beachboard, D.C.; Anderson-Daniels, J.M.; Denison, M.R. Mutations across Murine Hepatitis Virus Nsp4 Alter Virus Fitness and Membrane Modifications. J. Virol. 2015, 89, 2080–2089. [Google Scholar] [CrossRef]
- Oudshoorn, D.; Rijs, K.; Limpens, R.W.A.L.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus Nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, e01658-17. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A Contemporary View of Coronavirus Transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef]
- Sawicki, S.G.; Sawicki, D.L. Coronaviruses Use Discontinuous Extension for Synthesis of Subgenome-Length Negative Strands. Adv. Exp. Med. Biol. 1995, 380, 499–506. [Google Scholar] [CrossRef]
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R. The Coronavirus Proofreading Exoribonuclease Mediates Extensive Viral Recombination. PLoS Pathog. 2021, 17, e1009226. [Google Scholar] [CrossRef]
- Lai, M.M.; Baric, R.S.; Makino, S.; Keck, J.G.; Egbert, J.; Leibowitz, J.L.; Stohlman, S.A. Recombination between Nonsegmented RNA Genomes of Murine Coronaviruses. J. Virol. 1985, 56, 449–456. [Google Scholar] [CrossRef] [PubMed]
- de Haan, C.A.M.; Rottier, P.J.M. Molecular Interactions in the Assembly of Coronaviruses. Adv. Virus Res. 2005, 64, 165–230. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14. [Google Scholar] [CrossRef]
- Buchrieser, J.; Dufloo, J.; Hubert, M.; Monel, B.; Planas, D.; Rajah, M.M.; Planchais, C.; Porrot, F.; Guivel-Benhassine, F.; Van der Werf, S.; et al. Syncytia Formation by SARS-CoV-2-Infected Cells. EMBO J. 2020, 39, e106267. [Google Scholar] [CrossRef]
- Anka, A.U.; Tahir, M.I.; Abubakar, S.D.; Alsabbagh, M.; Zian, Z.; Hamedifar, H.; Sabzevari, A.; Azizi, G. Coronavirus Disease 2019 (COVID-19): An Overview of the Immunopathology, Serological Diagnosis and Management. Scand. J. Immunol. 2021, 93, e12998. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Huang, Y.; Wu, Z.; Sun, H.; Ma, W.; Fang, T. A Case of Coronavirus HKU1 Encephalitis. Acta Virol. 2020, 64, 261–263. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Tse, H.; Tsoi, H.; Cheng, V.C.C.; Lee, P.; Tang, B.S.F.; Cheung, C.H.Y.; Lee, R.A.; et al. Coronavirus HKU1 and Other Coronavirus Infections in Hong Kong. J. Clin. Microbiol. 2006, 44, 2063–2071. [Google Scholar] [CrossRef]
- Le Coupanec, A.; Desforges, M.; Kaufer, B.; Dubeau, P.; Côté, M.; Talbot, P.J. Potential Differences in Cleavage of the S Protein and Type 1 Interferon Together Control Human Coronavirus Infection, Propagation, and Neuropathology within the Central Nervous System. J. Virol. 2021, 95, e00140-21. [Google Scholar] [CrossRef]
- Talbot, P.J.; Ekandé, S.; Cashman, N.R.; Mounir, S.; Stewart, J.N. Neurotropism of Human Coronavirus 229E. Adv. Exp. Med. Biol. 1993, 342, 339–346. [Google Scholar] [CrossRef]
- Warnes, S.L.; Little, Z.R.; Keevil, C.W. Human Coronavirus 229E Remains Infectious on Common Touch Surface Materials. mBio 2015, 6, e01697-15. [Google Scholar] [CrossRef] [PubMed]
- Ayouni, I.; Maatoug, J.; Dhouib, W.; Zammit, N.; Fredj, S.B.; Ghammam, R.; Ghannem, H. Effective Public Health Measures to Mitigate the Spread of COVID-19: A Systematic Review. BMC Public Health 2021, 21, 1015. [Google Scholar] [CrossRef] [PubMed]
- Hellewell, J.; Abbott, S.; Gimma, A.; Bosse, N.I.; Jarvis, C.I.; Russell, T.W.; Munday, J.D.; Kucharski, A.J.; Edmunds, W.J.; Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group; et al. Feasibility of Controlling COVID-19 Outbreaks by Isolation of Cases and Contacts. Lancet Glob. Health 2020, 8, e488–e496. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, A.J.; Klepac, P.; Conlan, A.J.K.; Kissler, S.M.; Tang, M.L.; Fry, H.; Gog, J.R.; Edmunds, W.J.; CMMID COVID-19 Working Group. Effectiveness of Isolation, Testing, Contact Tracing, and Physical Distancing on Reducing Transmission of SARS-CoV-2 in Different Settings: A Mathematical Modelling Study. Lancet Infect. Dis. 2020, 20, 1151–1160. [Google Scholar] [CrossRef]
