Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts
Abstract
1. Introduction
2. Materials and Methods
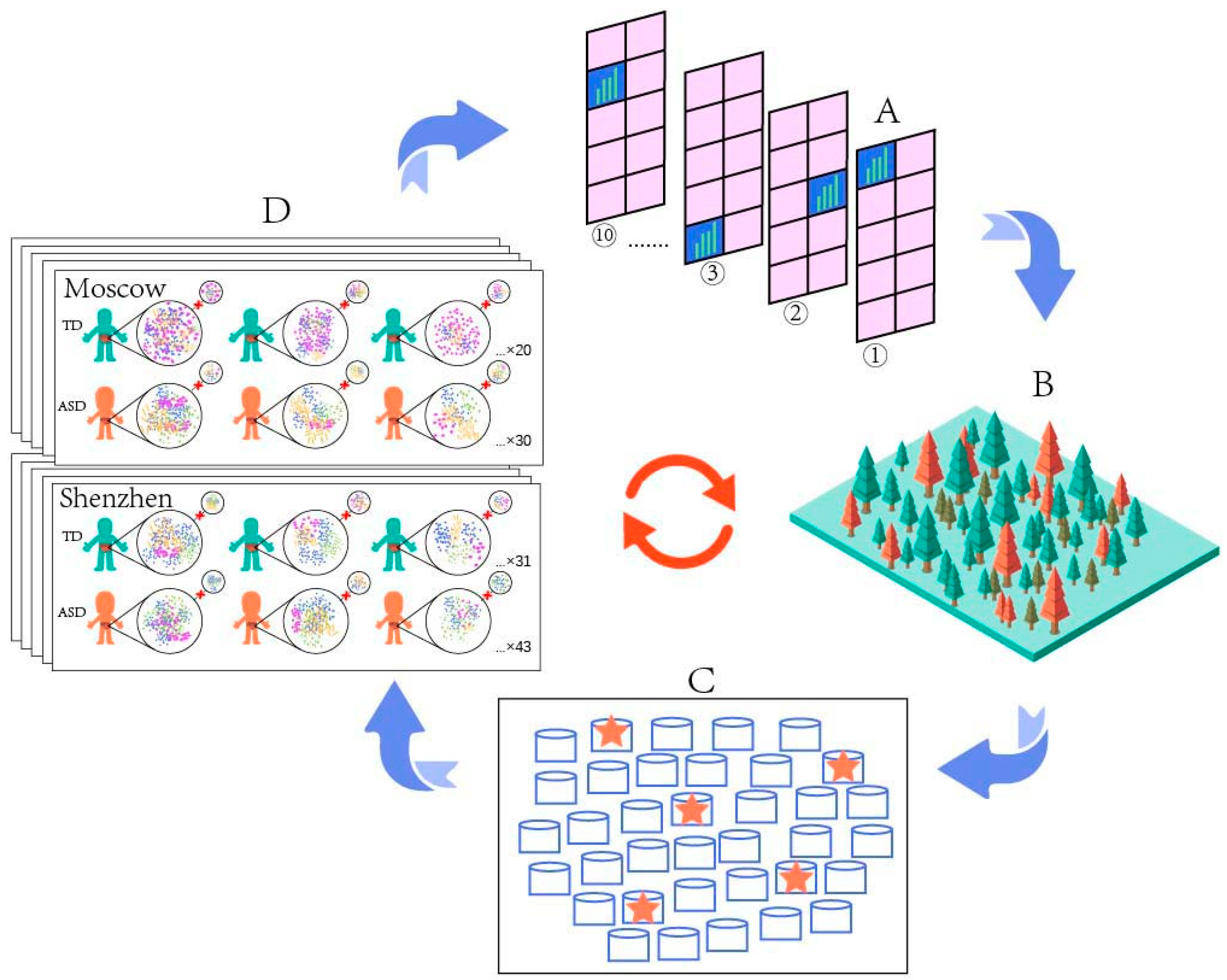
2.1. Gut Microbiome Data Acquisition
2.2. Microbiome Bioinformatics
2.3. Microbiome Data Analysis
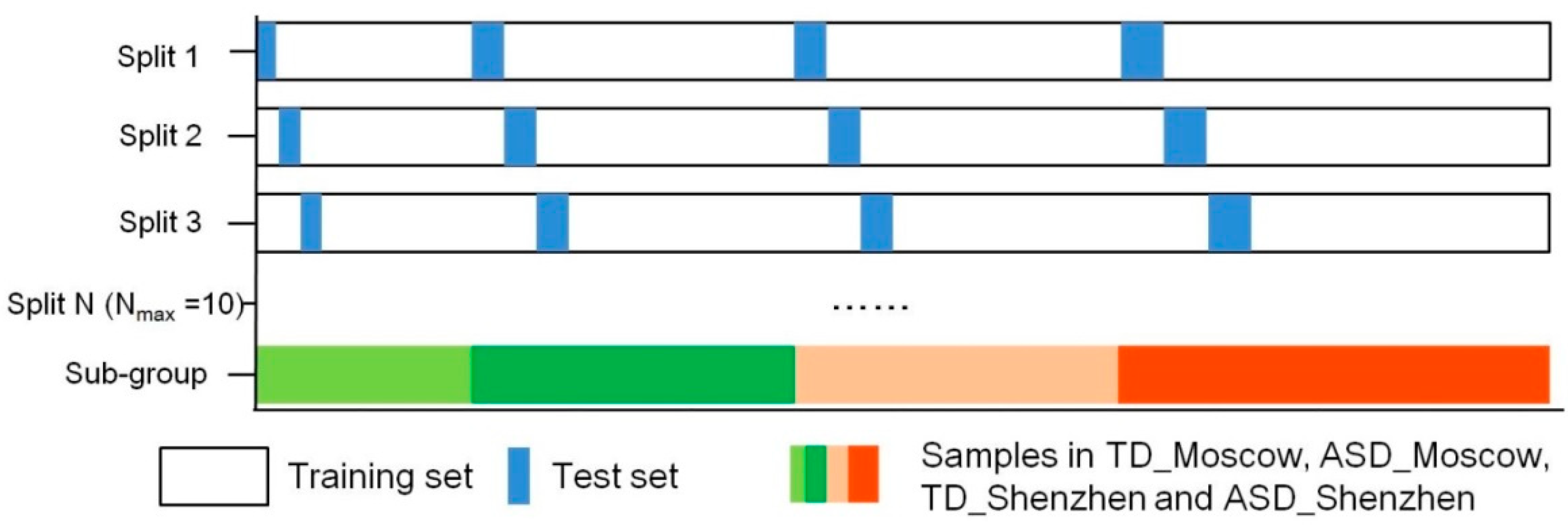
2.4. Prediction
- Processing input data. The input data of this paper are the microbial species and their relative abundance information from each sample generated after species annotation, where “features” are each microbial species and their relative abundance, and “labels” is the category of each sample, including neurotypical individuals and those with ASD.
