Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment †
Abstract
1. Introduction
2. Materials and Methods
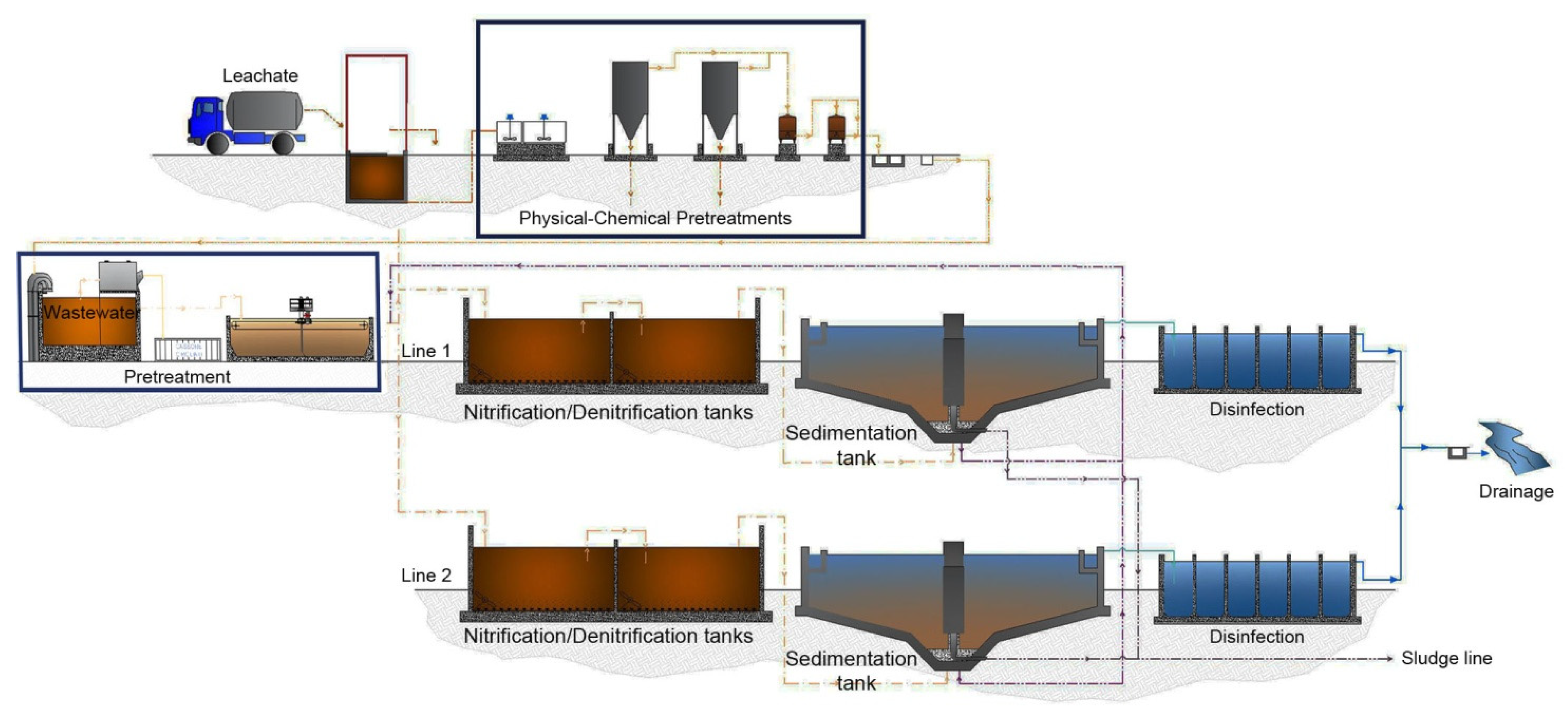
2.1. Description of Porto Sant’Elpidio Municipal Wastewater Treatment Plant
2.2. Media and Cell Growth Conditions
2.3. Analytical Methods
2.4. Analysis of Microbial Communities by 16S rRNA Gene Next-Generation Sequencing
2.5. Isolates Identification by 16S rRNA Gene Sanger Sequencing
3. Results and Discussion
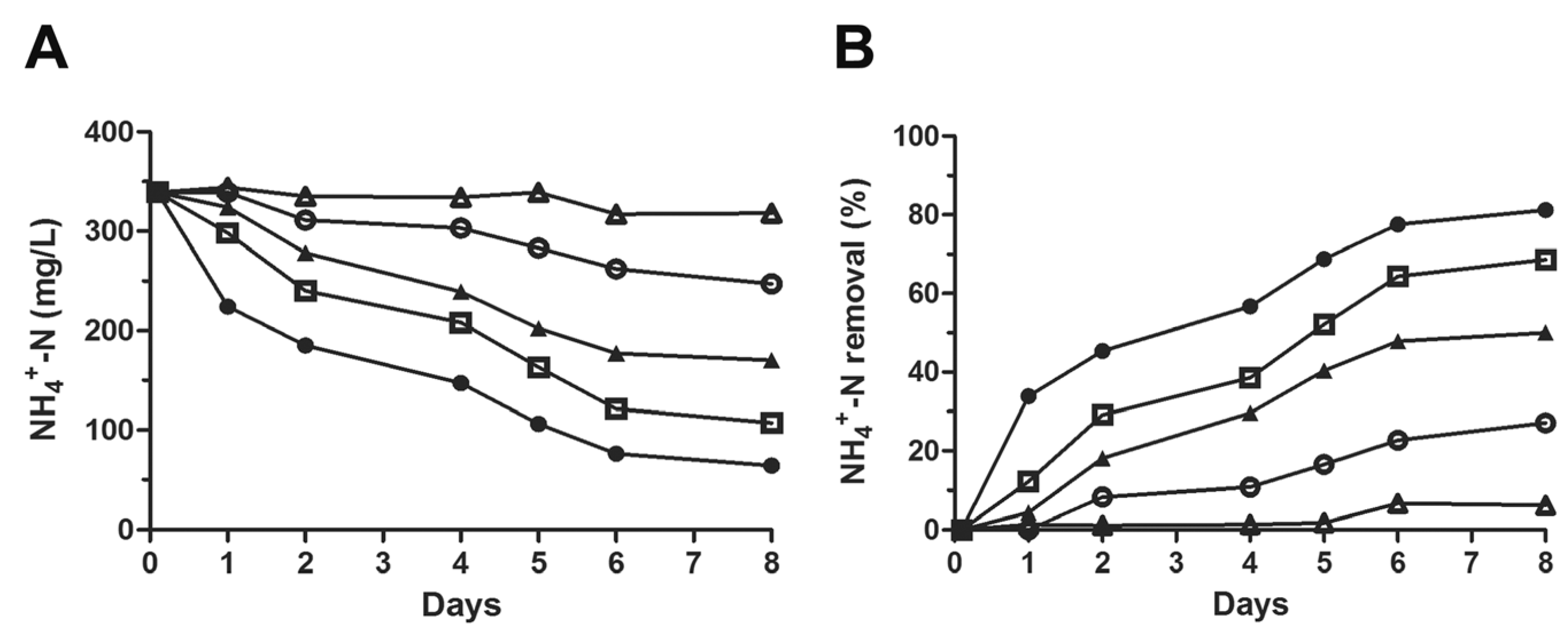
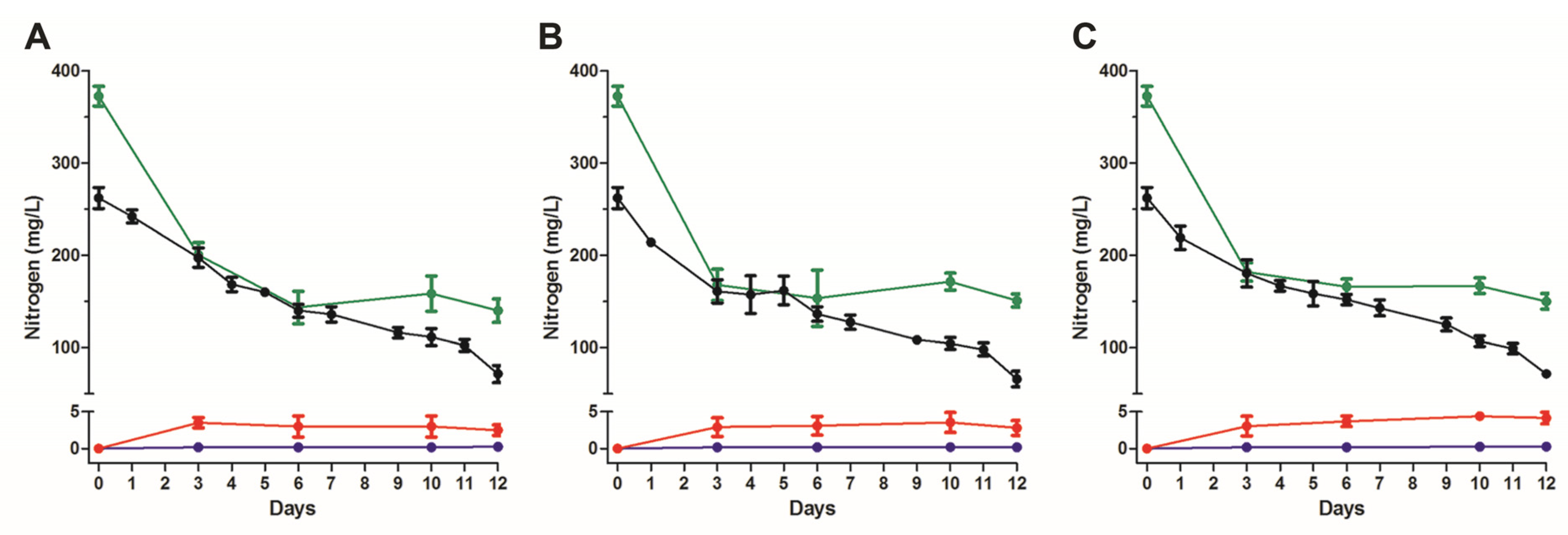
3.1. Nitrogen Removal Rate by Activated Sludge under Ammonia and Salt Stresses
3.2. Repetitive Re-Inoculum Assay
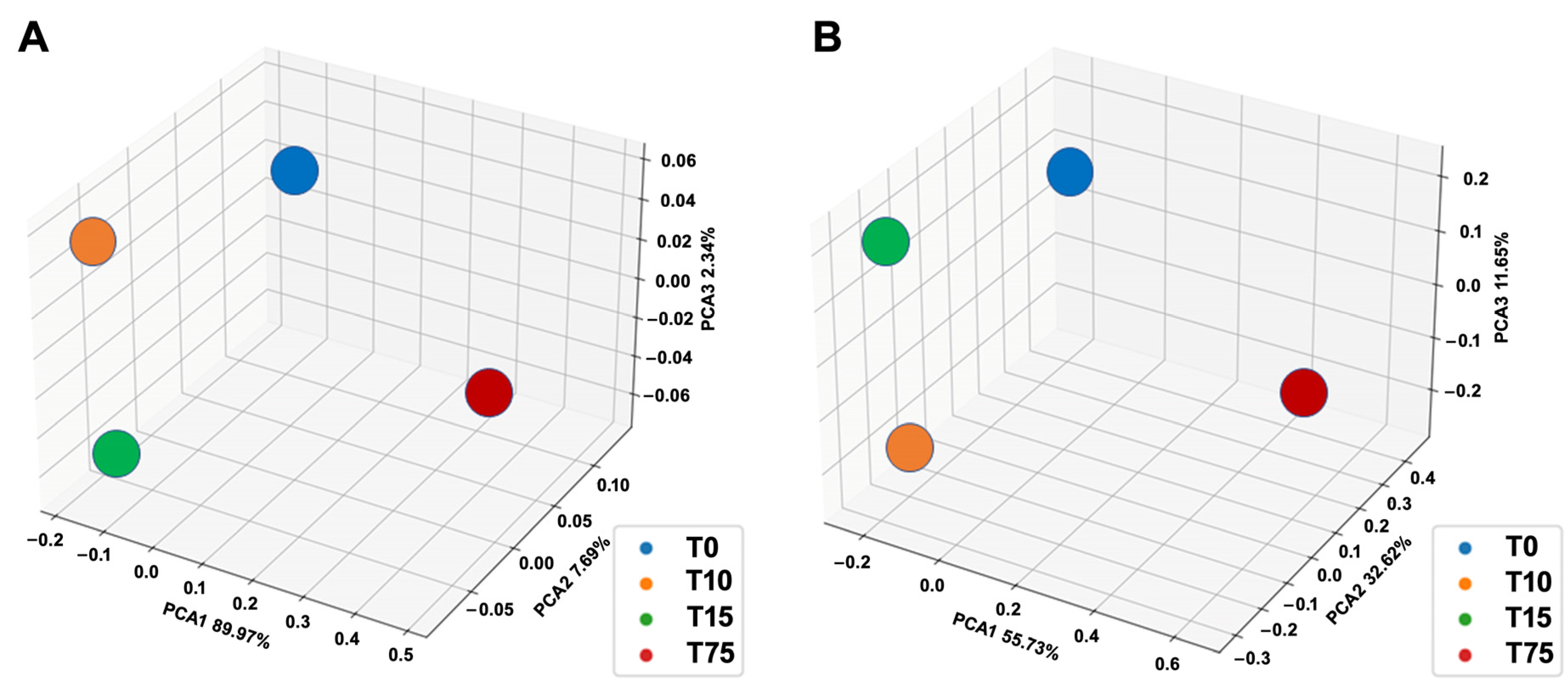
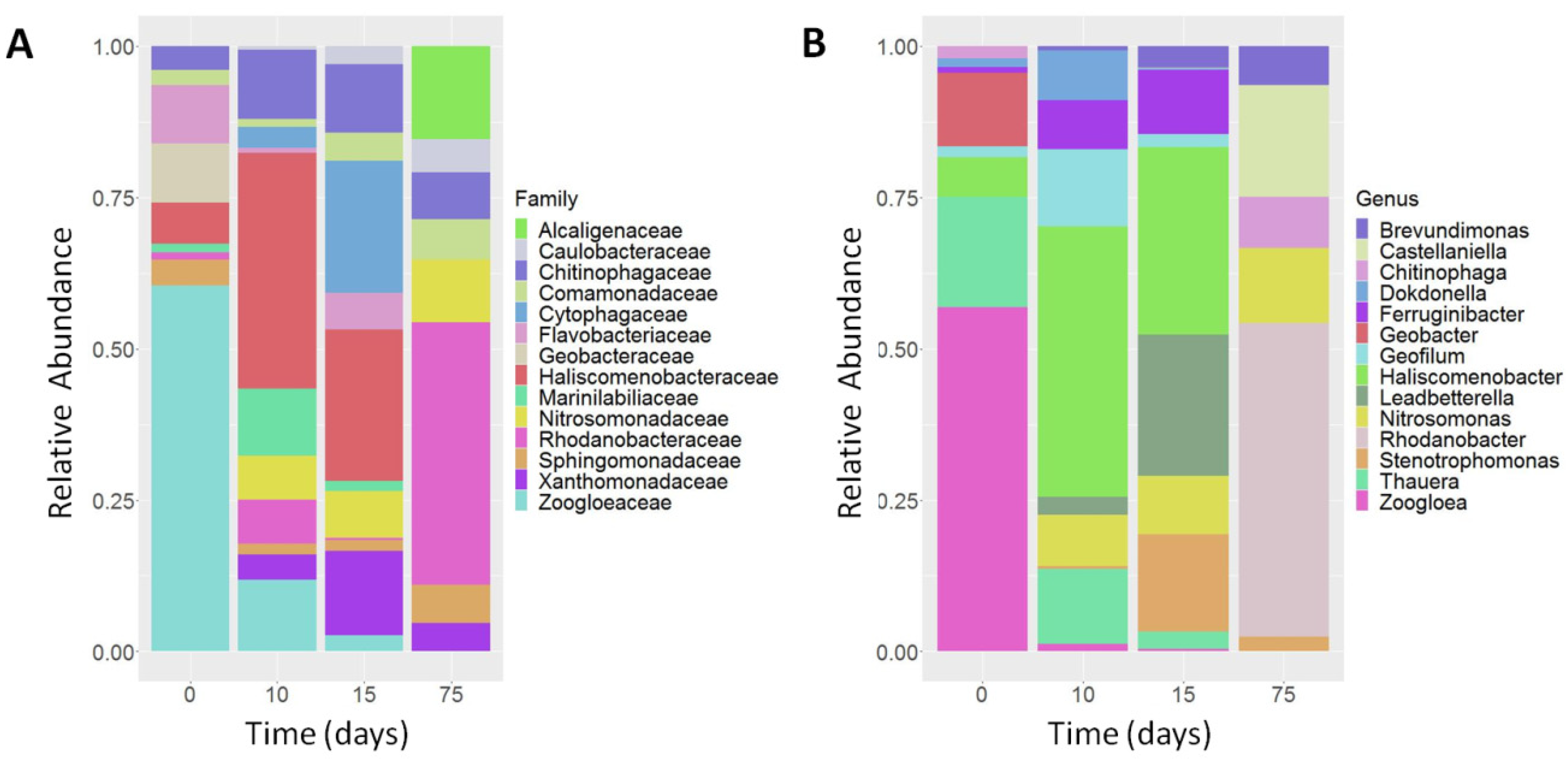
3.3. Determination of Ammonia-Degrading Microbial Communities by 16S rRNA Amplicon NGS
3.4. Heterotrophic Nitrification and Aerobic Denitrification by Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iskander, S.M.; Zhao, R.; Pathak, A.; Gupta, A.; Pruden, A.; Novak, J.T.; He, Z. A review of landfill leachate induced ultraviolet quenching substances: Sources, characteristics, and treatment. Water Res. 2018, 145, 297–311. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Xie, G.; Li, Z.; Fan, X.; Zhang, W.; Xing, F.; Tang, L.; Ren, J. Resource utilization of municipal solid waste incieration fly ash-cement and alkali-activated cementitious materials: A review. Sci. Total. Environ. 2022, 852, 158254. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.M.; Alfaia, R.G.S.M.; Campos, J.C. Landfill leachate treatment in Brazil—An overview. J. Environ. Manag. 