Soil Bacterial Diversity Responds to Long-Term Establishment of Perennial Legumes in Warm-Season Grassland at Two Soil Depths
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Soil Sampling and Analysis
2.3. Bioinformatic and Statistical Analysis
3. Results
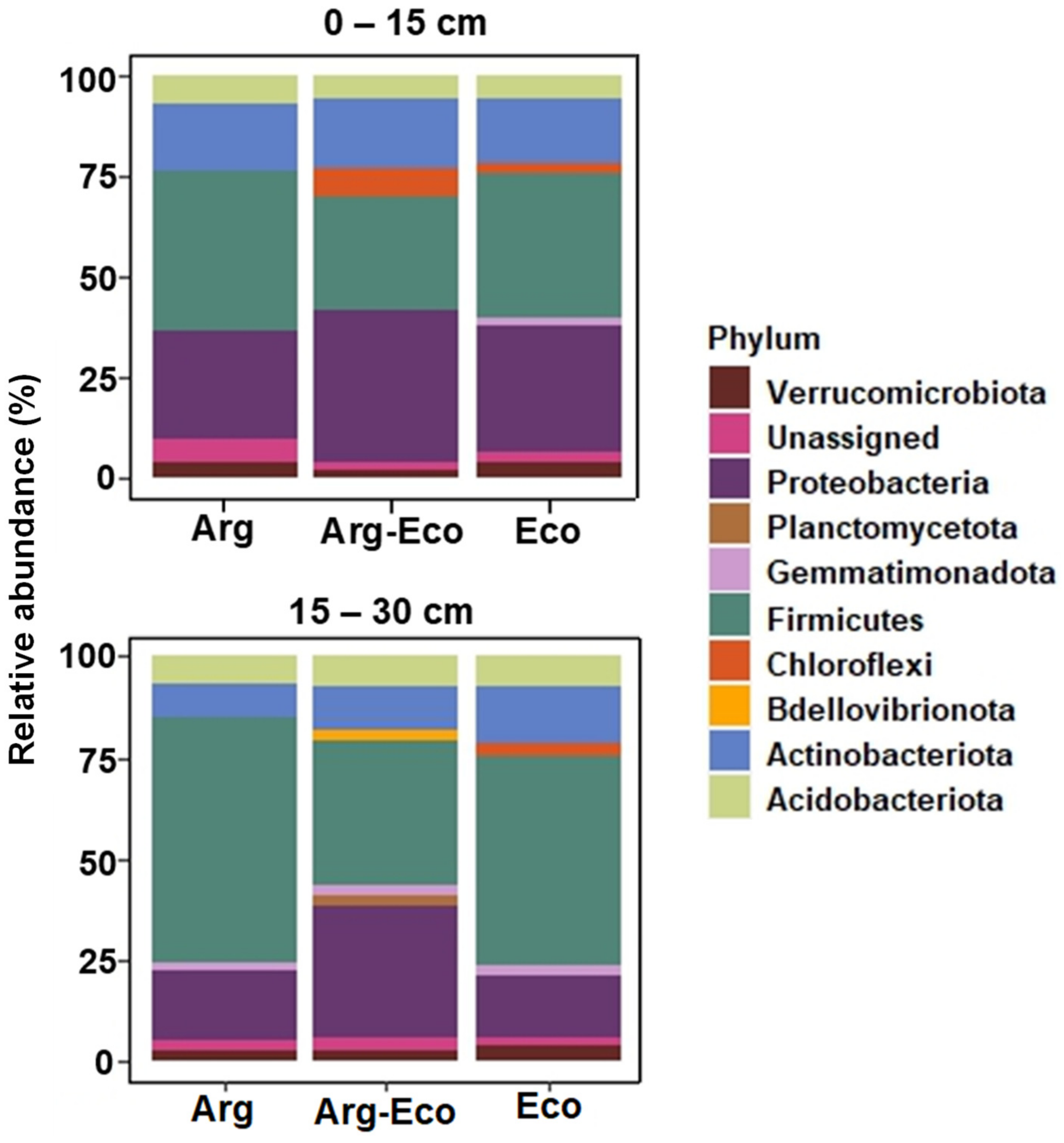
3.1. Soil Bacterial Community Composition
3.2. Soil Bacterial Diversity in Response to Forage Treatment and Soil Depth
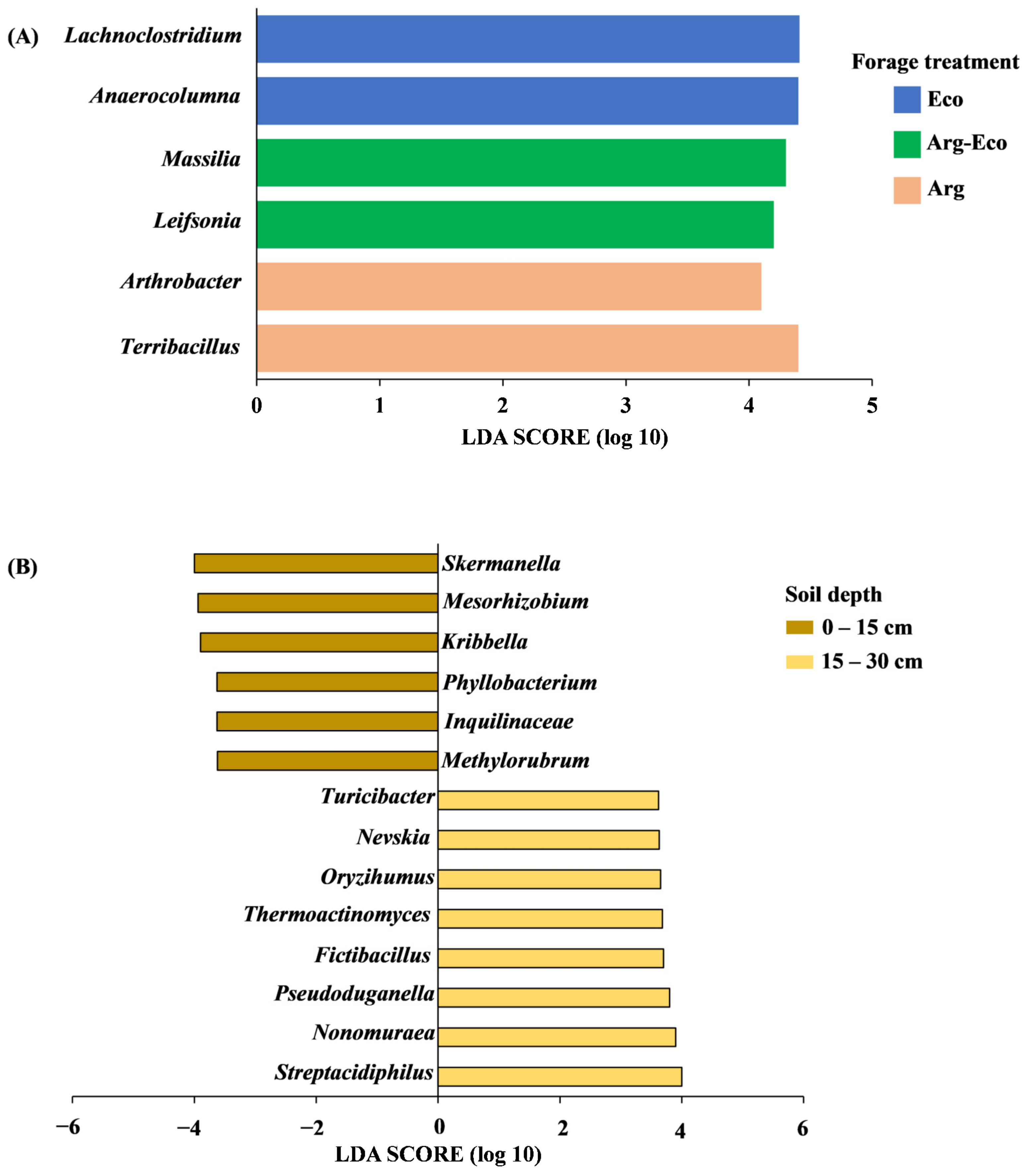
3.3. Core Microbiome and Soil Bacterial Biomarkers
4. Discussion
4.1. Soil Bacterial Diversity and Community Response
4.2. Soil Depth Influences Variation in Soil Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macharia, P.N.; Kinyamario, J.I.; Ekaya, W.N.; Gachene, C.K.K.; Mureithi, J.G.; Thuranira, E.G. Evaluation of Forage Legumes for Introduction into Natural Pastures of Semi-Arid Rangelands of Kenya. Grass Forage Sci. 2010, 65, 456–462. [Google Scholar] [CrossRef]
- Villegas, D.M.; Velasquez, J.; Arango, J.; Obregon, K.; Rao, I.M.; Rosas, G.; Oberson, A. Urochloa Grasses Swap Nitrogen Source When Grown in Association with Legumes in Tropical Pastures. Diversity 2020, 12, 419. [Google Scholar] [CrossRef]
- Sollenberger, L.E.; Dubeux Junior, J.C.B. Warm-Climate, Legume-Grass Forage Mixtures versus Grass-Only Swards: An Ecosystem Services Comparison. Rev. Bras. De Zootec. 2022, 51, e20210198. [Google Scholar] [CrossRef]
- Ortega-S, J.A.; Sollenberger, L.E.; Quesenberry, K.H.; Jones, C.S., Jr.; Cornell, J.A. Productivity and Persistence of Rhizoma Peanut Pastures under Different Grazing Managements. Agron. J. 1992, 84, 799–804. [Google Scholar] [CrossRef]