- Learning or training model. This step is mainly to find the optimal parameters of the model by repeating the sub-steps of “parameter estimation,” “model performance evaluation,” and “error identification and correction”.
- Once the optimal parameters are determined in step 2, the model with the optimal parameters is used to predict with the new input data.
3. Results
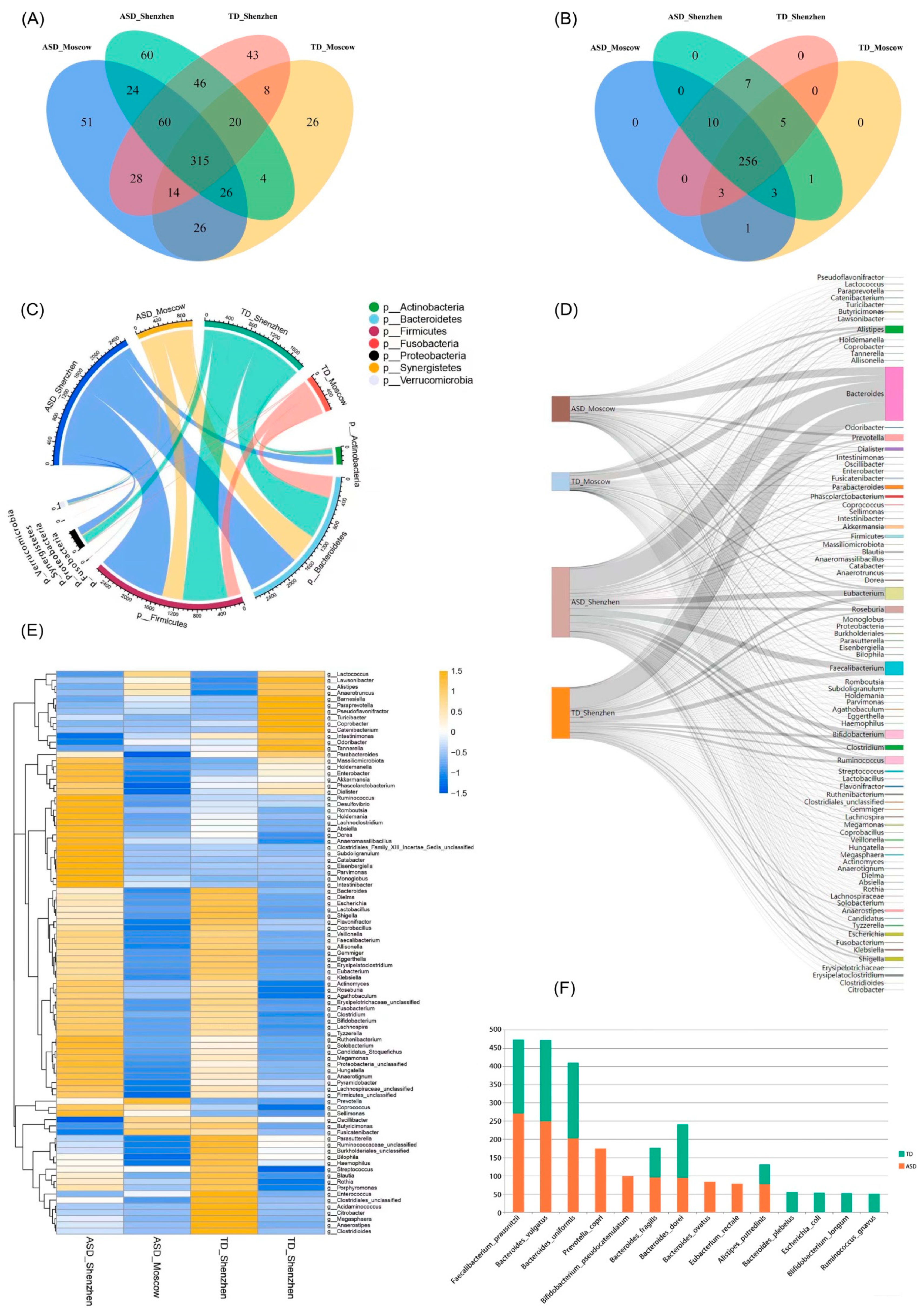
3.1. Species Composition of Gut Microbiota
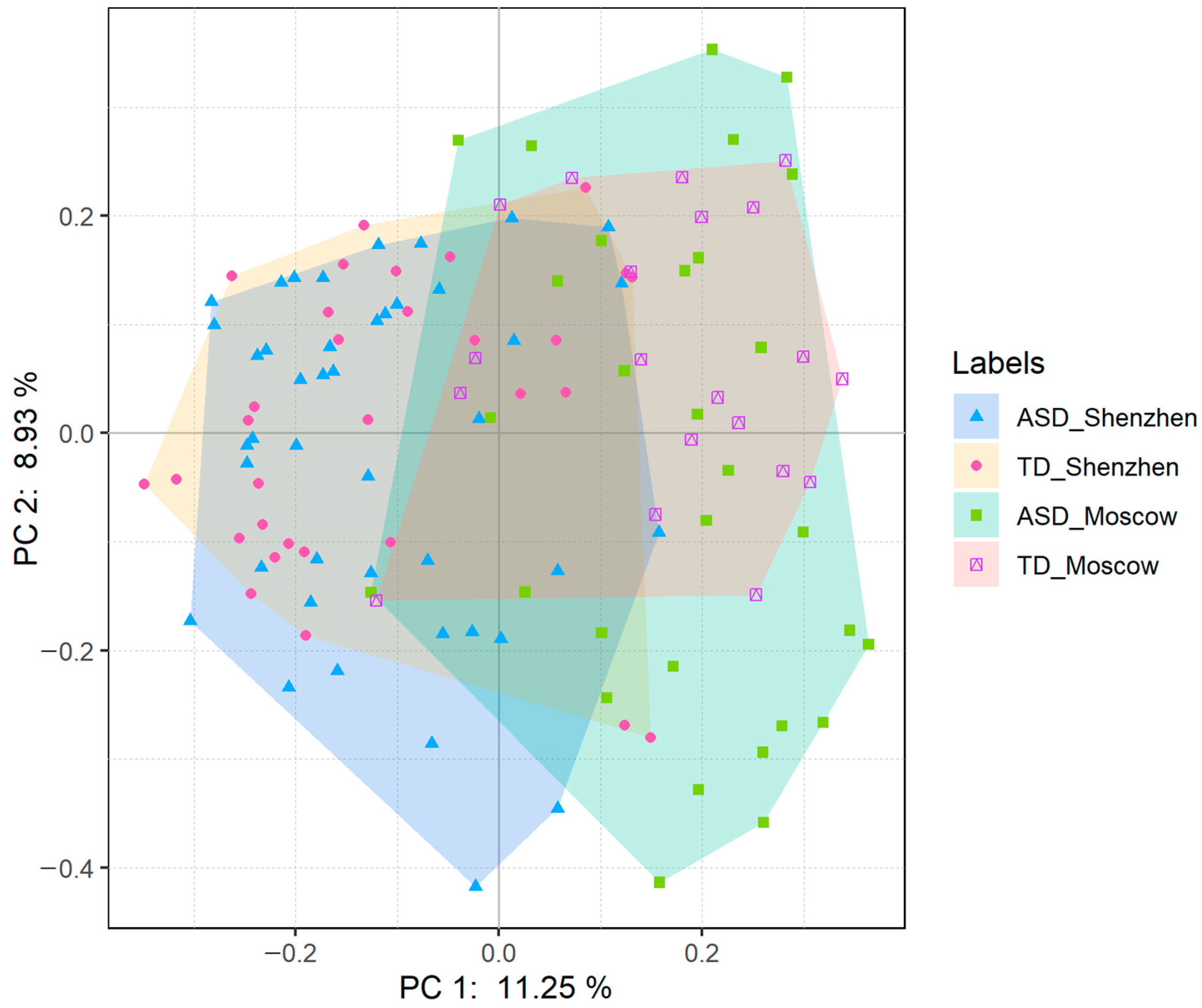
3.2. The ASD Group Was More Heterogeneous than the TD Group
3.3. No Biomarker Was Observed in the Species with Low Abundance
3.4. Correlations in the ASD Group Were More Complex than Those in the TD Group
3.5. Prediction Model Based on Random Forest Algorithm
3.6. Potential Biomarkers of ASD Diagnosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Battle, D.E. Diagnostic and Statistical Manual of Mental Disorders (DSM). CoDAS 2013, 25, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.L.; Braun, K.V.N.; Baio, J.; Bilder, D.; Charles, J.; Constantino, J.N.; Daniels, J.; Durkin, M.S.; Fitzgerald, R.T.; Kurzius-Spencer, M.; et al. Prevalence and Characteristics of Autism Spectrum Disorder among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill. Summ. 2018, 65, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wucailu Autism Research Institute. Report on the Industry Development of Autism Education and Rehabilitation in China II; Huaxia Publishing House: Beijing, China, 2017. (In Chinese) [Google Scholar]
- Buescher, A.V.S.; Cidav, Z.; Knapp, M.; Mandell, D.S. Costs of Autism Spectrum Disorders in the United Kingdom and the United States. JAMA Pediatr. 2014, 168, 721. [Google Scholar] [CrossRef] [PubMed]
- China Association of Persons with Psychiatric Disability and Their Relatives. Blue Papers on Needs of Parents with Autistic Children in China; China Association of Persons with Psychiatric Disability and Their Relatives: Beijing, China, 2014. (In Chinese) [Google Scholar]
- Piven, J.; Arndt, S.; Bailey, J.; Havercamp, S.; Andreasen, N.C.; Palmer, P. An MRI Study of Brain Size in Autism. Am. J. Psychiatry 1995, 152, 1145–1149. [Google Scholar] [CrossRef]
- Hazlett, H.C.; Gu, H.; Munsell, B.C.; Kim, S.H.; Styner, M.; Wolff, J.J.; Elison, J.T.; Swanson, M.R.; Zhu, H.; Botteron, K.N.; et al. Early Brain Development in Infants at High Risk for Autism Spectrum Disorder. Nature 2017, 542, 348–351. [Google Scholar] [CrossRef]
- Bosl, W.; Tierney, A.; Tager-Flusberg, H.; Nelson, C. EEG Complexity as a Biomarker for Autism Spectrum Disorder Risk. BMC Med. 2011, 9, 18. [Google Scholar] [CrossRef]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic Analysis of Autistic Brain Reveals Convergent Molecular Pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Cao, W.; Lin, S.; Xia, Q.; Du, Y.; Yang, Q.; Zhang, M.; Lu, Y.; Xu, J.; Duan, S.; Xia, J.; et al. Gamma Oscillation Dysfunction in MPFC Leads to Social Deficits in Neuroligin 3 R451C Knockin Mice. Neuron 2018, 97, 1253–1260.e7. [Google Scholar] [CrossRef]
