Fungal Glycoside Hydrolases Display Unique Specificities for Polysaccharides and Staphylococcus aureus Biofilms
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Commercial Enzymes
2.2. Cloning of an α-Xylosidase from Different Species of Fungi
2.3. Agar Plate GH Assay
2.4. Expression of GH Enzymes
2.5. GH Purification (Affinity Chromatography)
2.6. Mass Spectrometry
2.7. GH Cytotoxicity Assay
2.8. Xylosidase Activity Assay
2.9. Confirmation of the α-Xylosidase Gene
2.10. In Vitro Polystyrene Biofilm Model
2.11. The In Vitro Lubbock Chronic Wound Biofilm Model
3. Results
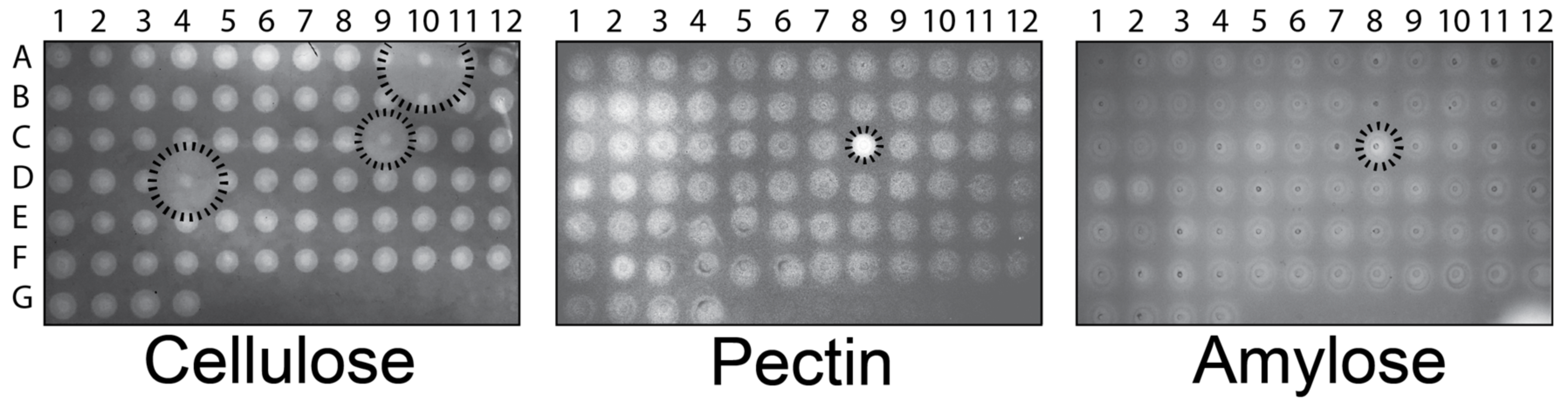
3.1. The Degradation of Carboxymethylcellulose, Amylose, and Pectin by Fungal GHs
3.2. GHs Active on Pure Polysaccharide Substrates Do Not Disperse S. aureus Biofilms
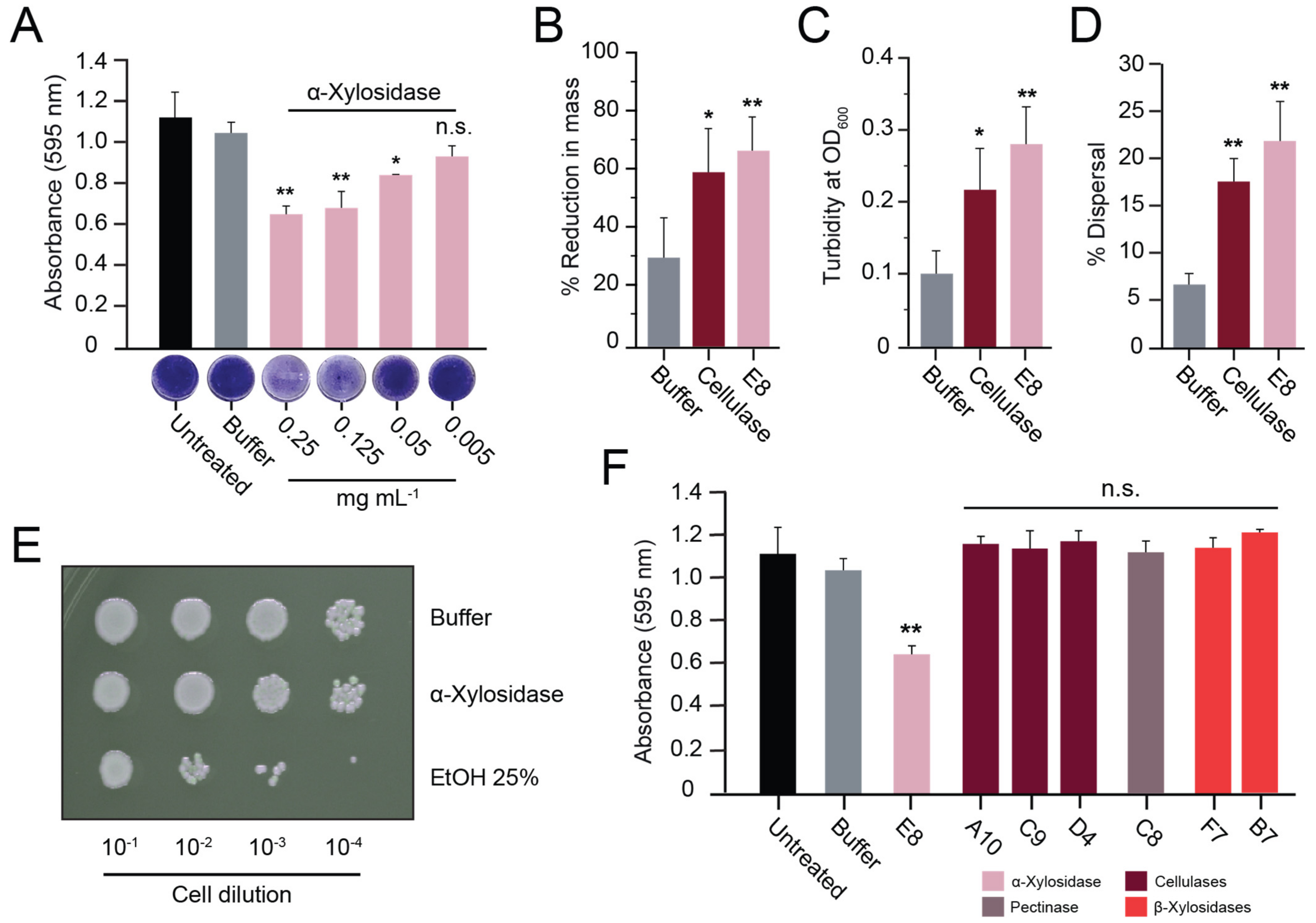
3.3. α-Xylosidase Disperses Biofilms
3.4. Purified α-Xylosidase Also Exhibits Biofilm-Degrading Activity
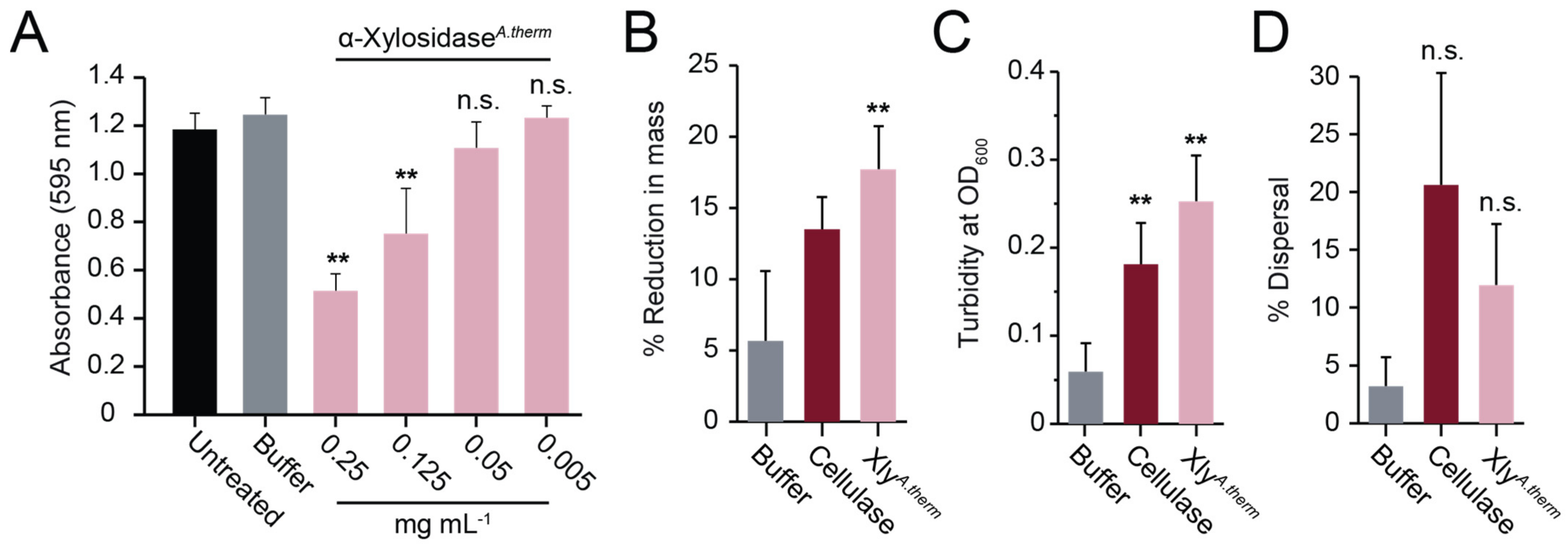
3.5. The Activities of Different Fungal Xylosidases against S. aureus Biofilms
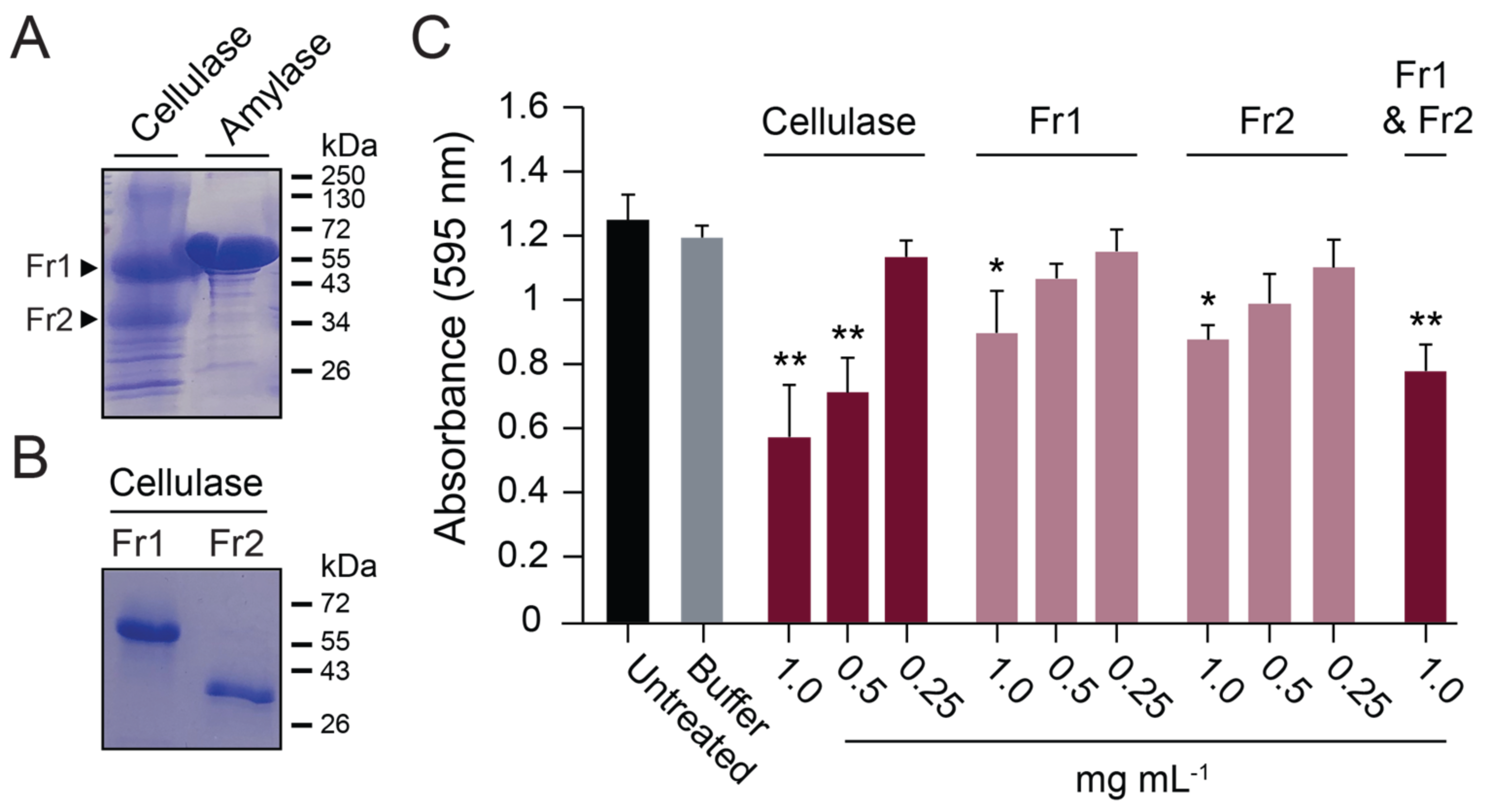
3.6. Analysis of Commercial Cellulase Preparations Revealed a Mixture of GH Enzymes
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Carvalho, C.C.C.R. Marine Biofilms: A Successful Microbial Strategy with Economic Implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D. Medical Biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Cugini, C.; Shanmugam, M.; Landge, N.; Ramasubbu, N. The Role of Exopolysaccharides in Oral Biofilms. J. Dent. Res. 2019, 98, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Karatan, E.; Watnick, P. Signals, Regulatory Networks, and Materials That Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. R 2009, 73, 310–347. [Google Scholar] [CrossRef]