2019, 232, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yang, G.; Tao, T.; Peng, Y. Recent advances in nitrogen removal from landfill leachate using biological treatments—A review. J. Environ. Manag. 2019, 235, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Wijekoon, P.; Koliyabandara, P.A.; Cooray, A.T.; Lam, S.S.; Athapattu, B.C.L.; Vithanage, M. Progress and prospects in mitigation of landfill leachate pollution: Risk, pollution potential, treatment and challenges. J. Hazard Mater. 2022, 421, 126627. [Google Scholar] [CrossRef]
- Foo, K.Y.; Hameed, B.H. An overview of landfill leachate treatment via activated carbon adsorption process. J. Hazard. Mater. 2009, 171, 54–60. [Google Scholar] [CrossRef]
- Lebron, Y.A.R.; Moreira, V.R.; Brasil, Y.L.; Silva, A.F.R.; Santos, L.V.S.; Lange, L.C.; Amaral, M.C.S. A survey on experiences in leachate treatment: Common practices, differences worldwide and future perspectives. J. Environ. Manag. 2021, 288, 112475. [Google Scholar] [CrossRef]
- de Almeida, R.; Porto, R.F.; Quintaes, B.R.; Bila, D.M.; Lavagnolo, M.C.; Campos, J.C. A review on membrane concentrate management from landfill leachate treatment plants: The relevance of resource recovery to close the leachate treatment loop. Waste Manag. Res. 2022, 734242X221116212. [Google Scholar] [CrossRef]
- Ma, S.; Zhou, C.; Pan, J.; Yang, G.; Sun, C.; Liu, Y.; Chen, X.; Zhao, Z. Leachate from municipal solid waste landfills in a global perspective: Characteristics, influential factors and environmental risks. J. Clean. Prod. 2022, 333, 130234. [Google Scholar] [CrossRef]