- Russo, F.B.; Freitas, B.C.; Pignatari, G.C.; Fernandes, I.R.; Sebat, J.; Muotri, A.R.; Beltrão-Braga, P.C.B. Modeling the Interplay between Neurons and Astrocytes in Autism Using Human Induced Pluripotent Stem Cells. Biol. Psychiatry 2018, 83, 569–578. [Google Scholar] [CrossRef]
- Cai, Y.; Tang, X.; Chen, X.; Li, X.; Wang, Y.; Bao, X.; Wang, L.; Sun, D.; Zhao, J.; Xing, Y.; et al. Liver X Receptor β Regulates the Development of the Dentate Gyrus and Autistic-like Behavior in the Mouse. Proc. Natl. Acad. Sci. USA 2018, 115, E2725–E2733. [Google Scholar] [CrossRef]
- Fernandez, A.; Meechan, D.W.; Karpinski, B.A.; Paronett, E.M.; Bryan, C.A.; Rutz, H.L.; Radin, E.A.; Lubin, N.; Bonner, E.R.; Popratiloff, A.; et al. Mitochondrial Dysfunction Leads to Cortical Under-Connectivity and Cognitive Impairment. Neuron 2019, 102, 1127–1142.e3. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.-M.; Li, J.-R.; Huang, Y.; Zhao, M.; Tang, X.; Wei, L. AutismKB: An Evidence-Based Knowledgebase of Autism Genetics. Nucleic Acids Res. 2012, 40, D1016–D1022. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Kim, J.; Zhou, L.; Wengert, E.; Zhang, L.; Wu, Z.; Carromeu, C.; Muotri, A.R.; Marchetto, M.C.N.; Gage, F.H.; et al. KCC2 Rescues Functional Deficits in Human Neurons Derived from Patients with Rett Syndrome. Proc. Natl. Acad. Sci. USA 2016, 113, 751–756. [Google Scholar] [CrossRef]
- Xu, X.; Li, C.; Gao, X.; Xia, K.; Guo, H.; Li, Y.; Hao, Z.; Zhang, L.; Gao, D.; Xu, C.; et al. Excessive UBE3A Dosage Impairs Retinoic Acid Signaling and Synaptic Plasticity in Autism Spectrum Disorders. Cell Res. 2018, 28, 48–68. [Google Scholar] [CrossRef] [PubMed]
- Brandler, W.M.; Antaki, D.; Gujral, M.; Kleiber, M.L.; Whitney, J.; Maile, M.S.; Hong, O.; Chapman, T.R.; Tan, S.; Tandon, P.; et al. Paternally Inherited Cis-Regulatory Structural Variants Are Associated with Autism. Science 2018, 360, 327–331. [Google Scholar] [CrossRef]
- Hannon, E.; Schendel, D.; Ladd-Acosta, C.; Grove, J.; iPSYCH-Broad ASD Group; Hansen, C.S.; Andrews, S.V.; Hougaard, D.M.; Bresnahan, M.; Mors, O.; et al. Elevated Polygenic Burden for Autism Is Associated with Differential DNA Methylation at Birth. Genome Med. 2018, 10, 19. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Walters, R.K.; Singh, T.; Wigdor, E.M.; Lescai, F.; Demontis, D.; Kosmicki, J.A.; Grove, J.; Stevens, C.; Bybjerg-Grauholm, J.; et al. Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder Have a Similar Burden of Rare Protein-Truncating Variants. Nat. Neurosci. 2019, 22, 1961–1965. [Google Scholar] [CrossRef]
- Bachmann, S.O.; Sledziowska, M.; Cross, E.; Kalbassi, S.; Waldron, S.; Chen, F.; Ranson, A.; Baudouin, S.J. Behavioral Training Rescues Motor Deficits in Cyfip1 Haploinsufficiency Mouse Model of Autism Spectrum Disorders. Transl. Psychiatry 2019, 9, 29. [Google Scholar] [CrossRef]