- Williams, M.J.; Hammond, A.C.; Kunkle, W.E.; Spreen, T.H. Stocker Performance on Continuously Grazed Mixed Grass-Rhizoma Peanut and Bahiagrass Pastures. J. Prod. Agric. 1991, 4, 19–24. [Google Scholar] [CrossRef]
- Santos, E.R.S.; Dubeux, J.C.B., Jr.; Sollenberger, L.E.; Blount, A.R.S.; Mackowiak, C.; DiLorenzo, N.; Jaramillo, D.M.; Garcia, L.; Pereira, T.P.; Ruiz-Moreno, M. Herbage Responses and Biological N2 Fixation of Bahiagrass and Rhizoma Peanut Monocultures Compared with Their Binary Mixtures. Crop Sci. 2018, 58, 2149–2163. [Google Scholar] [CrossRef]
- Jaramillo, D.M.; Dubeux, J.C.B.; Sollenberger, L.E.; Vendramini, J.M.B.; Mackowiak, C.; DiLorenzo, N.; Garcia, L.; Queiroz, L.M.D.; Santos, E.R.S.; Homem, B.G.C.; et al. Water Footprint, Herbage, and Livestock Responses for Nitrogen-fertilized Grass and Grass–legume Grazing Systems. Crop Sci. 2021, 61, 3844–3858. [Google Scholar] [CrossRef]
- Kohmann, M.M.; de Oliveira Bauer, M.; Sollenberger, L.E.; Moreno, L.S.B.; da Silva, L.S.; Saraiva, S.; Dubeux, J.C.B. Legume Proportion Affects Bahiagrass–rhizoma Peanut Mixture Production and Nutritive Value and Legume Composition of Cattle Diets. Appl. Anim. Sci. 2022, 38, 560–569. [Google Scholar] [CrossRef]
- Wang, X.-B.; Hsu, C.-M.; Dubeux, J.C.B., Jr.; Mackowiak, C.; Blount, A.; Han, X.-G.; Liao, H.-L. Effects of Rhizoma Peanut Cultivars (Arachis glabrata Benth.) on the Soil Bacterial Diversity and Predicted Function in Nitrogen Fixation. Ecol. Evol. 2019, 9, 12676–12687. [Google Scholar] [CrossRef]
- Guerra, V.A.; Beule, L.; Mackowiak, C.L.; Dubeux, J.C.B., Jr.; Blount, A.R.S.; Wang, X.-B.; Rowland, D.L.; Liao, H.-L. Soil Bacterial Community Response to Rhizoma Peanut Incorporation into Florida Pastures. J. Environ. Qual. 2022, 51, 55–65. [Google Scholar] [CrossRef]