- de Almeida, R.; Campos, J.; Oroski, F.d.A. Techno-economic evaluation of landfill leachate treatment by hybrid lime application and nanofiltration process. Multidiscip. J. Waste Resour. Residues 2020, 10, 170–181. [Google Scholar] [CrossRef]
- Dos Santos, H.A.P.; de Castilhos Júnior, A.B.; Nadaleti, W.C.; Lourenço, V.A. Ammonia recovery from air stripping process applied to landfill leachate treatment. Environ. Sci. Pollut. Res. Int. 2020, 27, 45108–45120. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.; Pimenta de Oliveira, T.J.; Maurício Gouvea, R.; Carbonelli Campos, J. Technical and economic aspects of a sequential MF + NF + zeolite system treating landfill leachate. J. Environ. Sci. Health. A Tox. Hazard Subst. Environ. Eng. 2022, 57, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Mai, W.; Chen, J.; Liu, H.; Liang, J.; Tang, J.; Wei, Y. Advances in Studies on Microbiota Involved in Nitrogen Removal Processes and Their Applications in Wastewater Treatment. Front. Microbiol. 2021, 12, 746293. [Google Scholar] [CrossRef] [PubMed]
- Bove, D.; Merello, S.; Frumento, D.; Al Arni, S.; Aliakbarian, B.; Converti, A. A critical review of biological processes and technologies for landfill leachate treatment. Chem. Eng. Technol. 2015, 38, 2115–2126. [Google Scholar] [CrossRef]
- Daims, H.; Lücker, S.; Wagner, M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef]
- Lehtovirta-Morley, L.E. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 2018, 365, fny058. [Google Scholar] [CrossRef]