- Gazestani, V.H.; Pramparo, T.; Nalabolu, S.; Kellman, B.P.; Murray, S.; Lopez, L.; Pierce, K.; Courchesne, E.; Lewis, N.E. A Perturbed Gene Network Containing PI3K–AKT, RAS–ERK and WNT–β-Catenin Pathways in Leukocytes Is Linked to ASD Genetics and Symptom Severity. Nat. Neurosci. 2019, 22, 1624–1634. [Google Scholar] [CrossRef]
- Endo, T.; Shioiri, T.; Kitamura, H.; Kimura, T.; Endo, S.; Masuzawa, N.; Someya, T. Altered Chemical Metabolites in the Amygdala-Hippocampus Region Contribute to Autistic Symptoms of Autism Spectrum Disorders. Biol. Psychiatry 2007, 62, 1030–1037. [Google Scholar] [CrossRef]
- Kang, D.-W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in Fecal Microbial Metabolites and Microbiota of Children with Autism Spectrum Disorders. Anaerobe 2018, 49, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4, e00321-18. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered Gut Microbial Profile Is Associated with Abnormal Metabolism Activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef] [PubMed]
- Vargason, T.; Roth, E.; Grivas, G.; Ferina, J.; Frye, R.E.; Hahn, J. Classification of Autism Spectrum Disorder from Blood Metabolites: Robustness to the Presence of Co-Occurring Conditions. Res. Autism Spectr. Disord. 2020, 77, 101644. [Google Scholar] [CrossRef]
- Al-Ayadhi, L.; Zayed, N.; Bhat, R.S.; Moubayed, N.M.S.; Al-Muammar, M.N.; El-Ansary, A. The Use of Biomarkers Associated with Leaky Gut as a Diagnostic Tool for Early Intervention in Autism Spectrum Disorder: A Systematic Review. Gut Pathog. 2021, 13, 54. [Google Scholar] [CrossRef]
- Kovtun, A.S.; Averina, O.V.; Alekseeva, M.G.; Danilenko, V.N. Antibiotic Resistance Genes in the Gut Microbiota of Children with Autistic Spectrum Disorder as Possible Predictors of the Disease. Microb. Drug Resist. 2020, 26, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Reichenberg, A.; Willfors, C.; Austin, C.; Gennings, C.; Berggren, S.; Lichtenstein, P.; Anckarsäter, H.; Tammimies, K.; Bölte, S. Fetal and Postnatal Metal Dysregulation in Autism. Nat. Commun. 2017, 8, 15493. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Iosif, A.-M.; Guerrero Angel, E.; Ozonoff, S. Association of Maternal Prenatal Vitamin Use With Risk for Autism Spectrum Disorder Recurrence in Young Siblings. JAMA Psychiatry 2019, 76, 391. [Google Scholar] [CrossRef]
- Piao, H.H.; Tam, V.T.M.; Na, H.S.; Kim, H.J.; Ryu, P.Y.; Kim, S.Y.; Rhee, J.H.; Choy, H.E.; Kim, S.W.; Hong, Y. Immunological Responses Induced by Asd and Wzy/Asd Mutant Strains of Salmonella Enterica Serovar Typhimurium in BALB/c Mice. J. Microbiol. 2010, 48, 486–495. [Google Scholar] [CrossRef]
