Abstract
Rhodococcus has been extensively studied for its excellent ability to degrade artificial chemicals and its capability to synthesize biosurfactants and antibiotics. In recent years, studies have attempted to use Rhodococcus as a gene expression host. Various genetic tools, such as plasmid vectors, transposon mutagenesis, and gene disruption methods have been developed for use in Rhodococcus; however, no effective method has been reported for performing large-size genome reduction. Therefore, the present study developed an effective plasmid-curing method using the levansucrase-encoding sacB gene and a simple two-step genome-reduction method using a modified Cre/loxP system. For the results, R. erythropolis JCM 2895 was used as the model; a mutant strain that cured all four plasmids and deleted seven chromosomal regions was successfully obtained in this study. The total DNA deletion size was >600 kb, which corresponds mostly to 10% of the genome size. Using this method, a genome-structure-stabilized and unfavorable gene/function-lacking host strain can be created in Rhodococcus. This genetic tool will help develop and improve Rhodococcus strains for various industrial and environmental applications.
1. Introduction
The genus Rhodococcus is a Gram-positive bacterium with a high G+C content belonging to the phylum Actinobacteria and has been evaluated in various aspects, such as ecology, pathogenicity, biosurfactant production, and degradation of chemical compounds [1,2,3,4,5,6]. In particular, the ability to degrade chemicals, especially aromatic compounds, has been extensively studied, and this is the most distinguishing feature of Rhodococcus [7,8,9]. In recent years, antibiotic-producing strains and novel bioactive compounds have been obtained from this genus [10,11,12].
At present, Rhodococcus has been isolated from a wide range of environments, including freshwater, seawater, soil, deep-sea floors, and polar regions as well as from animals, plants, and insects [5,13,14,15,16,17]. The diverse activities of this genus are associated with the wide range of environments colonized. The diverse activities of Rhodococcus are attributed to the genes possessed. Recently, many draft and complete genomes have been analyzed, and the chromosome sizes have been reported to be 6–7 Mb; the presence of linear and/or circular plasmids results in an increase in genome size of 1–2 Mb [18,19,20]. The large-sized genome structure can accommodate non-essential genes, and it makes this genus highly versatile.
A characteristic feature of the genomic structure of Rhodococcus involves its topology; certain species have circular chromosomes, and others have linear chromosomes. Four strains of this genus, including R. jostii RHA1 [20], R. opacus B4 [21], Rhodococcus sp. Strain R79, and R. koreensis strain R85 [19], have been clearly shown to possess a linear chromosome on the basis of complete genome sequencing analysis. Most of the other genomes analyzed in strains including R. opacus PD630 [18], R. fascians D188 [22], R. rhodochrous EP4 [23], R. equi strain DSSKP-R-001 [24], R. ruber strain C1 [25], R. triatomae DSM 44,893 [26], R. globerulus strain D757 [27], and R. erythropolis JCM 2895 [28] have been shown to possess a circular chromosome. The presence/emergence of a linear chromosome in Actinobacteria has been attributed to recombination between the circular chromosome and linear plasmid [29,30]. In this theory, the whole chromosomal DNA is integrated into the linear plasmid, and it behaves as a single linear replicon in this irreversible transformation. Therefore, the difference in linear or circular chromosomes is not related to the evolutionary lineage, and it most probably occurs individually in each strain. This phenomenon may occur at any time in strains containing linear plasmids, between both circular and linear replicons.
The presence of plasmids in Rhodococcus is of great ecological and functional importance. In R. equi and R. fascians, genes related to pathogenicity and infection are located on plasmids [3,31,32]; in R. jostii RHA1, R. erythropolis TA421, and other strains, the aromatic compound degradation genes are located on a large linear plasmid in addition to the chromosome [33,34,35]. In R. erythropolis BD2, the genes associated with metal tolerance are located on a linear plasmid [36]. In addition, R. erythropolis JCM 6824 contains antibiotic biosynthesis genes, which are located on the chromosome [37,38]. In contrast, antibiotic biosynthesis genes are encoded in a small circular plasmid in R. erythropolis JCM 2895 [28,39]. Regardless of the topology and size, plasmids are important for various functions in this genus.
Based on the activities of Rhodococcus, it has been developed and used as a microbial host, especially for mediating the biodegradation and bioproduction of certain compounds. Several genetic tools have been developed at present [40,41,42], and highly effective codon optimization techniques have been demonstrated in the genus to increase the expression level of cloned genes [43]. The advantages of using Rhodococcus as a microbial host, in addition to its activities mentioned above, are listed as follows: it grows quickly among Actinomycetes species and shows optimal growth on conventional nutrient medium such as Luria-Bertani (LB) agar. It does not have a life cycle (differentiation, such as sporulation) which is generally observed in Streptomyces, and it can be handled in the same manner as Escherichia coli. Since Rhodococcus is GC-rich, it is naturally suitable for expressing genes of Streptomyces and other Actinomycetes, which possess several genes that encode secondary metabolites. Useful genetic tools/methods have been established, including multiple plasmid vectors [44], gene-disruption systems via homologous recombination [40], transposon mutagenesis [45], and effective electrotransformation [46].
