Dynamic Interactions between Diarrhoeagenic Enteroaggregative Escherichia coli and Presumptive Probiotic Bacteria: Implications for Gastrointestinal Health
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Cell Culturing and Maintenance Conditions
2.3. Preparation of Epithelial Cells for Inflammation Assays
2.4. The Effects of EAEC and LAB on Epithelial Barrier Integrity
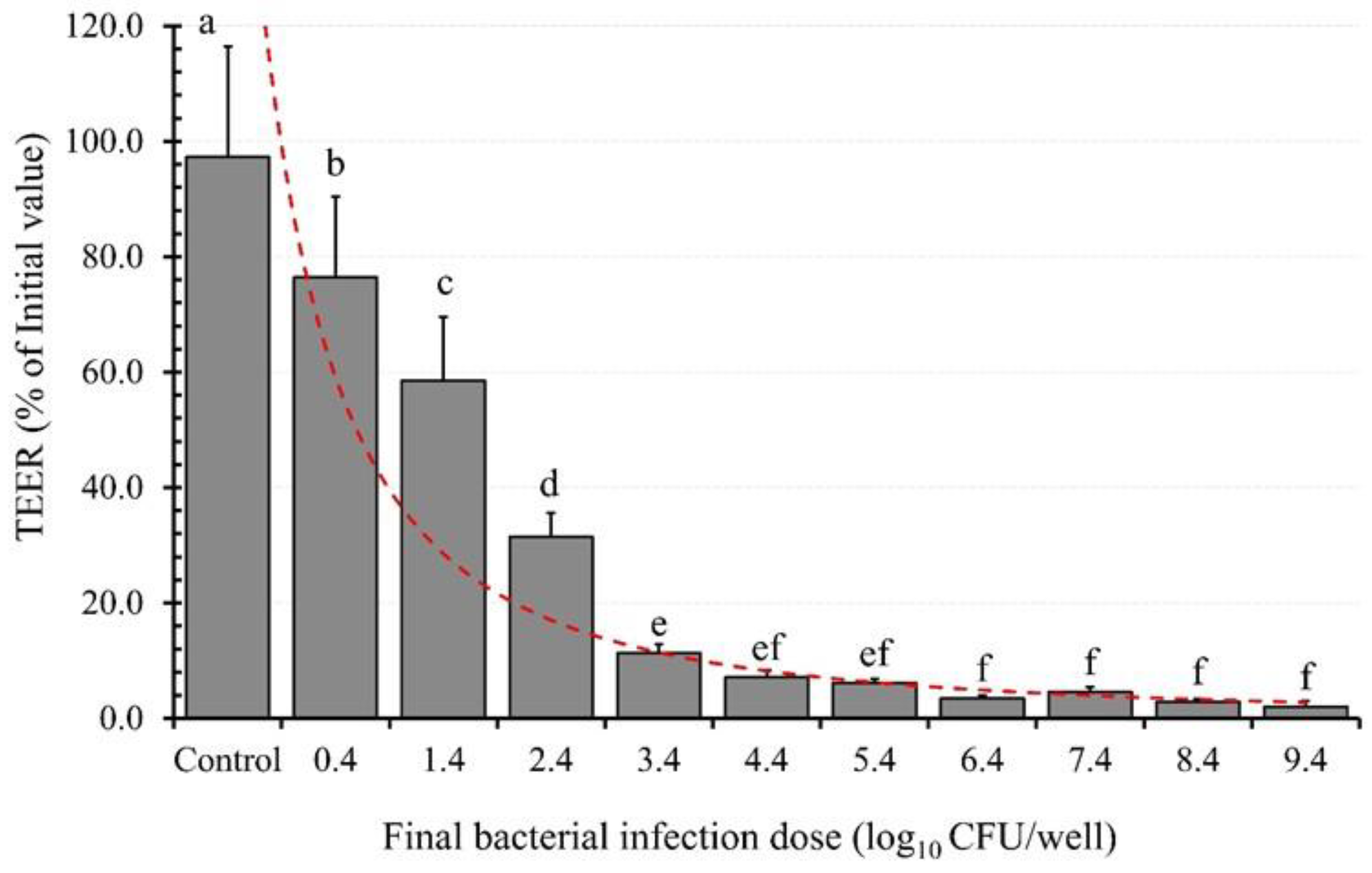
2.5. Effect of the Bacterial Infection Dose (BID) on Epithelial Barrier Integrity
2.6. Bacterial Infection Mode and Treatment Time (TT) Effects on Epithelial Barrier Integrity
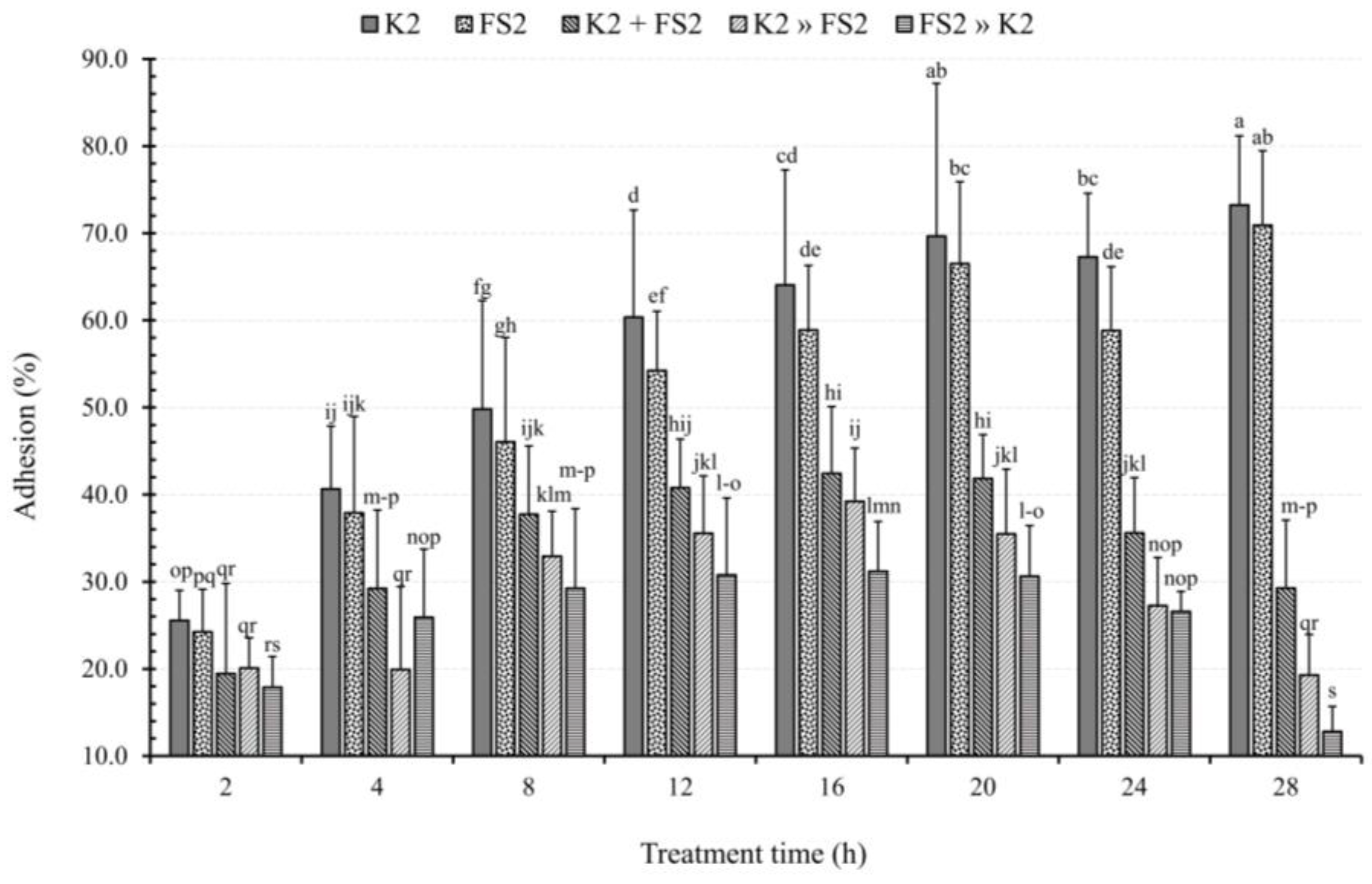
2.7. Bacterial Mode of Infection and TT Effects on Adhesion
2.8. Interleukin 8 (IL-8) Assay
2.9. Trans-Epithelial Electrical Resistance (TEER) Assay
2.10. Statistical Analysis
3. Results and Discussion
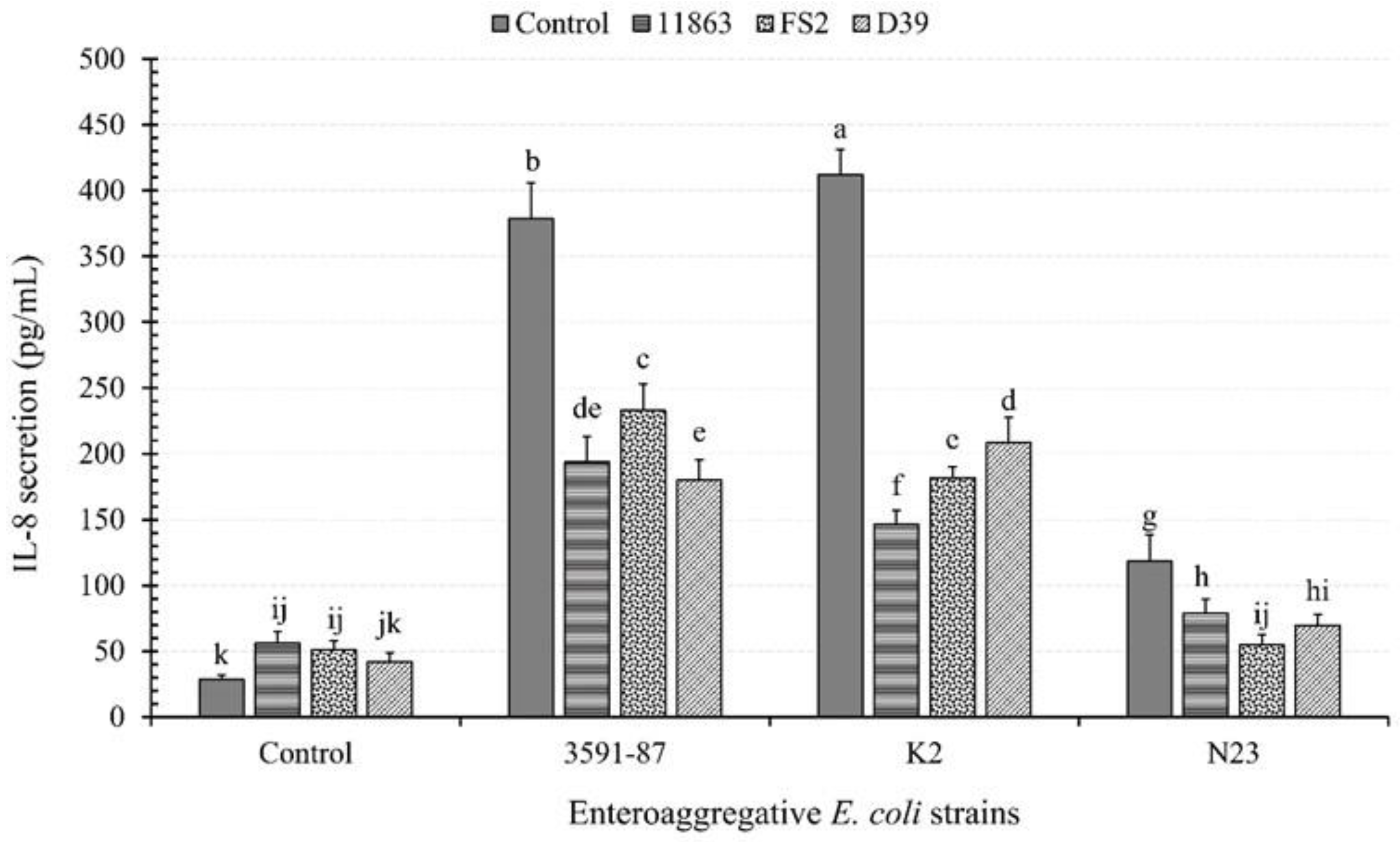
3.1. Cytokine Secretion from Caco-2 Monolayers in the Presence or Absence of EAEC and LAB
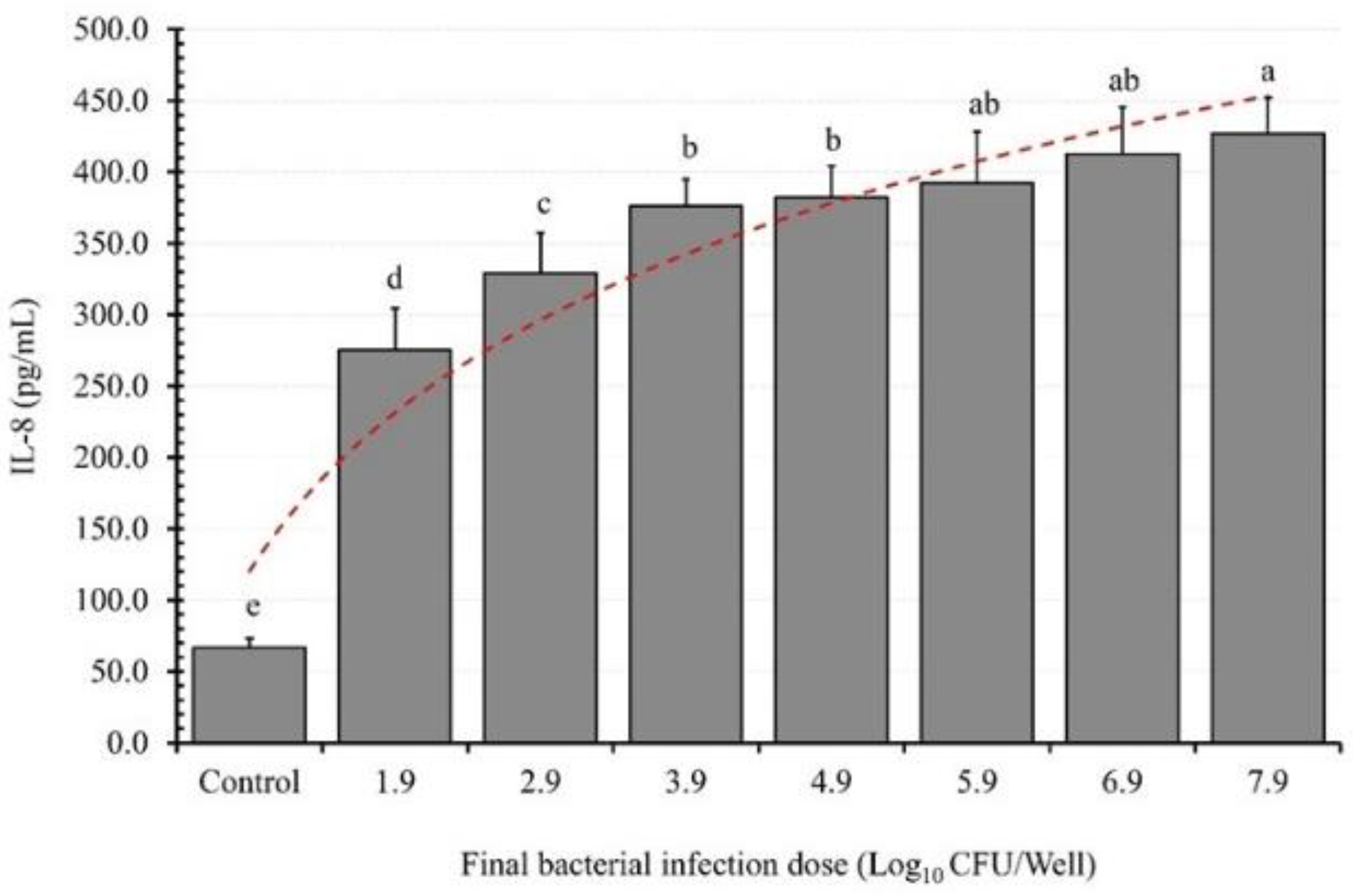
3.2. BID Effect on IL-8 Induction
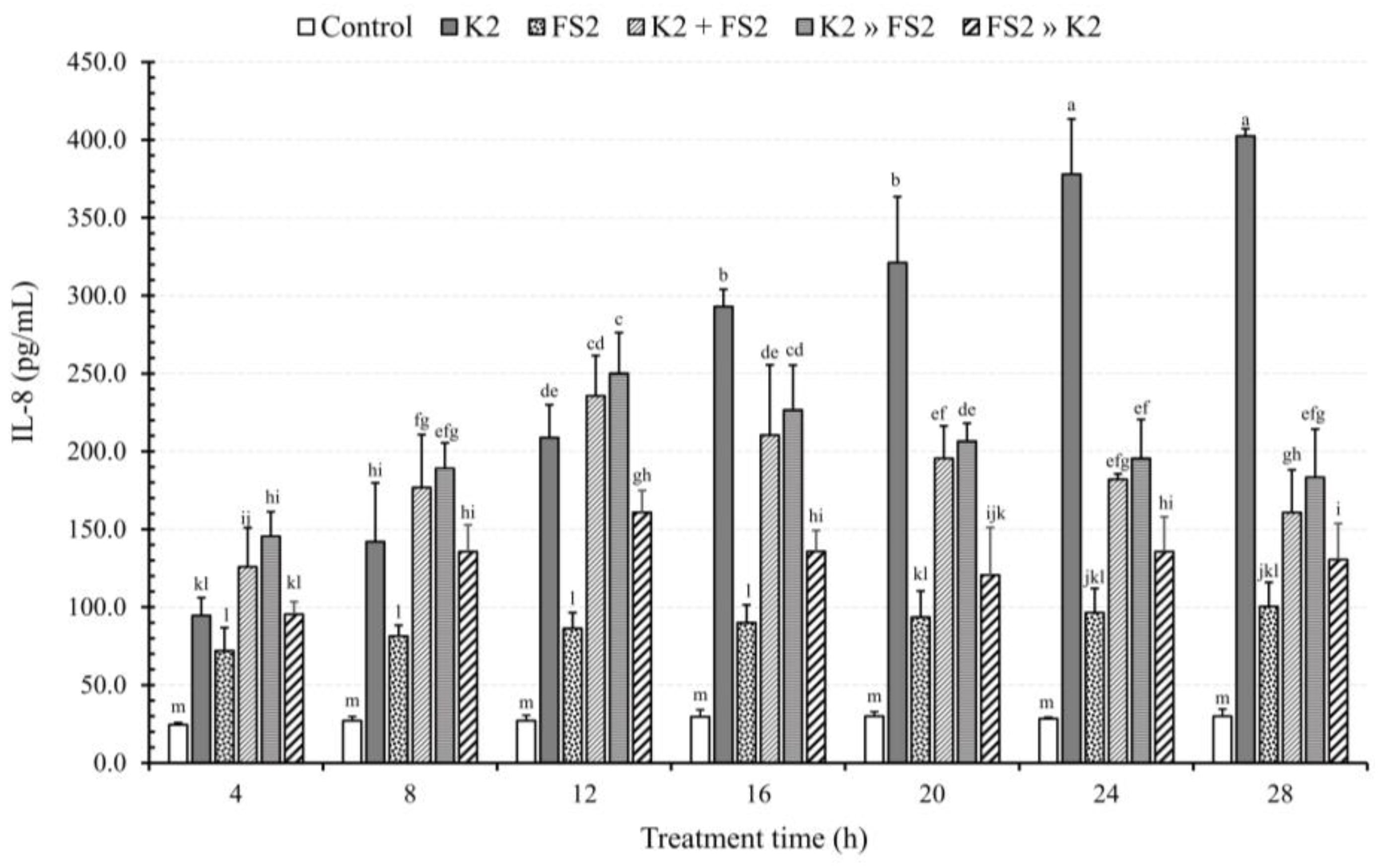
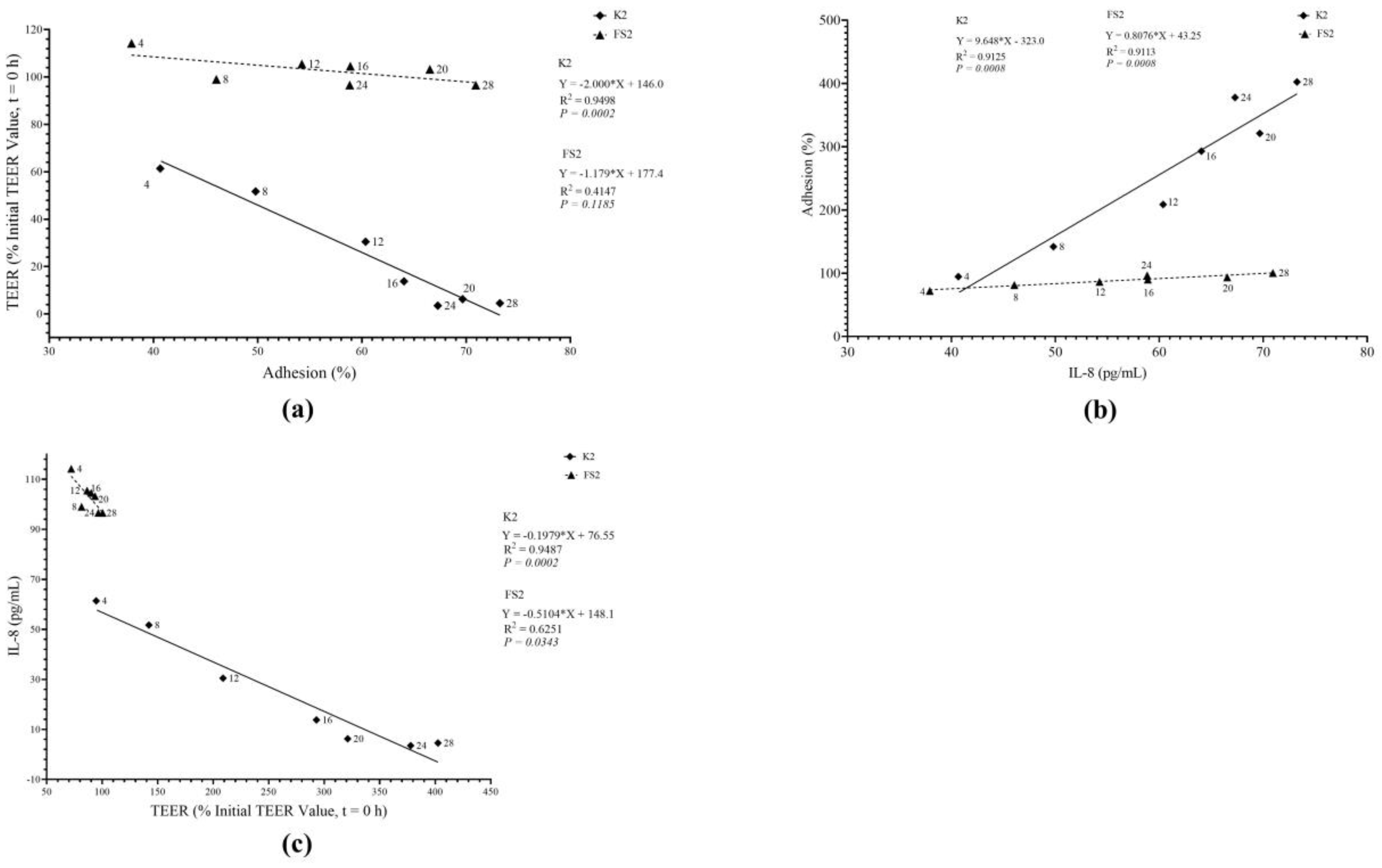
3.3. Bacterial Infection Mode and TT Effects on IL-8 Secretion
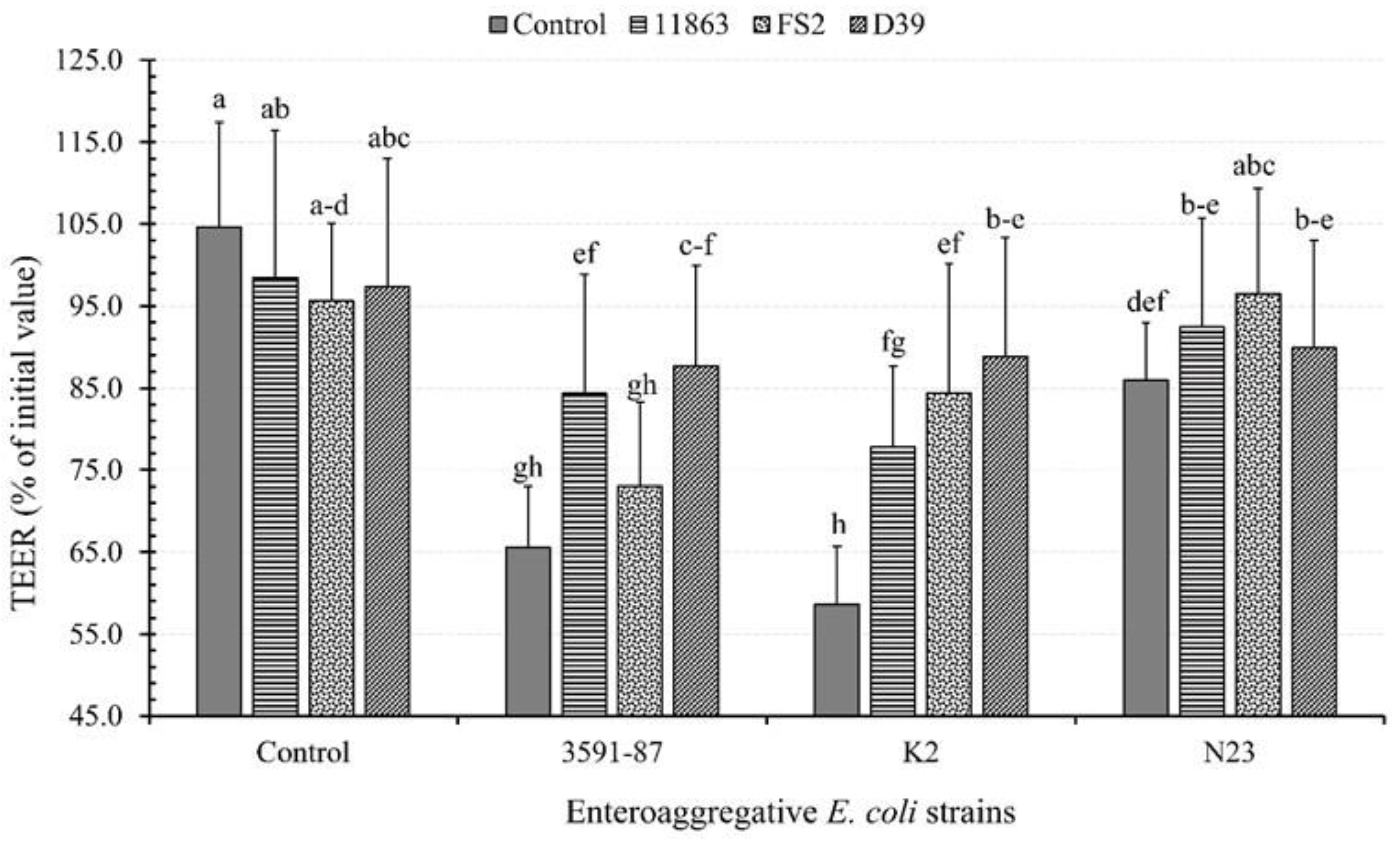
3.4. Effects of Bacterial Monoinfection on TEER
3.5. Effects of LAB and EAEC Coinfection on TEER
3.6. BID Effect on TEER
3.7. Bacterial Mode of Infection and TT Effects on TEER
3.8. Bacterial Mode of Infection and TT Effects on TEER
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajoka, M.S.R.; Shi, J.; Mehwish, H.M.; Zhu, J.; Li, Q.; Shao, D.; Huang, Q.; Yang, H. Interaction between diet composition and gut microbiota and its impact on gastrointestinal tract health. Food Sci. Hum. Wellness 2017, 6, 121–130. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef] [PubMed]