- Erhunmwunse, A.S.; Queiroz, L.M.D.; Zhang, K.; Mackowiak, C.L.; Blount, A.R.S.; Dubeux, J.C.B.; Liao, H.-L. Changes in Soil Microbial Diversity and Community Composition across Bahiagrass and Rhizoma Peanut Pastures. Biol. Fertil. Soils 2023, 59, 285–300. [Google Scholar] [CrossRef]
- Erhunmwunse, A.S.; Mackowiak, C.L.; Blount, A.R.S.; Dubeux, J.C.B.; Ogram, A.; Liao, H.-L. Short-Term Perennial Peanut Integration into Bahiagrass System Influence on Soil Microbial-Mediated Nitrogen Cycling Activities and Microbial Co-Occurrence Networks. Eur. J. Soil Biol. 2023, 119, 103566. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; van der Putten, W.H.; Wall, D.H. Ecological Linkages between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Kardol, P.; De Deyn, G.B.; Laliberté, E.; Mariotte, P.; Hawkes, C.V. Biotic Plant-Soil Feedbacks across Temporal Scales. J. Ecol. 2013, 101, 309–315. [Google Scholar] [CrossRef]
- Schmid, M.W.; van Moorsel, S.J.; Hahl, T.; De Luca, E.; De Deyn, G.B.; Wagg, C.; Niklaus, P.A.; Schmid, B. Effects of Plant Community History, Soil Legacy and Plant Diversity on Soil Microbial Communities. J. Ecol. 2021, 109, 3007–3023. [Google Scholar] [CrossRef]
- Kulmatiski, A.; Beard, K.H. Long-Term Plant Growth Legacies Overwhelm Short-Term Plant Growth Effects on Soil Microbial Community Structure. Soil Biol. Biochem. 2011, 43, 823–830. [Google Scholar] [CrossRef]
- Hannula, S.E.; Heinen, R.; Huberty, M.; Steinauer, K.; De Long, J.R.; Jongen, R.; Bezemer, T.M. Persistence of Plant-Mediated Microbial Soil Legacy Effects in Soil and inside Roots. Nat. Commun. 2021, 12, 5686. [Google Scholar] [CrossRef]