Genomic instability, which is an undesirable trait for a host microorganism, has been observed in Rhodococcus. For example, the emergence of alternative-sized linear plasmids has been observed in R. jostti RHA1 and R. opacus DSM 43,250 during laboratory cultivation [47,48]. The aforementioned chromosomal linearization is also associated with genomic instability. In addition, size reduction of circular plasmid vectors introduced in genetic experiments has been observed in several species (laboratory knowledge). This genomic instability may affect research experiments. Recent genome sequencing analysis of Rhocococcus identified several linear/circular plasmids; however, these cryptic plasmids are not essential in Rhodococcus used as a host microorganism based on compatibility with the vector to be introduced and the possibility of intra-genome recombination, as described above. Few studies reported that deletion of the plasmid improves the growth of the strain [49,50,51] or improves the expression of the gene in the vector [52]. Therefore, deletion of these cryptic plasmids is strongly desired.
The development of sophisticated host–vector systems using this genus remains an important area of study. An additional concern of using Rhodococcus as a host includes its innate abilities, such as bioproduction and degradation. Undesired background activity is anticipated when using the host for these purposes. In order to cancel the background activity, genes/gene clusters responsible for the activity must be deleted. These sizes are sometimes very large; therefore, conventional gene-disruption methods based on homologous recombination are not applicable. Genome engineering methods for large-scale deletion of chromosomal DNA, which greatly affects the genome structure, have not been established in Rhodococcus so far, and an effective method is desired [53,54].
This study reports an effective method for curing a cryptic plasmid and an effective genome-reduction method in R. erythropolis. The former contributes to increased genomic and introduced-vector stability, while the latter enables the removal of undesirable host innate genes and activities.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Primers, and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table S1 and primers in Table S2. For both Rhodococcus and E. coli cultivation, LB medium was used and kanamycin (200 µg/mL for Rhodococcus, 25 µg/mL for E. coli), chloramphenicol (34 µg/mL), apramycin (50 µg/mL), ampicillin (100 µg/mL), and thiostrepton (0.5 µg/mL) were added when necessary. W-minimal medium [55] supplemented with succinate (0.2%, w/v), sucrose (0.2%, w/v), casamino acids (0.2%, w/v), and thiamine (0.1%, w/v) was used for preparation of electrocompetent Rhodococcus.
2.2. Electrotransformation
Electrocompetent cells were prepared as follows: Rhodococcus cells grown in the liquid medium were collected via centrifugation (at 8000× g, 5 min, 4 °C), and the cell pellets were washed twice with sterilized 10% glycerol, after which the cells were finally resuspended in 10% glycerol. The cells were directly used for the next step or stored in −80 °C. The plasmids were introduced into the cell using the Gene Pulser Xcell Electroporation System (Bio-Rad, Hercules, CA, USA) (setting parameters: exponential protocol, voltage: 1.6 kV/cm, resistance: 500–800 Ω, time constant: 9–11 ms). Recovering culture was statically incubated in liquid LB medium containing 10 mM glucose for 4 h at 28 °C. Cells were then spread on LB agar containing an appropriate antibiotic to select the transformants.
2.3. Plasmid Curing Using sucB Gene
The pK18mobsacB vector [40,56] (suicide vector in Rhodococcus) containing a homologous DNA region (approximately 3 kb, PCR-amplified) of curing target plasmid was introduced into Rhodococcus cells via electrotransformation. Subsequently, kanamycin-resistant colonies were selected on agar medium and confirmed for homologous recombination (single crossover) using PCR. Further single colony isolation on the kanamycin media was conducted when necessary. Subsequently, the mutant was cultivated in liquid LB without kanamycin, followed by spreading on LB agar containing 20% sucrose without kanamycin. The loss of target plasmid was confirmed via colony PCR using two or three independent primer sets, which amplified different parts of the target plasmid (Figure 1). The PrimeSTAR GXL DNA polymerase and Tks Gflex DNA Polymerase (Takara Bio, Ootu, Japan) were used for DNA cloning and colony PCR, respectively.
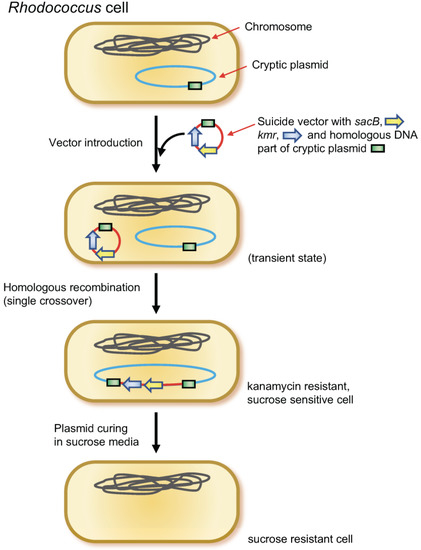
Figure 1.
Plasmid-curing method using sacB gene. Suicide vector with sacB gene was integrated to the cryptic plasmid to be cured by homologous recombination. Subsequently, the plasmid was cured by cultivating in a media containing 20% sucrose.