- Nsenga Kumwimba, M.; Meng, F. Roles of ammonia-oxidizing bacteria in improving metabolism and cometabolism of trace organic chemicals in biological wastewater treatment processes: A review. Sci. Total Environ. 2019, 659, 419–441. [Google Scholar] [CrossRef]
- Ren, Y.; Hao Ngo, H.; Guo, W.; Wang, D.; Peng, L.; Ni, B.J.; Wei, W.; Liu, Y. New perspectives on microbial communities and biological nitrogen removal processes in wastewater treatment systems. Bioresour. Technol. 2020, 297, 122491. [Google Scholar] [CrossRef]
- Yuan, C.; Lu, C.; Ma, Y.; Wang, Y.; Xie, Y.; Zhang, K.; Wang, Y.; Lv, L.; Feng, X.; Zhu, T. A novel method to treat old landfill leachate combining multi-stage biological contact oxidation (MBCO) and single-stage autotrophic nitrogen removal using anammox and partial nitrification (SNAP). Chem. Eng. J. 2019, 359, 1635–1643. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- 16s Metagenomic Library Prep Guide. Part # 15044223 B. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 1 November 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Sun, H.W.; Peng, Y.Z.; Shi, X.N. Advanced treatment of landfill leachate using anaerobic-aerobic process: Organic removal by simultaneous denitritation and methanogenesis and nitrogen removal via nitrite. Bioresour. Technol. 2015, 177, 337–345. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, H.; Wu, P.; Wang, H.; Deng, L.; Wang, W. Nitrification performance evaluation of activated sludge under high potassium ion stress during high-ammonia nitrogen organic wastewater treatment. J. Environ. Sci. 2022, 111, 84–92. [Google Scholar] [CrossRef]