- Williams, M.J.; Kelly-Begazo, C.A.; Stanley, R.L., Jr.; Quesenberry, K.H.; Prine, G.M. Establishment of Rhizoma Peanut: Interaction of Cultivar, Planting Date, and Location on Emergence and Rate of Cover. Agron. J. 1997, 89, 981–987. [Google Scholar] [CrossRef]
- Castillo, M.S.; Sollenberger, L.E.; Blount, A.R.; Ferrell, J.A.; Williams, M.J.; Mackowiak, C.L. Strip Planting a Legume into Warm-season Grass Pasture: Defoliation Effects during the Year of Establishment. Crop Sci. 2013, 53, 724–731. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in Microbial Community Composition through Two Soil Depth Profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Allison, V.J.; Yermakov, Z.; Miller, R.M.; Jastrow, J.D.; Matamala, R. Using Landscape and Depth Gradients to Decouple the Impact of Correlated Environmental Variables on Soil Microbial Community Composition. Soil Biol. Biochem. 2007, 39, 505–516. [Google Scholar] [CrossRef]
- Hao, J.; Chai, Y.N.; Lopes, L.D.; Ordóñez, R.A.; Wright, E.E.; Archontoulis, S.; Schachtman, D.P. The Effects of Soil Depth on the Structure of Microbial Communities in Agricultural Soils in Iowa (United States). Appl. Environ. Microbiol. 2021, 87, e02673-20. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The Unseen Majority: Soil Microbes as Drivers of Plant Diversity and Productivity in Terrestrial Ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Maron, P.-A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High Microbial Diversity Promotes Soil Ecosystem Functioning. Appl. Environ. Microbiol. 2018, 84, e02738-17. [Google Scholar] [CrossRef] [PubMed]
- Soil Survey Staff, Natural Resources Conservation Service, United States Department of Agriculture. Web Soil Survey. 2007. Available online: https://websoilsurvey.sc.egov.usda.gov/ (accessed on 10 May 2020).