- Marchezan, J.; Winkler dos Santos, E.G.A.; Deckmann, I.; Riesgo, R. dos S. Immunological Dysfunction in Autism Spectrum Disorder: A Potential Target for Therapy. Neuroimmunomodulation 2018, 25, 300–319. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Gut Instincts: Microbiota as a Key Regulator of Brain Development, Ageing and Neurodegeneration: Microbiota-Gut-Brain Axis across the Lifespan. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L.H. The Role of Microbiome in Central Nervous System Disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severia, C. The Gut-Brain Axis: Interactions between Enteric Microbiota, Central and Enteric Nervous Systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- Agustí, A.; García-Pardo, M.P.; López-Almela, I.; Campillo, I.; Maes, M.; Romaní-Pérez, M.; Sanz, Y. Interplay between the Gut-Brain Axis, Obesity and Cognitive Function. Front. Neurosci. 2018, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Alharthi, A.; Alhazmi, S.; Alburae, N.; Bahieldin, A. The Human Gut Microbiome as a Potential Factor in Autism Spectrum Disorder. Int. J. Mol. Sci. 2022, 23, 1363. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal Microbiota in Children with Autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Ma, B.; Liang, J.; Dai, M.; Wang, J.; Luo, J.; Zhang, Z.; Jing, J. Altered Gut Microbiota in Chinese Children with Autism Spectrum Disorders. Front. Cell. Infect. Microbiol. 2019, 9, 40. [Google Scholar] [CrossRef]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, W.; Zhang, J.; He, Y.; Wang, J. Analysis of Gut Microbiota Profiles and Microbe-Disease Associations in Children with Autism Spectrum Disorders in China. Sci. Rep. 2018, 8, 13981. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New Evidences on the Altered Gut Microbiota in Autism Spectrum Disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, R.; Cela, D.; Swann, J.R.; Vulevic, J.; Gibson, G.R.; Tzortzis, G.; Costabile, A. In Vitro Fermentation of B-GOS: Impact on Faecal Bacterial Populations and Metabolic Activity in Autistic and Non-Autistic Children. FEMS Microbiol. Ecol. 2017, 93, fiw233. [Google Scholar] [CrossRef] [PubMed]
- Newell, C.; Bomhof, M.R.; Reimer, R.A.; Hittel, D.S.; Rho, J.M.; Shearer, J. Ketogenic Diet Modifies the Gut Microbiota in a Murine Model of Autism Spectrum Disorder. Mol. Autism 2016, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, J.; Zhang, J.; Liang, C.; Wang, Y.; Chen, B.; Zhao, C.; Wang, J.; Zhang, G.; Zhao, D.; et al. Correlation of Gut Microbiome between ASD Children and Mothers and Potential Biomarkers for Risk Assessment. Genom. Proteom. Bioinform. 2019, 17, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Xu, J.; Song, Y.; Liu, B.; Xiong, Z. Leveraging Existing 16SrRNA Microbial Data to Define a Composite Biomarker for Autism Spectrum Disorder. Microbiol. Spectr. 2022, 10, e00331-22. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The Vaginal Microbiome and Preterm Birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Midani, F.S.; Weil, A.A.; Chowdhury, F.; Begum, Y.A.; Khan, A.I.; Debela, M.D.; Durand, H.K.; Reese, A.T.; Nimmagadda, S.N.; Silverman, J.D.; et al. Human Gut Microbiota Predicts Susceptibility to Vibrio Cholerae Infection. J. Infect. Dis. 2018, 218, 645–653. [Google Scholar] [CrossRef]