- Li, M.-Y.; Li, L.; Zhang, Y.; Wang, X.-S. Expression of the SARS-CoV-2 Cell Receptor Gene ACE2 in a Wide Variety of Human Tissues. Infect. Dis. Poverty 2020, 9, 45. [Google Scholar] [CrossRef]
- Vabret, A.; Dina, J.; Gouarin, S.; Petitjean, J.; Tripey, V.; Brouard, J.; Freymuth, F. Human (Non-Severe Acute Respiratory Syndrome) Coronavirus Infections in Hospitalised Children in France. J. Paediatr. Child Health 2008, 44, 176–181. [Google Scholar] [CrossRef]
- McFadden, M.J.; Gokhale, N.S.; Horner, S.M. Protect This House: Cytosolic Sensing of Viruses. Curr. Opin. Virol. 2017, 22, 36–43. [Google Scholar] [CrossRef]
- Akerlund, A.; Greiff, L.; Andersson, M.; Bende, M.; Alkner, U.; Persson, C.G. Mucosal Exudation of Fibrinogen in Coronavirus-Induced Common Colds. Acta Otolaryngol. 1993, 113, 642–648. [Google Scholar] [CrossRef]
- Linden, M.; Greiff, L.; Andersson, M.; Svensson, C.; Åkerlund, A.; Bende, M.; Andersson, E.; Persson, C.G.A. Nasal Cytokines in Common Cold and Allergic Rhinitis. Clin. Exp. Allergy 1995, 25, 166–172. [Google Scholar] [CrossRef]
- Beachboard, D.C.; Horner, S.M. Innate Immune Evasion Strategies of DNA and RNA Viruses. Curr. Opin. Microbiol. 2016, 32, 113–119. [Google Scholar] [CrossRef]
- Loo, S.-L.; Wark, P.A.B.; Esneau, C.; Nichol, K.S.; Hsu, A.C.-Y.; Bartlett, N.W. Human Coronaviruses 229E and OC43 Replicate and Induce Distinct Antiviral Responses in Differentiated Primary Human Bronchial Epithelial Cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 319, L926–L931. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The Papain-like Protease of Severe Acute Respiratory Syndrome Coronavirus Has Deubiquitinating Activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.A.; Fotouhi-Ardakani, N.; Lytvyn, V.; Lachance, P.; Sulea, T.; Ménard, R. The Papain-like Protease from the Severe Acute Respiratory Syndrome Coronavirus Is a Deubiquitinating Enzyme. J. Virol. 2005, 79, 15199–15208. [Google Scholar] [CrossRef]
- Lindner, H.A.; Lytvyn, V.; Qi, H.; Lachance, P.; Ziomek, E.; Ménard, R. Selectivity in ISG15 and Ubiquitin Recognition by the SARS Coronavirus Papain-like Protease. Arch. Biochem. Biophys. 2007, 466, 8–14. [Google Scholar] [CrossRef]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Takayama, J.; Ghosh, A.K.; et al. Deubiquitinating and Interferon Antagonism Activities of Coronavirus Papain-like Proteases. J. Virol. 2010, 84, 4619–4629. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus Papain-like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef]
- Aguirre, S.; Luthra, P.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Patel, J.; Lamothe, F.; Fredericks, A.C.; Tripathi, S.; Zhu, T.; Pintado-Silva, J.; et al. Dengue Virus NS2B Protein Targets CGAS for Degradation and Prevents Mitochondrial DNA Sensing during Infection. Nat. Microbiol. 2017, 2, 17037. [Google Scholar] [CrossRef]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and Nucleocapsid Proteins of SARS-CoV-2 Inhibit Type I Interferon Signaling Pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural Basis for Translational Shutdown and Immune Evasion by the Nsp1 Protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Blanco, M.R.; Bruce, E.A.; Honson, D.D.; Chen, L.M.; Chow, A.; Bhat, P.; Ollikainen, N.; Quinodoz, S.A.; Loney, C.; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325–1339.e21. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 Nsp13, Nsp14, Nsp15 and Orf6 Function as Potent Interferon Antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Swaim, C.D.; Canadeo, L.A.; Monte, K.J.; Khanna, S.; Lenschow, D.J.; Huibregtse, J.M. Modulation of Extracellular ISG15 Signaling by Pathogens and Viral Effector Proteins. Cell Rep. 2020, 31, 107772. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Junqueira, C.; Crespo, Â.; Ranjbar, S.; de Lacerda, L.B.; Lewandrowski, M.; Ingber, J.; Parry, B.; Ravid, S.; Clark, S.; Schrimpf, M.R.; et al. FcγR-Mediated SARS-CoV-2 Infection of Monocytes Activates Inflammation. Nature 2022, 606, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Edridge, A.W.D.; Kaczorowska, J.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Loens, K.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; et al. Seasonal Coronavirus Protective Immunity Is Short-Lasting. Nat. Med. 2020, 26, 1691–1693. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Juno, J.A.; Khoury, D.; Reynaldi, A.; Wheatley, A.K.; Kent, S.J.; Davenport, M.P. Prospects for Durable Immune Control of SARS-CoV-2 and Prevention of Reinfection. Nat. Rev. Immunol. 2021, 21, 395–404. [Google Scholar] [CrossRef]
- Larson, D.; Brodniak, S.L.; Voegtly, L.J.; Cer, R.Z.; Glang, L.A.; Malagon, F.J.; Long, K.A.; Potocki, R.; Smith, D.R.; Lanteri, C.; et al. A Case of Early Reinfection With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, e2827–e2828. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Lung, D.C.; Wong, E.Y.M.; Aw-Yong, K.L.; Wong, A.C.P.; Luk, H.K.H.; Li, K.S.M.; Fung, J.; Chan, T.T.Y.; Tang, J.Y.M.; et al. Molecular Evolution of Human Coronavirus 229E in Hong Kong and a Fatal COVID-19 Case Involving Coinfection with a Novel Human Coronavirus 229E Genogroup. mSphere 2021, 6, e00819-20. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Hung, I.F.-N.; Ip, J.D.; Chu, A.W.-H.; Chan, W.-M.; Tam, A.R.; Fong, C.H.-Y.; Yuan, S.; Tsoi, H.-W.; Ng, A.C.-K.; et al. Coronavirus Disease 2019 (COVID-19) Re-Infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, e2946–e2951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Bevan, M.J. CD8(+) T Cells: Foot Soldiers of the Immune System. Immunity 2011, 35, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.H.; Bouvier, M. MHC Class I Antigen Presentation: Learning from Viral Evasion Strategies. Nat. Rev. Immunol. 2009, 9, 503–513. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through down-Regulating MHC-Ι. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Memish, Z.A. Lack of Seasonal Variation of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Travel Med. Infect. Dis. 2019, 27, 125–126. [Google Scholar] [CrossRef]
- Kissler, S.M.; Tedijanto, C.; Goldstein, E.; Grad, Y.H.; Lipsitch, M. Projecting the Transmission Dynamics of SARS-CoV-2 through the Postpandemic Period. Science 2020, 368, 860–868. [Google Scholar] [CrossRef]
- Antia, R.; Halloran, M.E. Transition to Endemicity: Understanding COVID-19. Immunity 2021, 54, 2172–2176. [Google Scholar] [CrossRef]
- Lavine, J.S.; Bjornstad, O.N.; Antia, R. Immunological Characteristics Govern the Transition of COVID-19 to Endemicity. Science 2021, 371, 741–745. [Google Scholar] [CrossRef]
- Rennert, L.; Ma, Z.; McMahan, C.S.; Dean, D. Effectiveness and Protection Duration of COVID-19 Vaccines and Previous Infection against Any SARS-CoV-2 Infection in Young Adults. Nat. Commun. 2022, 13, 3946. [Google Scholar] [CrossRef] [PubMed]