2.4. Genome Reduction Using Cre/loxP System
2.4.1. Vector Construction
pBS-aphII-loxLE (used for the first vector construction in Round A) was constructed on the basis of pBluescriptII by introducing kanamycin-resistant gene (aphII) from pK18mobsacB and synthesized loxLE sequence (TACCGTTCGTATAGCATACATTATACGAAGTTAT) [57]. pACYC-aac-loxRE (used for the second vector construction in Round A) was constructed on the basis of pACYC184 [58] by introducing apramycin-resistant gene (aac(3)IV) from pHN1237 [59], synthesized loxRE sequence (ATAACTTCGTATAGCATACATTATACGAACGGTA) [57] and by deleting the tetracycline-resistant gene. pTip-sacB-cre (used as the third vector in Round A) was constructed on the basis of pTip-QC2 vector (E. coli-Rhodococcus shuttle vector [44]) by introducing sacB gene from pK18mobsacB and Cre recombinase gene (cre). The cre gene was placed under the regulation of thiostrepton-inducible Tip promoter [44]. pK18mobsacB-loxLE (used for the first vector construction in Round B) was constructed by introducing loxLE sequence into pK18mobsacB. pCH-cre-loxRE (used for the second vector construction in Round B) was constructed by ligating p15A ori from pACYC184, thiostrepton-inducible protein gene (tipA) and thiostrepton-resistant gene (tsr) from pTip-QC2, aac(3)IV, cre, and thiostrepton-inducible CH2.2 promoter from pTip-CH2.2 [41]. The cre gene was placed under the regulation of CH2.2 promoter. For each deletion target, the first vector was constructed by ligating an upstream flanking part of deletion target region (left homolog, approximately 3 kb), whereas the second vector was constructed by ligating a downstream flanking part of the deletion target region (right homolog, approximately 3 kb) (Figure 2).
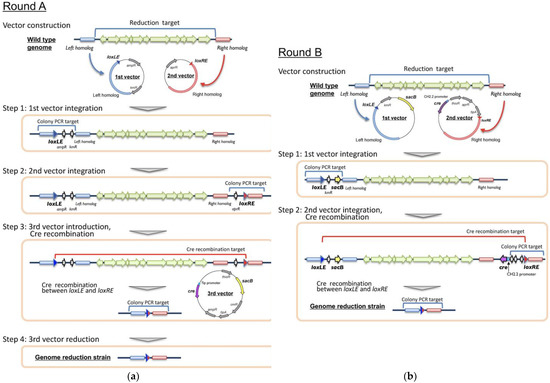
Figure 2.
Large-size genome-reduction method using Cre/loxP. (a) Four-step procedure requires three plasmid-introduction and one plasmid-reduction steps (Round A). (b) Two-step procedure requires only two plasmid-introduction steps (Round B). See details in the text.
2.4.2. Genome-Reduction Strategy
Round A (Figure 2A): The first plasmid vector was introduced into the cells and a single-crossover mutant (kanamycin-resistant) was selected as described above. Subsequently, the second plasmid vector was introduced into the mutant and a second single-crossover mutant (kanamycin- and apramycin-resistant, single crossover at two positions) was selected. Next, the second-single crossover mutant was transformed using the third vector, and a chloramphenicol-resistant colony was selected. cre recombinase expression (on the third vector) was induced by adding thiostrepton (0.5 µg/mL) in liquid medium without other antibiotics, followed by spreading on LB agar. The deletion of the target region between loxLE and loxRE was confirmed via colony PCR. Finally, the third vector was cured by cultivating cells on LB agar containing 20% (w/v) sucrose without antibiotics. Round B (Figure 2B): The first and second vectors were introduced, and second-single crossover mutant was selected as in Round A. cre recombinase expression (on the second vector, genome-integrated) was induced by adding thiostrepton. The deletion mutant was selected by cultivating colonies on LB agar containing 20% sucrose, and the deletion was confirmed via colony PCR.
3. Results
3.1. Isolation of Plasmid-Cured Strains
R. erythropolis JCM 2895, one of the strains used in the present study, produces a bacteriocin-like antibiotic protein [39]. Since JCM 2895 shows faster growth and less biofilm/aggregate formation compared with that of other R. erythropolis strains, we selected it as the candidate host strain. The complete genome has recently been decoded, and one linear plasmid (pR09L01, 227,989 bp) and three circular plasmids (pR09C01, 79,600 bp; pREC01, 5420 bp; and pREC02, 5444 bp) have been identified in addition to the one chromosome [28]. We first attempted to cure the four plasmids from the strain. Several plasmid-curing methods have been established, including repeating high-temperature culture [60], protoplast regeneration [61], and culture with sodium dodecyl sulphate [62,63] or acridine orange [63,64]. Among these methods, repeated application of high-temperature culture and protoplast regeneration were unsuccessful in curing plasmids from this strain (data not shown).
Since sacB is effective as a marker for counter selection in Rhodococcus [40,65], plasmid curing using sacB was attempted in the strain, as illustrated in Figure 1 (see also Materials and Methods). First, to obtain a circular plasmid pR09C01-cured mutant, a DNA fragment of partial pR09C01 was PCR-amplified using a primer set of R09CPD-F1 and R09CPD-R1 (amplify 3078 bp), which was inserted into pK18mobsacB at the EcoRI-XbaI site, resulting in a vector pK18R09CPD1. This vector was introduced into the wild-type strain, which successfully yielded kanamycin-resistant mutants. The appropriate single crossover was confirmed via colony PCR using a primer set, R09CPD-SCC1 and aphII-UR. The former hybridizes to the upstream flanking region of the homologous sequence, and the latter hybridizes to the kanamycin-resistant gene, which amplifies 3737 bp. The single-crossover mutant was then liquid-cultured without kanamycin and spread on LB agar containing sucrose without kanamycin. The colonies on the media were transferred to fresh media after which the kanamycin sensitivity and sucrose resistance were verified again. Plasmid deletion was confirmed via colony PCR using the R09CPD-F1 and R09CPD-R1 primer sets and was also confirmed using another primer set that amplifies another region of pR09C01 (R09CPD-F2 and R09CPD-R2, which amplify the 3006 bp). In this study, most of the candidates grown on sucrose-containing media were successfully cured of pR09C01, and the resultant strain was named strain R0901.