- Petrova, O.E.; Sauer, K. Escaping the Biofilm in More than One Way: Desorption, Detachment or Dispersion. Curr. Opin. Microbiol. 2016, 30, 67–78. [Google Scholar] [CrossRef]
- Fleming, D.; Rumbaugh, K.P. Approaches to Dispersing Medical Biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Sauer, K. Biofilm Dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Serra, D.O.; Hengge, R. Cellulose in Bacterial Biofilms. In Extracellular Sugar-Based Biopolymers Matrices; Biologically-Inspired Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 355–392. ISBN 9783030129187. [Google Scholar]
- Thongsomboon, W.; Serra, D.O.; Possling, A.; Hadjineophytou, C.; Hengge, R.; Cegelski, L. Phosphoethanolamine Cellulose: A Naturally Produced Chemically Modified Cellulose. Science 2018, 359, 334–338. [Google Scholar] [CrossRef]
- Wozniak, D.J.; Wyckoff, T.J.O.; Starkey, M.; Keyser, R.; Azadi, P.; O’Toole, G.A.; Parsek, M.R. Alginate Is Not a Significant Component of the Extracellular Polysaccharide Matrix of PA14 and PAO1 Pseudomonas Aeruginosa Biofilms. Proc. Natl. Acad. Sci. USA 2003, 100, 7907–7912. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and Resistance of Microbial Biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Rumbaugh, K.P.; James, G.; Schultz, G.; Phillips, P.; Yang, Q.; Watters, C.; Stewart, P.S.; Dowd, S.E. Biofilm Maturity Studies Indicate Sharp Debridement Opens a Time-Dependent Therapeutic Window. J. Wound Care 2010, 19, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Mauerhofer, S.; Schneider, J.; Maniura-Weber, K.; Rosenberg, U.; Ren, Q. Enzymes Enhance Biofilm Removal Efficiency of Cleaners. Antimicrob. Agents Chemother. 