- Ibrahimpasic, J.; Dragicevic, T.L.; Hren, M.Z.; Bacun-Druzina, V.; Curlin, M.; Vrcek, I.V. Nitrogen removal from municipal landfill leachate. Chem. Biochem. Eng. Q. 2010, 24, 453–458. [Google Scholar]
- Garland, J.L.; Lehman, R.M. Dilution/extinction of community phenotypic characters to estimate relative structural diversity in mixed communities. FEMS Microbiol. Ecol. 1999, 30, 333–343. [Google Scholar] [CrossRef]
- Franklin, R.B.; Garland, J.L.; Bolster, C.H.; Mills, A.L. Impact of dilution on microbial community structure and functional potential: Comparison of numerical simulations and batch culture experiments. Appl. Environ. Microbiol. 2001, 67, 702–712. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Cotto, I.; Dai, Z.; Huo, L.; Anderson, C.L.; Vilardi, K.J.; Ijaz, U.; Khunjar, W.; Wilson, C.; De Clippeleir, H.; Gilmore, K.; et al. Long solids retention times and attached growth phase favor prevalence of comammox bacteria in nitrogen removal systems. Water Res. 2020, 169, 115268. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, Y.; Steiner, N.; Cheng, H.; Wu, Y.; Woo, S.; Criddle, C.S. Impacts of nitrogen-containing coagulants on the nitritation/denitrification of anaerobic digester centrate. Environ. Sci. Water Res. Technol. 2020, 6, 3451–3459. [Google Scholar] [CrossRef]
- Yin, Z.; Bi, X.; Xu, C. Ammonia-Oxidizing Archaea (AOA) Play with Ammonia-Oxidizing Bacteria (AOB) in Nitrogen Removal from Wastewater. Archaea 2018, 2018, 8429145. [Google Scholar] [CrossRef] [PubMed]