Using strain R0901, curing of a linear plasmid pR09L01 was attempted in the same manner. The suicide vector containing a part of pR09L01, called pK18R09LPD01, was constructed and introduced into R0901. A pR09L01-cured mutant was obtained, confirmed, and named strain R0902. A pREC01-cured strain R0903 and pREC02-cured strain R0904 were also obtained in the same manner.
3.2. Development of Large Fragment Deletion (Genome-Reduction) Strategy Using the Cre/lox System
Various techniques have been developed for deleting single genes or short DNA regions (approximately > 10 kb) and have been widely used in Rhodococcus. The marker-less gene deletion system using sacB gene represents a major strategy for use in this genus [40,65]. However, no effective large-scale DNA deletion or introduction methods that enable genome-scale engineering has been successfully applied in Rhodococcus. The Cre/loxP-mediated genome-reduction method has already been successfully used in the Actinobacteria Streptomyces and Corynebacterium [57,66]. In the Cre/loxP system, Cre recombinase specifically recognizes the 34 nt lox sequence and catalyzes recombination. The basic reaction involves looping out (deleting) a DNA region between two distant lox sequences, leaving a single lox sequence [67]. It is a useful marker-less method that can be repeated many times (at many sites) at a high success rate. However, the procedure of the Cre/loxP system is laborious and time consuming; it involves a four-step procedure including three plasmid-introduction and one plasmid-reduction steps.
This four-step procedure, with a minor modification, was first attempted in R. erythropolis JCM 6824 [38,65], an aurachin RE antibiotic producer, to remove a 95 kb chromosomal DNA region containing its biosynthetic gene cluster (Round A, Figure 2A). The target region of 95 kb is not found in other strains of R. erythropolis; therefore, it has been regarded as a non-essential region and selected for deletion. The upstream flanking region of the deletion target (left homolog) was PCR-amplified using a primer set of 95k-LE-F2 and 95k-LE-R. The 3.1 kb amplicon was inserted into the XbaI-SphI site of pBS-aphII-loxLE, which yielded the first vector pBS-D95k-LE. Similarly, the downstream flanking region of the deletion target (right homolog) was PCR-amplified using a primer set of 95k-RE-F and 95k-RE-R2. The 3.2 kb amplicon was inserted into the SpeI-Sse8387I site of pACYC-aac-loxRE, which yielded the second vector pAC-D95k-RE. The first vector was introduced into wild-type JCM 6824 cells, and a kanamycin-resistant single-crossover mutant was obtained and confirmed via colony PCR, as performed in the plasmid-curing method (primer set; D95k-LE-CF and LE-DR, which amplify 3256 bp). The second vector was introduced into the single-crossover mutant cells of the first vector, and the apramycin-resistant mutant was obtained and crossover was confirmed via colony PCR (primer set; RE-UF and D95k-RE-CR, which amplify 3458 bp). Subsequently, the double mutant was transformed using the third vector, pTip-sacB-cre, which contained the cre recombinase and sacB genes. The resultant kanamycin/apramycin/chloramphenicol-resistant mutant was cultivated with thiostrepton to induce Cre recombinase expression. The recombination (genome reduction) between loxLE and loxRE was confirmed via colony PCR using a primer set comprising D95k-LE-CF and D95k-RE-CR. Using this primer set, a 6486 bp amplicon was obtained from the genome-reduction mutant, whereas no amplicon was obtained from the wild-type because the target size of >95 kb was too large to amplify via PCR. The third vector containing sacB was cured on sucrose-containing medium, as performed in the plasmid-curing method. Consequently, a genome-reduction strain was obtained. This is the first study to report a large-size genome-reduction method using the Cre/loxP system in Rhodococcus. The last vector deletion step was simplified owing to the use of the sacB system compared with that in the Streptomyces/Corynebacterium system; however, it is still a laborious method and must be further simplified.