- Florida Automated Weather Network (FAWN). 2020. Available online: https://fawn.ifas.ufl.edu/about.php (accessed on 30 October 2023).
- Santos, E.R.S.; Dubeux, J.C.B., Jr.; Menezes, R.C.; Mackowiak, C.L.; Sollenberger, L.E.; Ruiz-Moreno, M.; Jaramillo, D.M.; Garcia, L.; Queiroz, L.M.D. Particulate Soil Organic Matter in Bahiagrass-Rhizoma Peanut Mixtures and Their Monocultures. Soil Sci. Soc. Am. J. 2019, 83, 658–665. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-H.; Longley, R.; Bonito, G.; Liao, H.-L. A Two-Step PCR Protocol Enabling Flexible Primer Choice and High Sequencing Yield for Illumina MiSeq Meta-Barcoding. Agronomy 2021, 11, 1274. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Anderson, M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R. Vegan: Ecological Diversity R Package, Version 2.5-4. Available online: https://cran.r-project.org/web/packages/vegan/vignettes/diversity-vegan.pdf (accessed on 6 June 2023).
- Anderson, M.J. A New Method for Non-parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Millard, P.; Singh, B.K. Does Grassland Vegetation Drive Soil Microbial Diversity? Nutr. Cycl. Agroecosyst. 2010, 88, 147–158. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; Fu, S.; Yao, Q. Variation in Soil Microbial Community Structure Associated with Different Legume Species Is Greater than That Associated with Different Grass Species. Front. Microbiol. 2017, 8, 1007. [Google Scholar] [CrossRef]
- Philippot, L.; Spor, A.; Hénault, C.; Bru, D.; Bizouard, F.; Jones, C.M.; Sarr, A.; Maron, P.-A. Loss in Microbial Diversity Affects Nitrogen Cycling in Soil. ISME J. 2013, 7, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Girvan, M.S.; Campbell, C.D.; Killham, K.; Prosser, J.I.; Glover, L.A. Bacterial Diversity Promotes Community Stability and Functional Resilience after Perturbation. Environ. Microbiol. 2005, 7, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Tardy, V.; Mathieu, O.; Lévêque, J.; Terrat, S.; Chabbi, A.; Lemanceau, P.; Ranjard, L.; Maron, P.-A. Stability of Soil Microbial Structure and Activity Depends on Microbial Diversity. Environ. Microbiol. Rep. 2014, 6, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Habekost, M.; Eisenhauer, N.; Roscher, C.; Bessler, H.; Engels, C.; Oelmann, Y.; Scheu, S.; Wilcke, W.; Schulze, E.-D.; et al. Biotic and Abiotic Properties Mediating Plant Diversity Effects on Soil Microbial Communities in an Experimental Grassland. PLoS ONE 2014, 9, e96182. [Google Scholar] [CrossRef] [PubMed]
- Dassen, S.; Cortois, R.; Martens, H.; de Hollander, M.; Kowalchuk, G.A.; van der Putten, W.H.; De Deyn, G.B. Differential Responses of Soil Bacteria, Fungi, Archaea and Protists to Plant Species Richness and Plant Functional Group Identity. Mol. Ecol. 2017, 26, 4085–4098. [Google Scholar] [CrossRef]
- Guerra, V.A. Soil Microbial and Ecophysiological Aspects of Rhizoma Peanut Inclusion in Bahiagrass Pastures. Ph.D. Dissertation, University of Florida, Gainesville, FL, USA, 2021. [Google Scholar]
- Santos, E.R.S.; Dubeux, J.C.B., Jr.; Mackowiak, C.; Blount, A.; Sollenberger, L.E.; DiLorenzo, N.; Jaramillo, D.; Garcia, L.; Pereira-Neto, J.D. Root-rhizome Mass and Chemical Composition of Bahiagrass and Rhizoma Peanut Monocultures Compared with Their Binary Mixtures. Crop Sci. 2018, 58, 955–963. [Google Scholar] [CrossRef]