2016, 60, 3647–3652. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Chahin, L.; Rumbaugh, K. Glycoside Hydrolases Degrade Polymicrobial Bacterial Biofilms in Wounds. Antimicrob. Agents Chemother. 2017, 61, AAC.01998-16. [Google Scholar] [CrossRef]
- Lakshmi, S.A.; Alexpandi, R.; Shafreen, R.M.B.; Tamilmuhilan, K.; Srivathsan, A.; Kasthuri, T.; Ravi, A.V.; Shiburaj, S.; Pandian, S.K. Evaluation of Antibiofilm Potential of Four-Domain α-Amylase from Streptomyces griseus against Exopolysaccharides (EPS) of Bacterial Pathogens Using Danio rerio. Arch. Microbiol. 2022, 204, 243. [Google Scholar] [CrossRef]
- Kovach, K.N.; Fleming, D.; Wells, M.J.; Rumbaugh, K.P.; Gordon, V.D. Specific Disruption of Established Pseudomonas Aeruginosa Biofilms Using Polymer-Attacking Enzymes. Langmuir ACS J. Surf. Colloids 2020, 36, 1585–1595. [Google Scholar] [CrossRef]
- Redman, W.K.; Welch, G.S.; Rumbaugh, K.P. Differential Efficacy of Glycoside Hydrolases to Disperse Biofilms. Front. Cell Infect. Microbiol. 2020, 10, 379. [Google Scholar] [CrossRef]
- Kamali, E.; Jamali, A.; Izanloo, A.; Ardebili, A. In Vitro Activities of Cellulase and Ceftazidime, Alone and in Combination against Pseudomonas Aeruginosa Biofilms. BMC Microbiol. 2021, 21, 347. [Google Scholar] [CrossRef]
- Jee, S.-C.; Kim, M.; Sung, J.-S.; Kadam, A.A. Efficient Biofilms Eradication by Enzymatic-Cocktail of Pancreatic Protease Type-I and Bacterial α-Amylase. Polymers 2020, 12, 3032. [Google Scholar] [CrossRef]