Next, the two-step genome-reduction strategy needing only two vector-introduction steps without a plasmid-reduction step, was developed in R. erythropolis R0904, a plasmid-less derivative of R. erythropolis JCM 2895 constructed as described above (Round B, Figure 2B). Deletion target positions not conserved among other R. erythropolis strains were examined using GenomeMatcher software [68] (Figure S1). A total of seven regions, each having a size of >5 kb, were selected for deletion (Targets 1–7 = T1–T7). The left and right homolog sequences of T1 were PCR-amplified and introduced into the pK18mobsacB-loxLE and pCH-cre-loxRE vectors, respectively. The resultant vectors pSLE-R09T1 (first vector) and pCRE-R09T1 (second vector) were introduced into the R0904 cells, and single-crossover mutants were isolated as performed in Round A. The differences between Round A and Round B were as follows: sacB and cre recombinase genes in the third vector in Round A were separately integrated into the first and second vectors, respectively, in Round B. Further, sacB counter selection is used, not for plasmid reduction but for genome reduction. The kanamycin- and apramycin-resistant double mutant was then liquid cultured in the presence of thiostrepton, followed by spreading on agar plates containing sucrose. Genome reduction was confirmed via colony PCR similar to that in the Round A. We found that 100% of colonies on the plates had deleted T1 (approximately 117 kb in size, Table 1), and this strain was named R0905. Since the T2 size is relatively small (6.4 kb, Table 1), deletion was performed using a known marker-less double-crossover method [65]. Briefly, left and right homologs were tandemly inserted in the pK18mobsacB vector, and the resulting pK18-R09T2 vector was introduced into strain R0905. The double-crossover mutant was then isolated (independent of the Cre/loxP system) and named strain R0906. Using strain R0906, a T3 deletion mutant was isolated via a two-step procedure (Round B strategy) as in the T1 and named R0907. Similarly, all the other T4–T7 deletion mutants were isolated and named R0908–R0911, respectively (Table 1).

Table 1.
Deletion size and genome size of wild and mutant strains of R. erythropolis JCM 2895.
4. Discussion
The plasmid-curing method using sacB gene was first demonstrated in the genus Rhodococcus. Using sacB gene for curing cryptic plasmids have been tested in other bacteria [69,70]; however, they introduced the sacB gene by random transposon mutagenesis or using incompatible vectors to kick out the target cryptic plasmid. The former strategy required a screening step for the genuine mutant strain having transposon (sacB gene) on the target plasmid from the mutant library. The latter strategy required construction of a new vector incompatible with the target plasmid, that is, replication system of the target must be identified before the curing. After removing the target plasmid by incompatibility, the introduced vector must be cured via sacB selection system. In addition, multiple incompatible vectors have to be prepared when multiple plasmids are targeted. In our study, sacB gene was exclusively integrated into the target plasmid via homologous recombination using a suicide vector. Therefore, our procedure greatly reduced the laborious steps reported in the previous study. The successful isolation of strain R0904, which was cured of all the four plasmids, demonstrated that it is a highly effective method for use in R. erythropolis. In the process of curing pR09C01 and pR09L01 plasmid, a genuine single-crossover mutant strain was obtained without single colony isolation of the candidate colony. In contrast, for curing pREC01 and pREC02, the single colony isolation step was required on kanamycin-containing media. During the additional subculture, transient mutant cells harboring both original shape plasmid and single-crossover plasmid might be eliminated, and genuine single-crossover mutant cells were selected (not experimentally proven). The former two large plasmids, pR09C01 and pR09L01, are single copy or low copy, whereas the latter two small plasmids, pREC01 and pREC02, are multiple copy. The copy number difference might have affected the selection, i.e., the latter required the further selection. In this study, additional one-time single colony isolation was enough for the isolation of the mutant.
In the genome-reduction study, approx. 95 kb and 117 kb chromosomal deletions were carried out in R. erythropolis JCM 6825 and JCM 2895 (in the case of T1), respectively. These results demonstrated that the Cre/loxP system is useful for large-size genome reduction in Rhodococcus. In the strain JCM 2895, plasmid curing and genome reduction was performed repeatedly. The final deletion strain R. erythropolis R0911 was cured of all four linear and circular plasmids and showed seven deleted strain-specific DNA regions on the chromosome. The total DNA deletion size was >600 kb, which corresponds mostly to 10% of the genome size (Table 1). These results clearly demonstrated that the two-step procedure newly developed in this study represents a simple, effective, accurate, and repeatable tool for performing genome reduction. The new method greatly reduces the time and effort required because only two vector-introduction steps are needed, whereas three vector-introduction and one vector-reduction steps are required in the previous Cre/loxP method [57,66].
The key technology that enables this advanced technique involves the strictly regulated expression of cre recombinase present in the second vector. The CH2.2 promoter used in this study induces gene expression in the presence of thiostrepton; however, it strictly suppresses expression in the absence of the inducer with the TipA regulatory protein [41]. When a leaky promoter was used regardless of whether the level was low (such as Tip promoter recruited in the pTip-QC2 vector), the recombination between loxLE and loxRE occurred immediately after the introduction of the second vector and before the single-crossover event with the right homolog since Cre recombinase is highly active. In this case, the entire second vector DNA sequence would be integrated at the loxLE site and genome reduction would fail (Figure S2). Using 20% sucrose instead of 10% that is generally used is important for achieving sacB counter selection in this strain. Sucrose at 20% retards growth compared with that obtained at 10%; it eliminates false-positive clones efficiently, which may depend on the strain used.
The genome-reduction system greatly depends on single-crossover and Cre recombinase activity. Therefore, this system is applicable to any microorganism if single-crossover and Cre recombinase expression can be successfully used in the strain of interest.