- Daraz, U.; Erhunmwunse, A.S.; Dubeux, J.C.B., Jr.; Mackowiak, C.; Guerra, V.A.; Hsu, C.-M.; Ma, J.; Li, Y.; Yang, X.; Liao, H.-L.; et al. Soil Bacterial Communities Across Seven Rhizoma Peanut Cultivars (Arachis glabrata Benth.) Respond to Seasonal Variation. Microb. Ecol. 2023, 86, 2703–2715. [Google Scholar] [CrossRef]
- Peerawat, M.; Blaud, A.; Trap, J.; Chevallier, T.; Alonso, P.; Gay, F.; Thaler, P.; Spor, A.; Sebag, D.; Choosai, C.; et al. Rubber Plantation Ageing Controls Soil Biodiversity after Land Conversion from Cassava. Agric. Ecosyst. Environ. 2018, 257, 92–102. [Google Scholar] [CrossRef]
- Gonzalez-Pimentel, J.L.; Dominguez-Moñino, I.; Jurado, V.; Laiz, L.; Caldeira, A.T.; Saiz-Jimenez, C. The Rare Actinobacterium Crossiella Sp. Is a Potential Source of New Bioactive Compounds with Activity against Bacteria and Fungi. Microorganisms 2022, 10, 1575. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Guo, H.; Shafiqul Islam, M.; Zaki, H.E.M.; Wang, Z.; Wang, H.; Qi, X.; Guo, J.; Sun, L.; Wang, Q.; et al. Improvement Effect of Biochar on Soil Microbial Community Structure and Metabolites of Decline Disease Bayberry. Front. Microbiol. 2023, 14, 1154886. [Google Scholar] [CrossRef] [PubMed]
- Hellequin, E.; Binet, F.; Klarzynski, O.; Hallin, S.; Juhanson, J.; Daburon, V.; Monard, C. Shaping of Soil Microbial Communities by Plants Does Not Translate into Specific Legacy Effects on Organic Carbon Mineralization. Soil Biol. Biochem. 2021, 163, 108449. [Google Scholar] [CrossRef]
- Warnick, T.A.; Methé, B.A.; Leschine, S.B. Clostridium Phytofermentans Sp. Nov., a Cellulolytic Mesophile from Forest Soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Kadyan, S.; Panghal, M.; Kumar, S.; Singh, K.; Yadav, J.P. Assessment of Functional and Genetic Diversity of Aerobic Endospore Forming Bacilli from Rhizospheric Soil of Phyllanthus amarus L. World J. Microbiol. Biotechnol. 2013, 29, 1597–1610. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Mishra, V.; Rau, N.; Sharma, R.S. Increased Iron-Stress Resilience of Maize through Inoculation of Siderophore-Producing Arthrobacter Globiformis from Mine. J. Basic Microbiol. 2016, 56, 719–735. [Google Scholar] [CrossRef]
- Lugo, M.A.; Ferrero, M.; Menoyo, E.; Estévez, M.C.; Siñeriz, F.; Anton, A. Arbuscular Mycorrhizal Fungi and Rhizospheric Bacteria Diversity along an Altitudinal Gradient in South American Puna Grassland. Microb. Ecol. 2008, 55, 705–713. [Google Scholar] [CrossRef]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging Deeper to Find Unique Microbial Communities: The Strong Effect of Depth on the Structure of Bacterial and Archaeal Communities in Soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Rupela, O.P.; Toomsan, B.; Mittal, S.; Dart, P.J.; Thompson, J.A. Chickpea Rhizobium Populations: Survey of Influence of Season, Soil Depth and Cropping Pattern. Soil Biol. Biochem. 1987, 19, 247–252. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Li, X.; Li, X.; Wang, J.; Zhang, H. Changes of Microbial Population and N-Cycling Function Genes with Depth in Three Chinese Paddy Soils. PLoS ONE 2017, 12, e0189506. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current Knowledge and Perspectives of Paenibacillus: A Review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Kostka, J.E.; Green, S.J.; Rishishwar, L.; Prakash, O.; Katz, L.S.; Mariño-Ramírez, L.; Jordan, I.K.; Munk, C.; Ivanova, N.; Mikhailova, N.; et al. Genome Sequences for Six Rhodanobacter Strains, Isolated from Soils and the Terrestrial Subsurface, with Variable Denitrification Capabilities. J. Bacteriol. 2012, 194, 4461–4462. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, W.; Zhou, D.; Jing, T.; Li, K.; Zhao, Y.; Tang, W.; Qi, D.; Zhang, M.; Zang, X.; et al. Biodegradation of Lignocellulosic Agricultural Residues by a Newly Isolated Fictibacillus Sp. YS-26 Improving Carbon Metabolic Properties and Functional Diversity of the Rhizosphere Microbial Community. Bioresour. Technol. 2020, 310, 123381. [Google Scholar] [CrossRef]
- Sungthong, R.; Nakaew, N. The Genus Nonomuraea: A Review of a Rare Actinomycete Taxon for Novel Metabolites. J. Basic Microbiol. 2015, 55, 554–565. [Google Scholar] [CrossRef]
| Observed Features | Shannon Diversity | |
|---|---|---|
| Forage treatment (FT) | ||
| Arg | 353 c | 7.3 c |
| Arg-Eco | 481 b | 7.8 b |
| Eco | 508 a | 8.1 a |
| Soil depth (D) | ||
| 0 to 15 | 486 A | 8.0 A |
| 15 to 30 | 408 B | 7.5 B |
| Source of variation | p values 1 | |
| FT | 0.024 * | 0.046 * |
| D | 0.047 * | 0.049 * |
| FT × D | 0.439 | 0.643 |
| Source of Variation | Df | MS | Pseudo F | R2 | p |
|---|---|---|---|---|---|
| FT | 2 | 0.271 | 1.822 | 0.14 | 0.024 * |
| D | 1 | 1.097 | 7.362 | 0.28 | <0.001 *** |
| FT × D | 2 | 0.244 | 1.402 | 0.12 | 0.047 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erhunmwunse, A.S.; Guerra, V.A.; Liu, J.-C.; Mackowiak, C.L.; Blount, A.R.S.; Dubeux, J.C.B., Jr.; Liao, H.-L. Soil Bacterial Diversity Responds to Long-Term Establishment of Perennial Legumes in Warm-Season Grassland at Two Soil Depths. Microorganisms 2023, 11, 3002. https://doi.org/10.3390/microorganisms11123002
Erhunmwunse AS, Guerra VA, Liu J-C, Mackowiak CL, Blount ARS, Dubeux JCB Jr., Liao H-L. Soil Bacterial Diversity Responds to Long-Term Establishment of Perennial Legumes in Warm-Season Grassland at Two Soil Depths. Microorganisms. 2023; 11(12):3002. https://doi.org/10.3390/microorganisms11123002
Chicago/Turabian StyleErhunmwunse, Adesuwa Sylvia, Victor Alonso Guerra, Jung-Chen Liu, Cheryl L. Mackowiak, Ann Rachel Soffes Blount, José Carlos Batista Dubeux, Jr., and Hui-Ling Liao. 2023. "Soil Bacterial Diversity Responds to Long-Term Establishment of Perennial Legumes in Warm-Season Grassland at Two Soil Depths" Microorganisms 11, no. 12: 3002. https://doi.org/10.3390/microorganisms11123002
APA StyleErhunmwunse, A. S., Guerra, V. A., Liu, J.-C., Mackowiak, C. L., Blount, A. R. S., Dubeux, J. C. B., Jr., & Liao, H.-L. (2023). Soil Bacterial Diversity Responds to Long-Term Establishment of Perennial Legumes in Warm-Season Grassland at Two Soil Depths. Microorganisms, 11(12), 3002. https://doi.org/10.3390/microorganisms11123002