- Smirnova, E.; Puri, P.; Muthiah, M.D.; Daitya, K.; Brown, R.; Chalasani, N.; Liangpunsakul, S.; Shah, V.H.; Gelow, K.; Siddiqui, M.S.; et al. Fecal Microbiome Distinguishes Alcohol Consumption from Alcoholic Hepatitis but Does Not Discriminate Disease Severity. Hepatology 2020, 72, 271–286. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut Microbiome Development along the Colorectal Adenoma–Carcinoma Sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Galkin, F.; Aliper, A.; Putin, E.; Kuznetsov, I.; Gladyshev, V.N.; Zhavoronkov, A. Human Microbiome Aging Clocks Based on Deep Learning and Tandem of Permutation Feature Importance and Accumulated Local Effects. bioRxiv 2018. [Google Scholar] [CrossRef]
- Salosensaari, A.; Laitinen, V.; Havulinna, A.; Meric, G.; Cheng, S.; Perola, M.; Valsta, L.; Alfthan, G.; Inouye, M.; Watrous, J.D.; et al. Taxonomic Signatures of Long-Term Mortality Risk in Human Gut Microbiota. medRxiv 2020. [Google Scholar] [CrossRef]
- Chong, C.W.; Ahmad, A.F.; Lim, Y.A.L.; Teh, C.S.J.; Yap, I.K.S.; Lee, S.C.; Chin, Y.T.; Loke, P.; Chua, K.H. Effect of Ethnicity and Socioeconomic Variation to the Gut Microbiota Composition among Pre-Adolescent in Malaysia. Sci. Rep. 2015, 5, 13338. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Babraham, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. Erratum: MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2016, 13, 101. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International Conference on Weblogs and Social Mediam, San Jose, CA, USA, 17–20 May 2009; pp. 17–20. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine Learning Meta-Analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Yue, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; et al. Metagenomic Analysis of Colorectal Cancer Datasets Identifies Cross-cohort Microbial Diagnostic Signatures and a Link with Choline Degradation. Nat. Med. 2019, 12, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Huang, S.; Zhu, P.; Yue, F.; Zhao, H.; Yang, M.; Niu, Y.; Jing, G.; Su, X.; Li, H.; et al. A Microbiome-Based Index for Assessing Skin Health and Treatment Effects for Atopic Dermatitis in Children. mSystems 2019, 4, e00293-19. [Google Scholar] [CrossRef] [PubMed]
- Prasoodanan, P.K.V.; Sharma, A.K.; Mahajan, S.; Dhakan, D.B.; Maji, A.; Scaria, J.; Sharma, V.K. Western and Non-Western Gut Microbiomes Reveal New Roles of Prevotella in Carbohydrate Metabolism and Mouth–Gut Axis. Npj Biofilms Microbiomes 2021, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chu, Y.; Meng, Q.; Ding, R.; Shi, X.; Wang, Z.; He, Y.; Zhang, J.; Liu, J.; Zhang, J.; et al. A Quasi-Paired Cohort Strategy Reveals the Impaired Detoxifying Function of Microbes in the Gut of Autistic Children. Sci. Adv. 2020, 6, eaba3760. [Google Scholar] [CrossRef] [PubMed]