- Otieno, J.R.; Cherry, J.L.; Spiro, D.J.; Nelson, M.I.; Trovão, N.S. Origins and Evolution of Seasonal Human Coronaviruses. Viruses 2022, 14, 1551. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary History of the Closely Related Group 2 Coronaviruses: Porcine Hemagglutinating Encephalomyelitis Virus, Bovine Coronavirus, and Human Coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Moës, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.-M.; Van Ranst, M. Complete Genomic Sequence of Human Coronavirus OC43: Molecular Clock Analysis Suggests a Relatively Recent Zoonotic Coronavirus Transmission Event. J. Virol. 2005, 79, 1595–1604. [Google Scholar] [CrossRef]
- Brüssow, H.; Brüssow, L. Clinical Evidence That the Pandemic from 1889 to 1891 Commonly Called the Russian Flu Might Have Been an Earlier Coronavirus Pandemic. Microb. Biotechnol. 2021, 14, 1860–1870. [Google Scholar] [CrossRef]
- Corman, V.M.; Eckerle, I.; Memish, Z.A.; Liljander, A.M.; Dijkman, R.; Jonsdottir, H.; Juma Ngeiywa, K.J.Z.; Kamau, E.; Younan, M.; Al Masri, M.; et al. Link of a Ubiquitous Human Coronavirus to Dromedary Camels. Proc. Natl. Acad. Sci. USA 2016, 113, 9864–9869. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Crossley, B.M.; Mock, R.E.; Callison, S.A.; Hietala, S.K. Identification and Characterization of a Novel Alpaca Respiratory Coronavirus Most Closely Related to the Human Coronavirus 229E. Viruses 2012, 4, 3689–3700. [Google Scholar] [CrossRef]
- Pfefferle, S.; Oppong, S.; Drexler, J.F.; Gloza-Rausch, F.; Ipsen, A.; Seebens, A.; Müller, M.A.; Annan, A.; Vallo, P.; Adu-Sarkodie, Y.; et al. Distant Relatives of Severe Acute Respiratory Syndrome Coronavirus and Close Relatives of Human Coronavirus 229E in Bats, Ghana. Emerg. Infect. Dis. 2009, 15, 1377–1384. [Google Scholar] [CrossRef]
- Sabir, J.S.M.; Lam, T.T.-Y.; Ahmed, M.M.M.; Li, L.; Shen, Y.; Abo-Aba, S.E.M.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A.; et al. Co-Circulation of Three Camel Coronavirus Species and Recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Li, S.; Yount, B.; Smith, A.; Sturges, L.; Olsen, J.C.; Nagel, J.; Johnson, J.B.; Agnihothram, S.; Gates, J.E.; et al. Evidence Supporting a Zoonotic Origin of Human Coronavirus Strain NL63. J. Virol. 2012, 86, 12816–12825. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.F.; Haskew, A.N.; Gates, J.E.; Huynh, J.; Moore, C.J.; Frieman, M.B. Metagenomic Analysis of the Viromes of Three North American Bat Species: Viral Diversity among Different Bat Species That Share a Common Habitat. J. Virol. 2010, 84, 13004–13018. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Tang, K.; Shi, M.; Conrardy, C.; Li, K.S.M.; Lau, S.K.P.; Anderson, L.J.; Tong, S. Genomic Characterization of Seven Distinct Bat Coronaviruses in Kenya. Virus Res. 2012, 167, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, X.-D.; Zhang, H.-L.; Wang, M.-R.; Guan, X.-Q.; Holmes, E.C.; Zhang, Y.-Z. Extensive Genetic Diversity and Host Range of Rodent-Borne Coronaviruses. Virus Evol. 2020, 6, veaa078. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef]
- Becker, M.M.; Graham, R.L.; Donaldson, E.F.; Rockx, B.; Sims, A.C.; Sheahan, T.; Pickles, R.J.; Corti, D.; Johnston, R.E.; Baric, R.S.; et al. Synthetic Recombinant Bat SARS-like Coronavirus Is Infectious in Cultured Cells and in Mice. Proc. Natl. Acad. Sci. USA 2008, 105, 19944–19949. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef]
- Baric, R.S.; Fu, K.; Chen, W.; Yount, B. High Recombination and Mutation Rates in Mouse Hepatitis Virus Suggest That Coronaviruses May Be Potentially Important Emerging Viruses. Adv. Exp. Med. Biol. 1995, 380, 571–576. [Google Scholar] [CrossRef]