- Gnida, A.; Żabczyński, S.; Surmacz-Górska, J. Filamentous bacteria in the nitrifying activated sludge. Water Sci. Technol. 2018, 77, 2709–2713. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Shen, M.; Li, J.; Huang, S.; Li, Z.; Yan, Z.; Peng, Y. Cooperation between partial-nitrification, complete ammonia oxidation (comammox), and anaerobic ammonia oxidation (anammox) in sludge digestion liquid for nitrogen removal. Environ. Pollut. 2019, 254, 112965. [Google Scholar] [CrossRef] [PubMed]
- Ferrera, I.; Sánchez, O. Insights into microbial diversity in wastewater treatment systems: How far have we come? Biotechnol. Adv. 2016, 34, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Bock, E.; Wagner, M. Oxidation of inorganic nitrogen compounds as an energy source. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 457–495. [Google Scholar]
- Wang, Z.; Woo, S.; Yao, Y.; Cheng, H.; Wu, Y.; Criddle, C.S. Nitrogen removal as nitrous oxide for energy recovery: Increased process stability and high nitrous yields at short hydraulic residence times. Water Res. 2020, 173, 115575. [Google Scholar] [CrossRef]
- Chen, J.; Gu, S.; Hao, H.; Chen, J. Characteristics and metabolic pathway of Alcaligenes sp. TB for simultaneous heterotrophic nitrification-aerobic denitrification. Appl. Microbiol. Biotechnol. 2016, 100, 9787–9794. [Google Scholar] [CrossRef]
- Chen, Q.; Ni, J. Heterotrophic nitrification-aerobic denitrification by novel isolated bacteria. J. Ind. Microbiol. Biotechnol. 2011, 38, 1305–1310. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A review of research progress of heterotrophic nitrification and aerobic denitrification microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, T.; Song, Y.; Chen, H.; Lv, Y.K. Heterotrophic nitrogen removal by Acinetobacter sp. Y1 isolated from coke plant wastewater. J. Biosci. Bioeng. 