- Tierney, B.T.; Tan, Y.; Kostic, A.D.; Patel, C.J. Gene-Level Metagenomic Architectures across Diseases Yield High-Resolution Microbiome Diagnostic Indicators. Nat. Commun. 2021, 12, 2907. [Google Scholar] [CrossRef]
- El-Ansary, A.K.; Ben Bacha, A.G.; Al- Ayahdi, L.Y. Plasma Fatty Acids as Diagnostic Markers in Autistic Patients from Saudi Arabia. Lipids Health Dis. 2011, 10, 62. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, P.; Li, Y.; Wu, J.; Tan, X.; Zhou, J.; Sun, Z.; Chen, X.; Zhang, G.; Zhang, H.; et al. Landscapes of Bacterial and Metabolic Signatures and Their Interaction in Major Depressive Disorders. Sci. Adv. 2020, 6, eaba8555. [Google Scholar] [CrossRef]
- Yuan, Y.; Shi, Y.; Li, C.; Kim, J.; Cai, W.; Han, Z.; Feng, D.D. DeepGene: An Advanced Cancer Type Classifier Based on Deep Learning and Somatic Point Mutations. BMC Bioinform. 2016, 17, 476. [Google Scholar] [CrossRef]
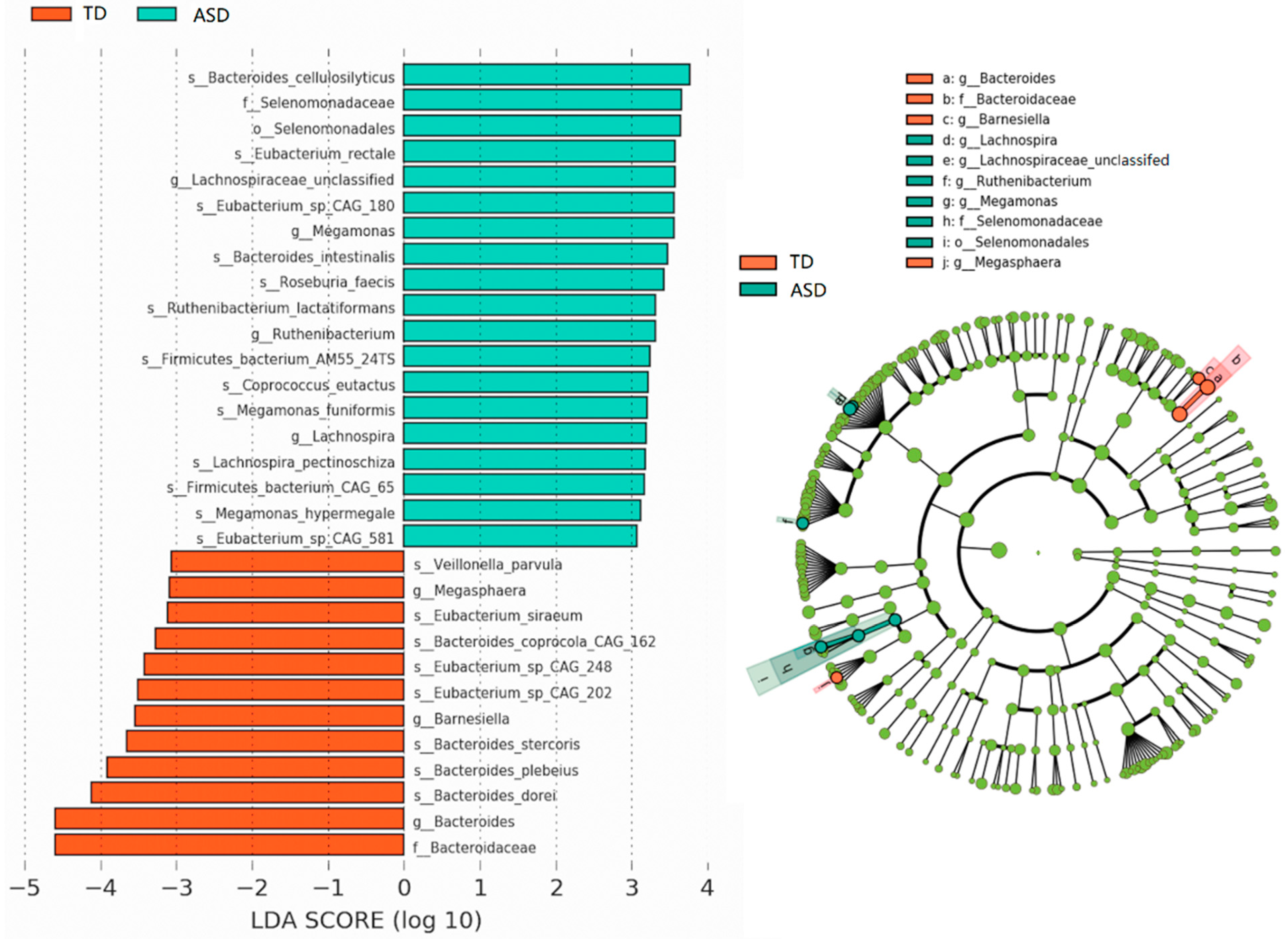

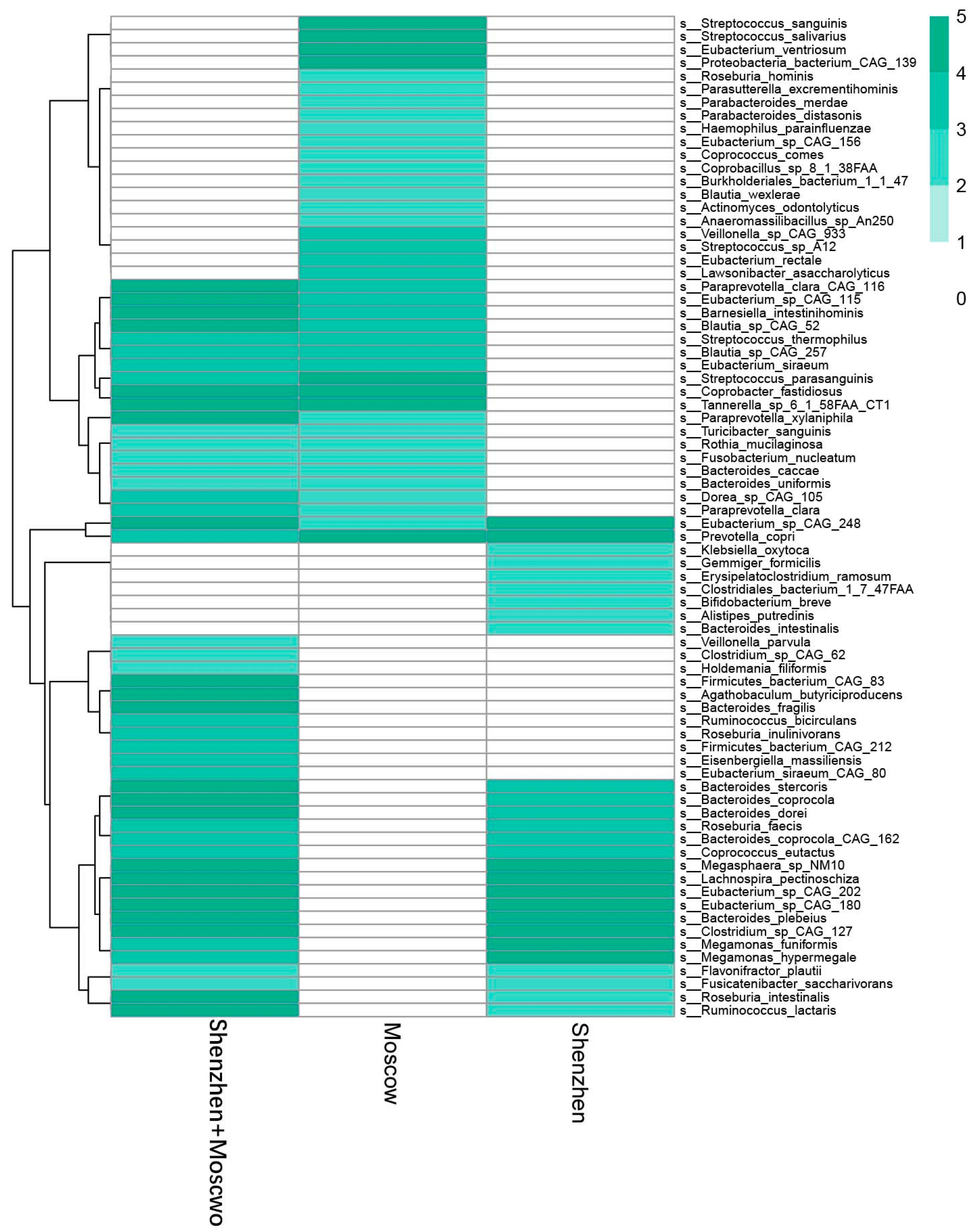
| Model | Number of Samples | Country or Region | Sequencing Methods | Manifestation of Species Disorder (ASD) | Reference |
|---|---|---|---|---|---|
| children | TD: 10; ASD: 10 | Slovakia | Realtime-PCR | phyla Bacteroidetes/Firmicutes: ↓ *; Lactobacillus: ↑; Bifidobacterium/Lactobacillus, Streptococcus thermophillus, the total bacteria content: - | [38] |
| children | TD: 45; ASD: 45 | China | 16S rRNA V3-V4 | At phylum level: -; genera Lachnoclostridium, Tyzzerella subgroup 4, Flavonifractor, unidentified_Lachnospiraceae: ↓ | [39] |
| children | TD: 20; ASD: 20 | - | 16S rRNA V2-V3 | genera Prevotella, Coprococcus, and unclassified_Veillonellaceae: ↓ | [40] |
| children | TD: 35; ASD: 6 | China | 16S rRNA V3-V4 | phyla Bacteroidetes/Firmicutes; genera Sutterella, Odoribacter and Butyricimonas: ↑ genera Veillonella and Streptococcuse: ↓ | [41] |
| children ASD | TD: 40; ASD: 40 | Italy | 16S rRNA V3-V5 | phylum Bacteroidetes, genera Alistipes, Bilophila, Dialister, Parabacteroides, Veillonella: ↓; phyla Firmicutes/Bacteroidetes; genera Collinsella, Corynebacterium, Dorea, and Lactobacillus; Escherichia/Shigella and Clostridium cluster XVII; fungal: genus Candida: ↑ | [42] |
| mice | TD: 10; ASD: 10 | USA | 16S rRNA V3-V5 | classes Bacteroidia, Clostridia: ↑ | [43] |
| children | TD: 3; ASD: 3 | UK | FISH-FCM | phyla Clostridium spp.: ↑ Bifidobacterial: ↓ | [44] |
| children | TD: 20; ASD: 18 | USA | 16S rRNA V4 | genera Bifidobacterium, Desulfovibrio: ↓ | [23] |
| mice | TD: 21; ASD: 25 | Canada | qRT-PCR | phyla Firmicutes: ↓ Bacteroidetes: ↑ | [45] |