- Ikumapayi, U.N.; Boisen, N.; Hossain, M.J.; Betts, M.; Lamin, M.; Saha, D.; Kwambana-Adams, B.; Dione, M.; Adegbola, R.A.; Roca, A. Identification of subsets of enteroaggregative Escherichia coli associated with diarrheal disease among under 5 years of age children from Rural Gambia. Am. J. Trop. Med. Hyg. 2017, 97, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Sosa, L.; Ochoa, T.J. Escherichia coli diarrhea. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 481–485. [Google Scholar]
- Aijuka, M.; Buys, E.M. Persistence of foodborne diarrheagenic Escherichia coli in the agricultural and food production environment: Implications for food safety and public health. Food Microbiol. 2019, 82, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Pakbin, B.; Bruck, W.M.; Rossen, J.W.A. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. Int. J. Mol. Sci. 2021, 22, 9922. [Google Scholar] [CrossRef]
- Modgil, V.; Mahindroo, J.; Narayan, C.; Kalia, M.; Yousuf, M.; Shahi, V.; Koundal, M.; Chaudhary, P.; Jain, R.; Sandha, K.S.; et al. Comparative analysis of virulence determinants, phylogroups, and antibiotic susceptibility patterns of typical versus atypical Enteroaggregative E. coli in India. PLoS Negl. Trop. Dis. 2020, 14, e0008769. [Google Scholar] [CrossRef]
- Miliwebsky, E.; Jure, M.Á.; Farfan, M.J.; Palermo, M.S. Interactions of Pathogenic Escherichia coli with Gut Microbiota. In Trending Topics in Escherichia coli Research: The Latin American Perspective; Springer: Berlin/Heidelberg, Germany, 2023; pp. 277–294. [Google Scholar]
- Woodward, S.E.; Krekhno, Z.; Finlay, B.B. Here, there, and everywhere: How pathogenic Escherichia coli sense and respond to gastrointestinal biogeography. Cell. Microbiol. 2019, 21, e13107. [Google Scholar] [CrossRef]