2015, 120, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ren, Y.; Liang, X.; Zhao, S.; Wang, J.; Xia, Z. Nitrogen removal characteristics of a heterotrophic nitrifier Acinetobacter junii YB and its potential application for the treatment of high-strength nitrogenous wastewater. Bioresour. Technol. 2015, 193, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Padhi, S.K.; Tripathy, S.; Sen, R.; Mahapatra, A.S.; Mohanty, S.; Maiti, N.K. Characterization of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater. Int. Biodeterior. Biodegradation. 2013, 78, 67–73. [Google Scholar] [CrossRef]
- He, T.; Xie, D.; Li, Z.; Ni, J.; Sun, Q. Ammonium stimulates nitrate reduction during simultaneous nitrification and denitrification process by Arthrobacter arilaitensis Y-10. Bioresour. Technol. 2017, 239, 66–73. [Google Scholar] [CrossRef] [PubMed]


| Parameter | Value ± SD |
|---|---|
| pH | 7.6 ± 0.25 |
| COD (mg/L) | 4066 ± 830 |
| BOD (mg/L O2) | 1207 ± 177 |
| BOD/COD | 0.30 ± 0.03 |
| NH4+-N (mg/L) | 1415 ± 166 |
| NO3− -N (mg/L) | 1.5 ± 1.2 |
| NO2− -N (mg/L) | traces |
| Cl− (mg/L) | 3253 ± 472 |
| Cu (mg/L) | 0.061 ± 0.04 |
| Pb (mg/L) | 0.005 ± 0.01 |
| Cr (mg/L) | 1.04 ± 0.53 |
| Ni (mg/L) | 0.234 ± 0.12 |
| Zn (mg/L) | 0.106 ± 0.11 |
| Sample | Raw Reads | Trimmed Reads | %_Trimmed_Reads_Over_Raw | Assembled Reads | %_Assembled Reads over Trimmed | Hit Reads | % Hit over Assembled | No Hit Reads |
|---|---|---|---|---|---|---|---|---|
| 0 days | 98,151 | 91,281 | 93 | 50,242 | 55 | 22,641 | 45 | 27,601 |