| children and mothers | TD: 30; ASD: 59 | China | 16S rRNA V1-V2 | Children: phylum Proteobacteria: ↑; genera Enhydrobacter, Chryseobacterium, Streptococcus, and Acinetobacter: ↑; species Acinetobacter rhizosphaerae, Acinetobacter johnsonii, Prevotella melaninogenica: ↓ Mother: families Moraxellaceae and Enterobacteriaceae, genus Faecalibacterium: ↓ | [46] |
| minors | TD: 450 ASD: 569 | China, Ecuador, Italy, Korean | 16S rRNA V3-V4, V4, V4-V5 | Results were variable according to different analysis methods and parameter settings. | [47] |
| children | TD: 31 ASD: 43 | China | Shotgun metagenomic sequencing | phylum Actinobacteria: ↑; three Clostridium taxons, two Eggerthella taxons, two Klebsiella taxons: ↑; taxons Bacteroides vulgatus, Betaproteobacteria, Campylobacter jejuni subsp. jejuni 81–176, Campylobacter jejuni subsp. jejuni ICDCCJ07001, Candidatus Chloracidobacterium thermophilum B, Coraliomargarita akajimensis DSM 45221, Proteus mirabilis, and HI4320 Spirochaeta thermophila DSM 6192: ↓ | [24] |
| children | TD: 20 ASD: 30 | Moscow | Shotgun metagenomic sequencing | species Enterococcus faecium, Megasphaera elsdenii, Bacteroides fragilis: ↑ | [28] |
| Characteristic | Moscow Cohort | Shenzhen Cohort |
|---|---|---|
| Subjects of ASD (n) | 30 | 43 |
| Subjects of TD (n) | 20 | 31 |
| Age (years) | 3–5 | 2–7 |
| Sequencing instruments | Illumina NovaSeq 6000 | Illumina HiSeq 4000 |
| Layout | PAIRED | PAIRED |
| AvgSpotLen | 300 | 300 |
| Bytes (Gb) | 1.92–4.08 | 0.526–4.09 |
| Model A | Model B | Model C | Model D | Model E | Average | |
|---|---|---|---|---|---|---|
| 1st iteration | 136 | 88 | 111 | 109 | 140 | 117 |
| 2nd iteration | 78 | 49 | 68 | 65 | 90 | 70 |
| 3rd iteration | 60 | 38 | 54 | 44 | 60 | 51 |
| 4th iteration | 48 | 34 | 46 | 38 | 38 | 41 |
| 5th iteration | 43 | 28 | 43 | 36 | 35 | 37 |
| 6th iteration | 36 | 24 | 34 | 35 | 30 | 32 |
| Model A | Model B | Model C | Model D | Model E | Average | |
|---|---|---|---|---|---|---|
| 1st iteration | 112 | 125 | 110 | 116 | 109 | 114 |
| 2nd iteration | 93 | 96 | 73 | 80 | 84 | 85 |
| 3rd iteration | 59 | 77 | 56 | 72 | 71 | 67 |
| 4th iteration | 51 | 61 | 50 | 62 | 53 | 55 |
| 5th iteration | 48 | 56 | 45 | 58 | 51 | 52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Fu, P. Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts. Microorganisms 2023, 11, 291. https://doi.org/10.3390/microorganisms11020291
Wang W, Fu P. Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts. Microorganisms. 2023; 11(2):291. https://doi.org/10.3390/microorganisms11020291
Chicago/Turabian StyleWang, Wenjuan, and Pengcheng Fu. 2023. "Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts" Microorganisms 11, no. 2: 291. https://doi.org/10.3390/microorganisms11020291
APA StyleWang, W., & Fu, P. (2023). Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts. Microorganisms, 11(2), 291. https://doi.org/10.3390/microorganisms11020291