- Ellis, S.J.; Crossman, L.C.; McGrath, C.J.; Chattaway, M.A.; Holken, J.M.; Brett, B.; Bundy, L.; Kay, G.L.; Wain, J.; Schuller, S. Identification and characterisation of enteroaggregative Escherichia coli subtypes associated with human disease. Sci. Rep. 2020, 10, 7475. [Google Scholar] [CrossRef]
- Dias, R.C.B.; Tanabe, R.H.S.; Vieira, M.A.; Cergole-Novella, M.C.; Dos Santos, L.F.; Gomes, T.A.T.; Elias, W.P.; Hernandes, R.T. Analysis of the Virulence Profile and Phenotypic Features of Typical and Atypical enteroaggregative Escherichia coli (EAEC) Isolated From Diarrheal Patients in Brazil. Front. Cell. Infect. Microbiol. 2020, 10, 144. [Google Scholar] [CrossRef]
- Govindarajan, D.K.; Viswalingam, N.; Meganathan, Y.; Kandaswamy, K. Adherence patterns of Escherichia coli in the intestine and its role in pathogenesis. Med. Microecol. 2020, 5, 100025. [Google Scholar] [CrossRef]
- Sauvaitre, T.; Van Landuyt, J.; Durif, C.; Roussel, C.; Sivignon, A.; Chalancon, S.; Uriot, O.; Van Herreweghen, F.; Van de Wiele, T.; Etienne-Mesmin, L. Role of mucus-bacteria interactions in Enterotoxigenic Escherichia coli (ETEC) H10407 virulence and interplay with human microbiome. Npj Biofilms Microbiomes 2022, 8, 86. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Talavera Rodríguez, A.; Roy, S.; Hossain, M.I.; Islam, M.A.; Lanza, V.F.; Julian, T.R. High genomic diversity and heterogenous origins of pathogenic and antibiotic-resistant Escherichia coli in household settings represent a challenge to reducing transmission in low-income settings. Msphere 2020, 5, 00704–00719. [Google Scholar] [CrossRef] [PubMed]
- Geurtsen, J.; de Been, M.; Weerdenburg, E.; Zomer, A.; McNally, A.; Poolman, J. Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol. Rev. 2022, 46, fuac031. [Google Scholar] [CrossRef] [PubMed]