5. Conclusions
In this study, an effective plasmid-curing method and an efficient large-size genome-deletion method was developed. The application of both methods will considerably advance genome manipulation in Rhodococcus, improving the stability of the genome and plasmids, and it will enable the elimination of unnecessary background host functions that retard the desirable activities of heterologous genes. These techniques may be applicable for strain improvement as well as for minimum genome studies on Rhodococcus, which have not been conducted previously.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020268/s1, Table S1: Bacterial strains and plasmids used in this study; Table S2: Primers used in this study; Figure S1: Determination of genome reduction target position (T1–T7) of R. erythropolis JCM 2895 by comparing genome sequence of four other R. erythropolis strains; Figure S2: Cre recombination between chromosomal loxLE and plasmid loxRE.
Author Contributions
Conceptualization, W.K.; methodology, W.K.; validation, W.K.; investigation, W.K. and M.H.; resources, W.K.; data curation, W.K. and M.H.; writing—original draft preparation, W.K.; writing—review and editing, W.K.; supervision, W.K.; project administration, W.K.; funding acquisition, W.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI, grant numbers 25108728 and 17H05457.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Haruo Ikeda and Mamoru Komatsu for their technical support on the Cre/loxP study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ikeda, Y.; Kishimoto, M.; Shintani, M.; Yoshida, N. Oligotrophic Gene Expression in Rhodococcus erythropolis N9T-4 under Various Nutrient Conditions. Microorganisms 2022, 10, 1725. [Google Scholar] [CrossRef] [PubMed]
- Goethals, K.; Vereecke, D.; Jaziri, M.; Van Montagu, M.; Holsters, M. Leafy gall formation by Rhodococcus fascians. Annu. Rev. Phytopathol. 2001, 39, 27–52. [Google Scholar] [CrossRef]
- Meijer, W.G.; Prescott, J.F. Rhodococcus equi. Vet. Res. 2004, 35, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Nishi, S.; Fukuoka, T.; Kitamoto, D.; Watsuji, T.O.; Nagano, Y.; Yabuki, A.; Nakagawa, S.; Hatada, Y.; Horiuchi, J. Deep-sea Rhodococcus sp. BS-15, lacking the phytopathogenic fas genes, produces a novel glucotriose lipid biosurfactant. Mar. Biotechnol. 2014, 16, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Roslee, A.F.A.; Zakaria, N.N.; Convey, P.; Zulkharnain, A.; Lee, G.L.Y.; Gomez-Fuentes, C.; Ahmad, S.A. Statistical optimisation of growth conditions and diesel degradation by the Antarctic bacterium, Rhodococcus sp. strain AQ507. Extremophiles 2020, 24, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.J.; Kulakov, L.A.; Allen, C.C.R. Biodegradation and Rhodococcus—Masters of catabolic versatility. Curr. Opin. Biotechnol. 2005, 16, 282–290. [Google Scholar] [CrossRef]
- Maeda, M.; Chung, S.Y.; Song, E.; Kudo, T. Multiple genes encoding 2,3-dihydroxybiphenyl 1,2-dioxygenase in the gram-positive polychlorinated biphenyl-degrading bacterium Rhodococcus erythropolis Ta421, isolated from a termite ecosystem. Appl. Environ. Microbiol. 1995, 61, 549–555. [Google Scholar] [CrossRef]
- Dabrock, B.; Kesseler, M.; Averhoff, B.; Gottschalk, G. Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolis BD2 that encodes isopropylbenzene and trichloroethene catabolism. Appl. Environ. Microbiol. 1994, 60, 853–860. [Google Scholar] [CrossRef]
- Kitagawa, W.; Kimura, N.; Kamagata, Y. A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 2004, 186, 4894–4902. [Google Scholar] [CrossRef]
- Kitagawa, W.; Tamura, T. Three types of antibiotics produced from Rhodococcus erythropolis strains. Microb. Environ. 2008, 23, 167–171. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Uchida, R.; Takakusagi, Y.; Matsumoto, A.; Jiang, C.L.; Takahashi, Y.; Arai, M.; Kobayashi, S.; Matsumoto, M.; Inokoshi, J.; et al. Lariatins, novel anti-mycobacterial peptides with a lasso structure, produced by Rhodococcus jostii K01-B0171. J. Antibiot. 2007, 60, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Harunari, E.; Bando, M.; Igarashi, Y. Rausuquinone, a non-glycosylated pluramycin-class antibiotic from Rhodococcus. J. Antibiot. 2022, 75, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A.F. Rhodococcus triatomae sp. nov., isolated from a blood-sucking bug. Int. J. Syst. Evol. Microbiol. 2005, 55, 1575–1579. [Google Scholar] [CrossRef]
- Hackbusch, S.; Noirungsee, N.; Viamonte, J.; Sun, X.; Bubenheim, P.; Kostka, J.E.; Muller, R.; Liese, A. Influence of pressure and dispersant on oil biodegradation by a newly isolated Rhodococcus strain from deep-sea sediments of the gulf of Mexico. Mar. Pollut. Bull. 2020, 150, 110683. [Google Scholar] [CrossRef]
- Ocampo-Sosa, A.A.; Lewis, D.A.; Navas, J.; Quigley, F.; Callejo, R.; Scortti, M.; Leadon, D.P.; Fogarty, U.; Vazquez-Boland, J.A. Molecular epidemiology of Rhodococcus equi based on traA, vapA, and vapB virulence plasmid markers. J. Infect. Dis. 2007, 196, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Tilford, P.E. Fasciation of sweet peas caused by Phytomonas fascians n. sp. J. Agric. Res. 1936, 53, 0383–0394. [Google Scholar]
- Watanabe, K.; Shimizu, H.; Aihara, H.; Nakamura, R.; Suzuki, K.; Komagata, K. Isolation and identification of cholesterol-degrading Rhodococcus strains from food of animal origin and their cholesterol oxidase activities. J. Gen. Appl. Microbiol. 1986, 32, 137–147. [Google Scholar] [CrossRef]
- Holder, J.W.; Ulrich, J.C.; DeBono, A.C.; Godfrey, P.A.; Desjardins, C.A.; Zucker, J.; Zeng, Q.; Leach, A.L.; Ghiviriga, I.; Dancel, C.; et al. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Benning, S.; Brugnone, N.; Siani, R.; Kublik, S.; Schloter, M.; Rad, V. Complete genome sequences of two Rhodococcus sp. strains with large and linear chromosomes, isolated from apple rhizosphere. Microbiol. Resour. Announc. 2021, 10, e0015921. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef]
- Rhodococcus opacus B4, Complete Sequence. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NC_012522.1 (accessed on 30 December 2022).