| 10 days | 82,295 | 76,807 | 93 | 46,538 | 61 | 30,319 | 65 | 16,219 |
| 15 days | 95,210 | 88,113 | 93 | 53,764 | 61 | 33,086 | 62 | 20,678 |
| 75 days | 18,060 | 16,920 | 94 | 12,487 | 74 | 7861 | 63 | 4626 |
| Sample | Shannon | Simpson | Chao1 | α-Diversity |
|---|---|---|---|---|
| 0 days | 4.808 | 0.937 | 87.5 | 80 |
| 10 days | 4.255 | 0.872 | 94.667 | 90 |
| 15 days | 5.104 | 0.943 | 107.333 | 98 |
| 75 days | 3.264 | 0.807 | 55.5 | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrilli, R.; Fabbretti, A.; Cerretani, A.; Pucci, K.; Pagliaretta, G.; Picciolini, M.; Napolioni, V.; Falconi, M. Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment. Microorganisms 2023, 11, 311. https://doi.org/10.3390/microorganisms11020311
Petrilli R, Fabbretti A, Cerretani A, Pucci K, Pagliaretta G, Picciolini M, Napolioni V, Falconi M. Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment. Microorganisms. 2023; 11(2):311. https://doi.org/10.3390/microorganisms11020311
Chicago/Turabian StylePetrilli, Rossana, Attilio Fabbretti, Alex Cerretani, Kathleen Pucci, Graziella Pagliaretta, Matteo Picciolini, Valerio Napolioni, and Maurizio Falconi. 2023. "Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment" Microorganisms 11, no. 2: 311. https://doi.org/10.3390/microorganisms11020311
APA StylePetrilli, R., Fabbretti, A., Cerretani, A., Pucci, K., Pagliaretta, G., Picciolini, M., Napolioni, V., & Falconi, M. (2023). Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment. Microorganisms, 11(2), 311. https://doi.org/10.3390/microorganisms11020311