- Modgil, V.; Narayan, C.; Kaur, H.; Yadav, V.K.; Chaudhary, N.; Kant, V.; Mohan, B.; Bhatia, A.; Taneja, N. Analysis of the virulence and inflammatory markers elicited by Enteroaggregative Escherichia coli isolated from clinical and non-clinical sources in an experimental infection model, India. Microbiol. Res. 2022, 13, 882–897. [Google Scholar] [CrossRef]
- Moran-Garcia, N.; Lopez-Saucedo, C.; Becerra, A.; Meza-Segura, M.; Hernandez-Cazares, F.; Guerrero-Baez, J.; Galindo-Gomez, S.; Tsutsumi, V.; Schnoor, M.; Mendez-Tenorio, A.; et al. A Novel Adult Murine Model of Typical Enteroaggregative Escherichia coli Infection Reveals Microbiota Dysbiosis, Mucus Secretion, and AAF/II-Mediated Expression and Localization of beta-Catenin and Expression of MUC1 in Ileum. Front. Cell. Infect. Microbiol. 2022, 12, 885191. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Renouf, M.J.; Omotoso, O.; McPhee, J.B. Inflammatory bowel disease-associated adherent-invasive Escherichia coli have elevated host-defense peptide resistance. FEMS Microbiol. Lett. 2022, 369, fnac098. [Google Scholar] [CrossRef] [PubMed]
- Denney, L.; Ho, L.P. The role of respiratory epithelium in host defence against influenza virus infection. Biomed. J. 2018, 41, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.M.; Bamashmous, S.; Darveau, R.P.; Rajapakse, S. An Ayurvedic herbal extract inhibits oral epithelial cell IL-8 responses to host and bacterial agonists. BMC Complement. Med. Ther. 2020, 20, 62. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, G.; Wang, Y.; He, M.; Xu, Q.; Lu, J.; Liu, H.; Xu, C. Tumour-associated neutrophils orchestrate intratumoural IL-8-driven immune evasion through Jagged2 activation in ovarian cancer. Br. J. Cancer 2020, 123, 1404–1416. [Google Scholar] [CrossRef]