- Loiselle, M.; Anderson, K.W. The Use of Cellulase in Inhibiting Biofilm Formation from Organisms Commonly Found on Medical Implants. Biofouling 2003, 19, 77–85. [Google Scholar] [CrossRef]
- Kaplan, J.B.; Mlynek, K.D.; Hettiarachchi, H.; Alamneh, Y.A.; Biggemann, L.; Zurawski, D.V.; Black, C.C.; Bane, C.E.; Kim, R.K.; Granick, M.S. Extracellular Polymeric Substance (EPS)-Degrading Enzymes Reduce Staphylococcal Surface Attachment and Biocide Resistance on Pig Skin in Vivo. PLoS ONE 2018, 13, e0205526. [Google Scholar] [CrossRef]
- Kaur, A.; Rishi, V.; Soni, S.K.; Rishi, P. A Novel Multi-Enzyme Preparation Produced from Aspergillus niger Using Biodegradable Waste: A Possible Option to Combat Heterogeneous Biofilms. AMB Express 2020, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Donelli, G.; Francolini, I.; Romoli, D.; Guaglianone, E.; Piozzi, A.; Ragunath, C.; Kaplan, J.B. Synergistic Activity of Dispersin B and Cefamandole Nafate in Inhibition of Staphylococcal Biofilm Growth on Polyurethanes. Antimicrob. Agents Chemother. 2007, 51, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Vasu, P.; Persson, S.; Mort, A.J.; Somerville, C.R. Development and Application of a Suite of Polysaccharide-Degrading Enzymes for Analyzing Plant Cell Walls. Proc. Natl. Acad. Sci. USA 2006, 103, 11417–11422. [Google Scholar] [CrossRef]
- Bauer, S.; Vasu, P.; Mort, A.J.; Somerville, C.R. Cloning, Expression, and Characterization of an Oligoxyloglucan Reducing End-Specific Xyloglucanobiohydrolase from Aspergillus nidulans. Carbohyd. Res. 2005, 340, 2590–2597. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 10738. [Google Scholar] [CrossRef]