- Makino, S.; Keck, J.G.; Stohlman, S.A.; Lai, M.M. High-Frequency RNA Recombination of Murine Coronaviruses. J. Virol. 1986, 57, 729–737. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, Y.; Li, Q.; Lu, G.; Li, C.; Jin, R.; Li, L.; Xu, B.; Gao, L.; Yin, J.; et al. Genetic Characteristics of Human Coronavirus HKU1 in Mainland China during 2018. Arch. Virol. 2022, 167, 2173–2180. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, W.; Zhang, S.; Wei, P.; Zhang, L.; Chen, D.; Qiu, S.; Li, X.; Zhao, J.; Shi, Y.; et al. Two Novel Human Coronavirus OC43 Genotypes Circulating in Hospitalized Children with Pneumonia in China. Emerg. Microbes Infect. 2022, 11, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Freuling, C.M.; Breithaupt, A.; Müller, T.; Sehl, J.; Balkema-Buschmann, A.; Rissmann, M.; Klein, A.; Wylezich, C.; Höper, D.; Wernike, K.; et al. Susceptibility of Raccoon Dogs for Experimental SARS-CoV-2 Infection. Emerg. Infect. Dis. J. 2020, 26, 12. [Google Scholar] [CrossRef]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The Origins of SARS-CoV-2: A Critical Review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, S.-Y.; Yang, X.-L.; Huang, H.-M.; Zhang, Y.-J.; Guo, H.; Luo, C.-M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat Coronaviruses Related to SARS-CoV-2 and Infectious for Human Cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef]
- Mascola, J.R.; Graham, B.S.; Fauci, A.S. SARS-CoV-2 Viral Variants—Tackling a Moving Target. JAMA 2021, 325, 1261–1262. [Google Scholar] [CrossRef]
- Singh, D.; Yi, S.V. On the Origin and Evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef]
- Fiorentini, S.; Messali, S.; Zani, A.; Caccuri, F.; Giovanetti, M.; Ciccozzi, M.; Caruso, A. First Detection of SARS-CoV-2 Spike Protein N501 Mutation in Italy in August, 2020. Lancet Infect. Dis. 2021, 21, e147. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Davis, B.D.; Chen, S.S.; Sincuir Martinez, J.M.; Plummer, J.T.; Vail, E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA 2021, 325, 1324–1326. [Google Scholar] [CrossRef]
- Dhar, M.S.; Marwal, R.; Vs, R.; Ponnusamy, K.; Jolly, B.; Bhoyar, R.C.; Sardana, V.; Naushin, S.; Rophina, M.; Mellan, T.A.; et al. Genomic Characterization and Epidemiology of an Emerging SARS-CoV-2 Variant in Delhi, India. Science 2021, 374, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kaushik, R.; Singh, A.; Uversky, V.N.; Zhang, K.Y.J.; Sahu, U.; Bhatia, S.; Sanyal, A. Bayesian Molecular Dating Analyses Combined with Mutational Profiling Suggest an Independent Origin and Evolution of SARS-CoV-2 Omicron BA.1 and BA.2 Sub-Lineages. Viruses 2022, 14, 2764. [Google Scholar] [CrossRef] [PubMed]
- Markotter, W.; Coertse, J.; De Vries, L.; Geldenhuys, M.; Mortlock, M. Bat-borne Viruses in Africa: A Critical Review. J. Zool. Lond. Engl. 1987 2020, 311, 77–98. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrison, C.M.; Doster, J.M.; Landwehr, E.H.; Kumar, N.P.; White, E.J.; Beachboard, D.C.; Stobart, C.C. Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era. Microorganisms 2023, 11, 445. https://doi.org/10.3390/microorganisms11020445
Harrison CM, Doster JM, Landwehr EH, Kumar NP, White EJ, Beachboard DC, Stobart CC. Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era. Microorganisms. 2023; 11(2):445. https://doi.org/10.3390/microorganisms11020445
Chicago/Turabian StyleHarrison, Cameron M., Jayden M. Doster, Emily H. Landwehr, Nidhi P. Kumar, Ethan J. White, Dia C. Beachboard, and Christopher C. Stobart. 2023. "Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era" Microorganisms 11, no. 2: 445. https://doi.org/10.3390/microorganisms11020445
APA StyleHarrison, C. M., Doster, J. M., Landwehr, E. H., Kumar, N. P., White, E. J., Beachboard, D. C., & Stobart, C. C. (2023). Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era. Microorganisms, 11(2), 445. https://doi.org/10.3390/microorganisms11020445