- Huang, F.C. The Interleukins Orchestrate Mucosal Immune Responses to Salmonella Infection in the Intestine. Cells 2021, 10, 3492. [Google Scholar] [CrossRef]
- Izquierdo, M.; Lopez, J.; Gallardo, P.; Vidal, R.M.; Ossa, J.C.; Farfan, M.J. Bacteria from gut microbiota associated with diarrheal infections in children promote virulence of Shiga toxin-producing and enteroaggregative Escherichia coli pathotypes. Front. Cell. Infect. Microbiol. 2022, 12, 867205. [Google Scholar] [CrossRef] [PubMed]
- Joon, A.; Chandel, S.; Ghosh, S. Enteroaggregative Escherichia coli induced activation of epidermal growth factor receptor contributes to IL-8 secretion by cultured human intestinal epithelial cells. Microbes Infect. 2023, 25, 105166. [Google Scholar] [CrossRef] [PubMed]
- Chandel, S.; Joon, A.; Kaur, S.; Ghosh, S. Role of ST6GAL1 and ST6GAL2 in subversion of cellular signaling during enteroaggregative Escherichia coli infection of human intestinal epithelial cell lines. Appl. Microbiol. Biotechnol. 2023, 107, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Heng, X.; Guo, L.; Lessing, D.J.; Chu, W. SCFAs improve disease resistance via modulate gut microbiota, enhance immune response and increase antioxidative capacity in the host. Fish. Shellfish. Immunol. 2022, 120, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-C.; Lu, Y.-T.; Liao, Y.-H. Beneficial effect of probiotics on Pseudomonas aeruginosa–infected intestinal epithelial cells through inflammatory IL-8 and antimicrobial peptide human beta-defensin-2 modulation. Innate Immun. 2020, 26, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Burgueño, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, L.S.; Graef, F.A.; Healey, G.R.; Bosman, E.S.; Jacobson, K.; Sly, L.M.; Vallance, B.A. Inflammatory bowel disease and immunonutrition: Novel therapeutic approaches through modulation of diet and the gut microbiome. Immunology 2018, 155, 36–52. [Google Scholar] [CrossRef]