- Ma, H.; Kunes, S.; Schatz, P.J.; Botstein, D. Plasmid Construction by Homologous Recombination in Yeast. Gene 1987, 58, 201–216. [Google Scholar] [CrossRef]
- Loke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of Genomic DNA from Yeasts for PCR-Based Applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Ademark, P.; Varga, A.; Medve, J.; Harjunpää, V.; Drakenberg, T.; Tjerneld, F.; Stålbrand, H. Softwood Hemicellulose-Degrading Enzymes from Aspergillus niger: Purification and Properties of a β-Mannanase. J. Biotechnol. 1998, 63, 199–210. [Google Scholar] [CrossRef]
- Sun, Y.; Dowd, S.E.; Smith, E.; Rhoads, D.D.; Wolcott, R.D. In Vitro Multispecies Lubbock Chronic Wound Biofilm Model. Wound Repair Regen. 2008, 16, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Larsbrink, J.; Izumi, A.; Ibatullin, F.M.; Nakhai, A.; Gilbert, H.J.; Davies, G.J.; Brumer, H. Structural and Enzymatic Characterization of a Glycoside Hydrolase Family 31 α-Xylosidase from Cellvibrio japonicus Involved in Xyloglucan Saccharification. Biochem. J. 2011, 436, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Lovering, A.L.; Lee, S.S.; Kim, Y.-W.; Withers, S.G.; Strynadka, N.C.J. Mechanistic and Structural Analysis of a Family 31 α-Glycosidase and Its Glycosyl-Enzyme Intermediate. J. Biol. Chem. 2005, 280, 2105–2115. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Walton, J.D.; Brumm, P.; Phillips, G.N. Crystal Structure of α-Xylosidase from Aspergillus niger in Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases. ACS Sustain. Chem. Eng. 2020, 8, 2540–2547. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Sieiro, C.; Revilla, G.; González-Villa, T.; Zarra, I. Cloning and Expression Pattern of a Gene Encoding an α-Xylosidase Active against Xyloglucan Oligosaccharides from Arabidopsis. Plant. Physiol. 2001, 126, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, M.; Kaneko, A.; Mori, H.; Chiba, S.; Kimura, A. Structural Elements to Convert Escherichia coli A-xylosidase (YicI) into A-glucosidase. FEBS Lett. 2006, 580, 2707–2711. [Google Scholar] [CrossRef]
- Rumbaugh, K.P. How Well Are We Translating Biofilm Research from Bench-Side to Bedside? Biofilm 2020, 2, 100028. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellis, J.R.; Bull, J.J.; Rowley, P.A. Fungal Glycoside Hydrolases Display Unique Specificities for Polysaccharides and Staphylococcus aureus Biofilms. Microorganisms 2023, 11, 293. https://doi.org/10.3390/microorganisms11020293
Ellis JR, Bull JJ, Rowley PA. Fungal Glycoside Hydrolases Display Unique Specificities for Polysaccharides and Staphylococcus aureus Biofilms. Microorganisms. 2023; 11(2):293. https://doi.org/10.3390/microorganisms11020293
Chicago/Turabian StyleEllis, Jeremy R., James J. Bull, and Paul A. Rowley. 2023. "Fungal Glycoside Hydrolases Display Unique Specificities for Polysaccharides and Staphylococcus aureus Biofilms" Microorganisms 11, no. 2: 293. https://doi.org/10.3390/microorganisms11020293
APA StyleEllis, J. R., Bull, J. J., & Rowley, P. A. (2023). Fungal Glycoside Hydrolases Display Unique Specificities for Polysaccharides and Staphylococcus aureus Biofilms. Microorganisms, 11(2), 293. https://doi.org/10.3390/microorganisms11020293