- Stamler, R.A.; Vereecke, D.; Zhang, Y.; Schilkey, F.; Devitt, N.; Randall, J.J. Complete genome and plasmid sequences for Rhodococcus fascians D188 and draft sequences for Rhodococcus Isolates PBTS 1 and PBTS 2. Genome Announc. 2016, 4, e00495-16. [Google Scholar] [CrossRef] [PubMed]
- Rhodococcus rhodochrous Strain EP4 Chromosome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP032221.1 (accessed on 30 December 2022).
- Prescottella equi Strain DSSKP-R-001 Chromosome, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP027793.1 (accessed on 30 December 2022).
- Rhodococcus ruber Strain C1 Chromosome, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP044211.1 (accessed on 30 December 2022).
- Rhodococcus triatomae Strain DSM 44893 Chromosome, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP048813.1 (accessed on 30 December 2022).
- Rhodococcus globerulus Strain D757 Chromosome, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP079698.1 (accessed on 30 December 2022).
- Kitagawa, W.; Hata, M. Complete genome sequence of Rhodococcus erythropolis JCM 2895, an antibiotic protein-producing strain. Microbiol. Resour. Announc. 2022, 11, e0068222. [Google Scholar] [CrossRef] [PubMed]
- Kirby, R. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 2011, 319, 1–10. [Google Scholar] [CrossRef]
- Chen, C.W.; Huang, C.H.; Lee, H.H.; Tsai, H.H.; Kirby, R. Once the circle has been broken: Dynamics and evolution of Streptomyces chromosomes. Trends Genet. 2002, 18, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Crespi, M.; Messens, E.; Caplan, A.B.; Vanmontagu, M.; Desomer, J. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992, 11, 795–804. [Google Scholar] [CrossRef]
- Stes, E.; Francis, I.; Pertry, I.; Dolzblasz, A.; Depuydt, S.; Vereecke, D. The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol. Lett. 2013, 342, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Kobayashi, H.; Masai, E.; Fukuda, M. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2001, 67, 2021–2028. [Google Scholar] [CrossRef]
- Kitagawa, W.; Miyauchi, K.; Masai, E.; Fukuda, M. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 2001, 183, 6598–6606. [Google Scholar] [CrossRef] [PubMed]
- Kosono, S.; Maeda, M.; Fuji, F.; Arai, H.; Kudo, T. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl. Environ. Microbiol. 1997, 63, 3282–3285. [Google Scholar] [CrossRef]
- Stecker, C.; Johann, A.; Herzberg, C.; Averhoff, B.; Gottschalk, G. Complete nucleotide sequence and genetic organization of the 210-kilobase linear plasmid of Rhodococcus erythropolis BD2. J. Bacteriol. 2003, 185, 5269–5274. [Google Scholar] [CrossRef]
- Kitagawa, W.; Tamura, T. A quinoline antibiotic from Rhodococcus erythropolis JCM 6824. J. Antibiot. 2008, 61, 680–682. [Google Scholar] [CrossRef]
- Kitagawa, W.; Hata, M.; Sekizuka, T.; Kuroda, M.; Ishikawa, J. Draft genome sequence of Rhodococcus erythropolis JCM 6824, an aurachin RE antibiotic producer. Genome Announ. 2014, 2, e01026-14. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, W.; Mitsuhashi, S.; Hata, M.; Tamura, T. Identification of a novel bacteriocin-like protein and structural gene from Rhodococcus erythropolis JCM 2895, using suppression-subtractive hybridization. J. Antibiot. 2018, 71, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001, 205, 197–202. [Google Scholar] [CrossRef]
- Nakashima, N.; Tamura, T. A novel system for expressing recombinant proteins over a wide temperature range from 4 to 35 degrees C. Biotechnol. Bioeng. 2004, 86, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Sallam, K.I.; Mitani, Y.; Tamura, T. Construction of random transposition mutagenesis system in Rhodococcus erythropolis using IS1415. J. Biotechnol. 2006, 121, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Kitagawa, W.; Kumagai, T.; Tajima, N.; Nishimiya, Y.; Tamano, K.; Yasutake, Y.; Tamura, T.; Kameda, T. Developing a codon optimization method for improved expression of recombinant proteins in actinobacteria. Sci. Rep. 2019, 9, 8338. [Google Scholar] [CrossRef]
- Nakashima, N.; Tamura, T. Isolation and characterization of a rolling-circle-type plasmid form Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 2004, 70, 5557–5568. [Google Scholar] [CrossRef] [PubMed]
- Sallam, K.I.; Tamura, N.; Tamura, T. A multipurpose transposon-based vector system mediates protein expression in Rhodococcus erythropolis. Gene 2007, 386, 173–182. [Google Scholar] [CrossRef]