- Aijuka, M.; Santiago, A.E.; Giron, J.A.; Nataro, J.P.; Buys, E.M. Enteroaggregative Escherichia coli is the predominant diarrheagenic E. coli pathotype among irrigation water and food sources in South Africa. Int. J. Food Microbiol. 2018, 278, 44–51. [Google Scholar] [CrossRef]
- Ntuli, V.; Njage, P.M.; Buys, E.M. Extended-spectrum β-lactamase, shigatoxin and haemolysis capacity of O157 and non-O157 E. coli serotypes from producer-distributor bulk milk. Int. Dairy J. 2017, 66, 126–134. [Google Scholar] [CrossRef]
- Fayemi, O.E.; Buys, E.M. Effect of Lactobacillus plantarum on the survival of acid-tolerant non-O157 Shiga toxin-producing E. coli (STEC) strains in fermented goat’s milk. Int. J. Dairy Technol. 2017, 70, 399–406. [Google Scholar] [CrossRef]
- Agbemavor, W.S.K.; Buys, E.M. Presumptive probiotic bacteria from traditionally fermented African food challenge the adhesion of enteroaggregative E. coli. J. Food Saf. 2021, 41, e12905. [Google Scholar] [CrossRef]
- Aijuka, M.; Santiago, A.E.; Girón, J.A.; Nataro, J.P.; Buys, E.M. Escherichia coli isolated from food sources and irrigation water: A potential risk for causing intestinal dysfunction? Food Control 2019, 102, 139–148. [Google Scholar] [CrossRef]
- Lodemann, U.; Strahlendorf, J.; Schierack, P.; Klingspor, S.; Aschenbach, J.R.; Martens, H. Effects of the probiotic Enterococcus faecium and pathogenic Escherichia coli strains in a pig and human epithelial intestinal cell model. Scientifica 2015, 2015, 235184. [Google Scholar] [CrossRef] [PubMed]
- Statpoint Technologies INC Statgraphics Centurion XIX, software version 19.1.1; Statpoint Technologies, Inc.: Warrenton, VA, USA, 2020.
- Cai, J.; Culley, M.K.; Zhao, Y.; Zhao, J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell. 2018, 9, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Díaz, C.; Varela-Trinidad, G.U.; Muñoz-Sánchez, G.; Solórzano-Castanedo, K.; Avila-Arrezola, K.E.; Iñiguez-Gutiérrez, L.; Delgado-Rizo, V.; Fafutis-Morris, M. To Trap a Pathogen: Neutrophil Extracellular Traps and Their Role in Mucosal Epithelial and Skin Diseases. Cells 2021, 10, 1469. [Google Scholar] [CrossRef] [PubMed]
- Soo, K.; Tham, C.; Khalid, B.; Basir, R.; Chee, H. IL-8 as a potential in-vitro severity biomarker for dengue. Trop. Biomed. 2019, 36, 1027–1037. [Google Scholar]
- Garcia-Gonzalez, N.; Prete, R.; Battista, N.; Corsetti, A. Adhesion Properties of Food-Associated Lactobacillus plantarum Strains on Human Intestinal Epithelial Cells and Modulation of IL-8 Release. Front. Microbiol. 2018, 9, 2392. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Bai, Y.; Zha, L.; Ullah, N.; Ullah, H.; Shah, S.R.H.; Sun, H.; Zhang, C. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front. Cell. Infect. Microbiol. 2021, 11, 716299. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Tesfay, S.; Tomson, F.L.; Kanteti, R.P.; Viswanathan, V.K.; Hecht, G. Balance of bacterial pro- and anti-inflammatory mediators dictates net effect of enteropathogenic Escherichia coli on intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G685–G694. [Google Scholar] [CrossRef]
- Yang, J.S.; Jeon, J.H.; Jang, M.S.; Kang, S.S.; Ahn, K.B.; Song, M.; Yun, C.H.; Han, S.H. Vibrio cholerae OmpU induces IL-8 expression in human intestinal epithelial cells. Mol. Immunol. 2018, 93, 47–54. [Google Scholar] [CrossRef]
- Grab, D.J.; Nyarko, E.; Barat, N.C.; Nikolskaia, O.V.; Dumler, J.S. Anaplasma phagocytophilum-Borrelia burgdorferi coinfection enhances chemokine, cytokine, and matrix metalloprotease expression by human brain microvascular endothelial cells. Clin. Vaccine Immunol. 2007, 14, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Guan, N.; Du, Y.; Zhang, X.; Li, J.; Xia, X. Cronobacter sakazakii ATCC 29544 Translocated Human Brain Microvascular Endothelial Cells via Endocytosis, Apoptosis Induction, and Disruption of Tight Junction. Front. Microbiol. 2021, 12, 675020. [Google Scholar] [CrossRef] [PubMed]
- Lehtoranta, L.; Latvala, S.; Lehtinen, M.J. Role of Probiotics in Stimulating the Immune System in Viral Respiratory Tract Infections: A Narrative Review. Nutrients 2020, 12, 3163. [Google Scholar] [CrossRef] [PubMed]
- Dewi, B.E.; Takasaki, T.; Kurane, I. In vitro assessment of human endothelial cell permeability: Effects of inflammatory cytokines and dengue virus infection. J. Virol. Methods 2004, 121, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Schavemaker, F.; Kosim, K.; Kurek, D.; Haarmans, M.; Bulst, M.; Lee, K.; Wegner, S.; Hankemeier, T.; Joore, J.; et al. High throughput transepithelial electrical resistance (TEER) measurements on perfused membrane-free epithelia. Lab Chip 2021, 21, 1676–1685. [Google Scholar] [CrossRef]
- Yang, Y.; Latorre, J.D.; Khatri, B.; Kwon, Y.M.; Kong, B.W.; Teague, K.D.; Graham, L.E.; Wolfenden, A.D.; Mahaffey, B.D.; Baxter, M.; et al. Characterization and evaluation of lactic acid bacteria candidates for intestinal epithelial permeability and Salmonella typhimurium colonization in neonatal turkey poults. Poult. Sci. 2018, 97, 724. [Google Scholar] [CrossRef] [PubMed]
- Cosme-Silva, L.; Dal-Fabbro, R.; Cintra, L.T.A.; Dos Santos, V.R.; Duque, C.; Ervolino, E.; Mogami Bomfim, S.; Gomes-Filho, J.E. Systemic administration of probiotics reduces the severity of apical periodontitis. Int. Endod. J. 2019, 52, 1738–1749. [Google Scholar] [CrossRef]
- Kazun, B.; Malaczewska, J.; Kazun, K.; Kaminski, R.; Adamek-Urbanska, D.; Zylinska-Urban, J. Dietary administration of beta-1,3/1,6-glucan and Lactobacillus plantarum improves innate immune response and increases the number of intestine immune cells in roach (Rutilus rutilus). BMC Vet. Res. 2020, 16, 216. [Google Scholar] [CrossRef]
- Han, S.-K.; Shin, Y.-J.; Lee, D.-Y.; Kim, K.M.; Yang, S.-J.; Kim, D.S.; Choi, J.-W.; Lee, S.; Kim, D.-H. Lactobacillus rhamnosus HDB1258 modulates gut microbiota-mediated immune response in mice with or without lipopolysaccharide-induced systemic inflammation. BMC Microbiol. 2021, 21, 146. [Google Scholar] [CrossRef]
- Delgado-Diaz, D.J.; Jesaveluk, B.; Hayward, J.A.; Tyssen, D.; Alisoltani, A.; Potgieter, M.; Bell, L.; Ross, E.; Iranzadeh, A.; Allali, I.; et al. Lactic acid from vaginal microbiota enhances cervicovaginal epithelial barrier integrity by promoting tight junction protein expression. Microbiome 2022, 10, 141. [Google Scholar] [CrossRef]