- Shao, Z.; Dick, W.A.; Behki, R.M. An improved Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. using electroporation. Lett. Appl. Microbiol. 1995, 21, 261–266. [Google Scholar] [CrossRef]
- Fukuda, M.; Shimizu, S.; Okita, N.; Seto, M.; Masai, E. Structural alteration of linear plasmids encoding the genes for polychlorinated biphenyl degradation in Rhodococcus strain RHA1. Antonie Van Leeuwenhoek 1998, 74, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Redenbach, M.; Scheel, J.; Schmidt, U. Chromosome topology and genome size of selected actinomycetes species. Antonie Van Leeuwenhoek 2000, 78, 227–235. [Google Scholar] [CrossRef]
- Takai, S.; Sugawara, T.; Watanabe, Y.; Sasaki, Y.; Tsubaki, S.; Sekizaki, T. Effect of growth temperature on maintenance of virulent Rhodococcus equi. Vet. Microbiol. 1994, 39, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Molinatto, G.; Franzil, L.; Steels, S.; Puopolo, G.; Pertot, I.; Ongena, M. Key impact of an uncommon plasmid on Bacillus amyloliquefaciens subsp. plantarum S499 developmental traits and lipopeptide production. Front. Microbiol. 2017, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.K.; Dubey, A.K. Metabolic burden as reflected by maintenance coefficient of recombinant Escherichia coli overexpressing target gene. Biotechnol. Lett. 1995, 17, 1155–1160. [Google Scholar] [CrossRef]
- Jang, Y.J.; Kim, S.A.; Seo, S.O.; Li, L.; Han, N.S. Plasmid curing resulted in improved heterologous gene expression in Leuconostoc citreum EFEL2700. Lett. Appl. Microbiol. 2019, 68, 430–436. [Google Scholar] [CrossRef]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannoni, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, H. Genetic toolkits for engineering Rhodococcus species with versatile applications. Biotechnol. Adv. 2021, 49, 107748. [Google Scholar] [CrossRef]
- Kitagawa, W.; Takami, S.; Miyauchi, K.; Masai, E.; Kamagata, Y.; Tiedje, J.M.; Fukuda, M. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 2002, 184, 509–518. [Google Scholar] [CrossRef]
- Schafer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Puhler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Suzuki, N.; Nonaka, H.; Tsuge, Y.; Inui, M.; Yukawa, H. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl. Environ. Microbiol. 2005, 71, 8472–8480. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Tamura, T. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res. 2009, 37, e103. [Google Scholar] [CrossRef] [PubMed]
- Takai, S.; Sekizaki, T.; Ozawa, T.; Sugawara, T.; Watanabe, Y.; Tsubaki, S. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect. Immun. 1991, 59, 4056–4060. [Google Scholar] [CrossRef]
- Gasson, M.J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1983, 154, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fazlurrahman; Batra, M.; Pandey, J.; Suri, C.R.; Jain, R.K. Isolation and characterization of an atrazine-degrading Rhodococcus sp. strain MB-P1 from contaminated soil. Lett. Appl. Microbiol. 2009, 49, 721–729. [Google Scholar] [CrossRef]
- Coleman, N.V.; Spain, J.C.; Duxbury, T. Evidence that RDX biodegradation by Rhodococcus strain DN22 is plasmid-borne and involves a cytochrome p-450. J. Appl. Microbiol. 2002, 93, 463–472. [Google Scholar] [CrossRef]
- Dodge, A.G.; Wackett, L.P.; Sadowsky, M.J. Plasmid localization and organization of melamine degradation genes in Rhodococcus sp. strain Mel. Appl. Environ. Microbiol. 2012, 78, 1397–1403. [Google Scholar] [CrossRef]
- Kitagawa, W.; Ozaki, T.; Nishioka, T.; Yasutake, Y.; Hata, M.; Nishiyama, M.; Kuzuyama, T.; Tamura, T. Cloning and heterologous expression of the aurachin RE biosynthesis gene cluster afford a new cytochrome P450 for quinoline N-hydroxylation. ChemBioChem 2013, 14, 1085–1093. [Google Scholar] [CrossRef]
- Komatsu, M.; Uchiyama, T.; Omura, S.; Cane, D.E.; Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 2646–2651. [Google Scholar] [CrossRef]
- Ghosh, K.; Van Duyne, G.D. Cre-loxP biochemistry. Methods 2002, 28, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinform. 2008, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Hale, L.; Lazos, O.; Haines, A.; Thomas, C. An efficient stress-free strategy to displace stable bacterial plasmids. Biotechniques 2010, 48, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M.F.; Quandt, J.; O’Connell, M.P.; Pühler, A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene 1989, 78, 111–120. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).