- Wang, C.; Cui, Y.; Qu, X. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch. Microbiol. 2018, 200, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; van der Mei, H.C.; Busscher, H.J.; Peterson, B.W. Two-Stage Interpretation of Changes in TEER of Intestinal Epithelial Layers Protected by Adhering Bifidobacteria During E. coli Challenges. Front. Microbiol. 2020, 11, 599555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Zhang, C.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Strain-specific effects of Akkermansia muciniphila on the regulation of intestinal barrier. Food Sci. Hum. Wellness 2023, 12, 1526–1537. [Google Scholar] [CrossRef]
- Brunner, J.; Ragupathy, S.; Borchard, G. Target specific tight junction modulators. Adv. Drug. Deliv. Rev. 2021, 171, 266–288. [Google Scholar] [CrossRef] [PubMed]
- Uotani, T.; Murakami, K.; Uchida, T.; Tanaka, S.; Nagashima, H.; Zeng, X.L.; Akada, J.; Estes, M.K.; Graham, D.Y.; Yamaoka, Y. Changes of tight junction and interleukin-8 expression using a human gastroid monolayer model of Helicobacter pylori infection. Helicobacter 2019, 24, e12583. [Google Scholar] [CrossRef] [PubMed]
- Gvoic, M.; Vukmirovic, S.; Al-Salami, H.; Mooranian, A.; Mikov, M.; Stankov, K. Bile acids as novel enhancers of CNS targeting antitumor drugs: A comprehensive review. Pharm. Dev. Technol. 2021, 26, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Beutel, O.; Maraspini, R.; Pombo-Garcia, K.; Martin-Lemaitre, C.; Honigmann, A. Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 2019, 179, 923–936.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Lo, C.; Zhuang, J.; Angsantikul, P.; Zhang, Q.; Wei, X.; Zhou, Z.; Obonyo, M.; Fang, R.H.; et al. Inhibition of Pathogen Adhesion by Bacterial Outer Membrane-Coated Nanoparticles. Angew. Chem. Int. Ed. Engl. 2019, 58, 11404–11408. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Bamford, C.G. SARS-CoV-2, bacterial co-infections, and AMR: The deadly trio in COVID-19? EMBO Mol. Med. 2020, 12, e12560. [Google Scholar] [CrossRef]
- Shigetomi, K.; Ikenouchi, J. Cell Adhesion Structures in Epithelial Cells Are Formed in Dynamic and Cooperative Ways. Bioessays 2019, 41, e1800227. [Google Scholar] [CrossRef]
- Lepine, A.F.P.; de Wit, N.; Oosterink, E.; Wichers, H.; Mes, J.; de Vos, P. Lactobacillus acidophilus Attenuates Salmonella-Induced Stress of Epithelial Cells by Modulating Tight-Junction Genes and Cytokine Responses. Front. Microbiol. 2018, 9, 1439. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, X.; Xu, Z. Identification of antibacterial substances of Lactobacillus plantarum DY-6 for bacteriostatic action. Food Sci. Nutr. 2020, 8, 2854–2863. [Google Scholar] [CrossRef] [PubMed]
- Vareille-Delarbre, M.; Miquel, S.; Garcin, S.; Bertran, T.; Balestrino, D.; Evrard, B.; Forestier, C. Immunomodulatory Effects of Lactobacillus plantarum on Inflammatory Response Induced by Klebsiella pneumoniae. Infect. Immun. 2019, 87, e00570-19. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lee, A.; Huang, S.; Gao, J.; Spence, J.R.; Owyang, C. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes 2019, 10, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ji, H.; Wang, S.; Liu, H.; Zhang, W.; Zhang, D.; Wang, Y. Probiotic Lactobacillus plantarum Promotes Intestinal Barrier Function by Strengthening the Epithelium and Modulating Gut Microbiota. Front. Microbiol. 2018, 9, 1953. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jiang, Y.; Xu, K.; Cui, M.; Ye, W.; Zhao, G.; Jin, L.; Chen, X. The progress of gut microbiome research related to brain disorders. J. Neuroinflammation 2020, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M. Nutraceuticals as modulators of gut microbiota: Role in therapy. Br. J. Pharmacol. 2020, 177, 1351–1362. [Google Scholar] [CrossRef]
- Pamer, E.G. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 2016, 352, 535–538. [Google Scholar] [CrossRef]
- Davoren, M.J.; Liu, J.; Castellanos, J.; Rodríguez-Malavé, N.I.; Schiestl, R.H. A novel probiotic, Lactobacillus johnsonii 456, resists acid and can persist in the human gut beyond the initial ingestion period. Gut Microbes 2019, 10, 458–480. [Google Scholar] [CrossRef]
- Gueimonde, M.; Jalonen, L.; He, F.; Hiramatsu, M.; Salminen, S. Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res. Int. 2006, 39, 467–471. [Google Scholar] [CrossRef]
- Pilkington, G.J.; Maherally, Z.; Jassam, S.; Barbu, E.; Fillmore, H. An all human 3d in vitro model of the blood brain barrier in nanoparticle delivery and cancer metastasis studies. Neuro-Oncol. 2014, 16, iii33. [Google Scholar] [CrossRef][Green Version]
- Morita, H.; He, F.; Fuse, T.; Ouwehand, A.C.; Hashimoto, H.; Hosoda, M.; Mizumachi, K.; Kurisaki, J.i. Adhesion of lactic acid bacteria to Caco-2 cells and their effect on cytokine secretion. Microbiol. Immunol. 2002, 46, 293–297. [Google Scholar] [CrossRef] [PubMed]
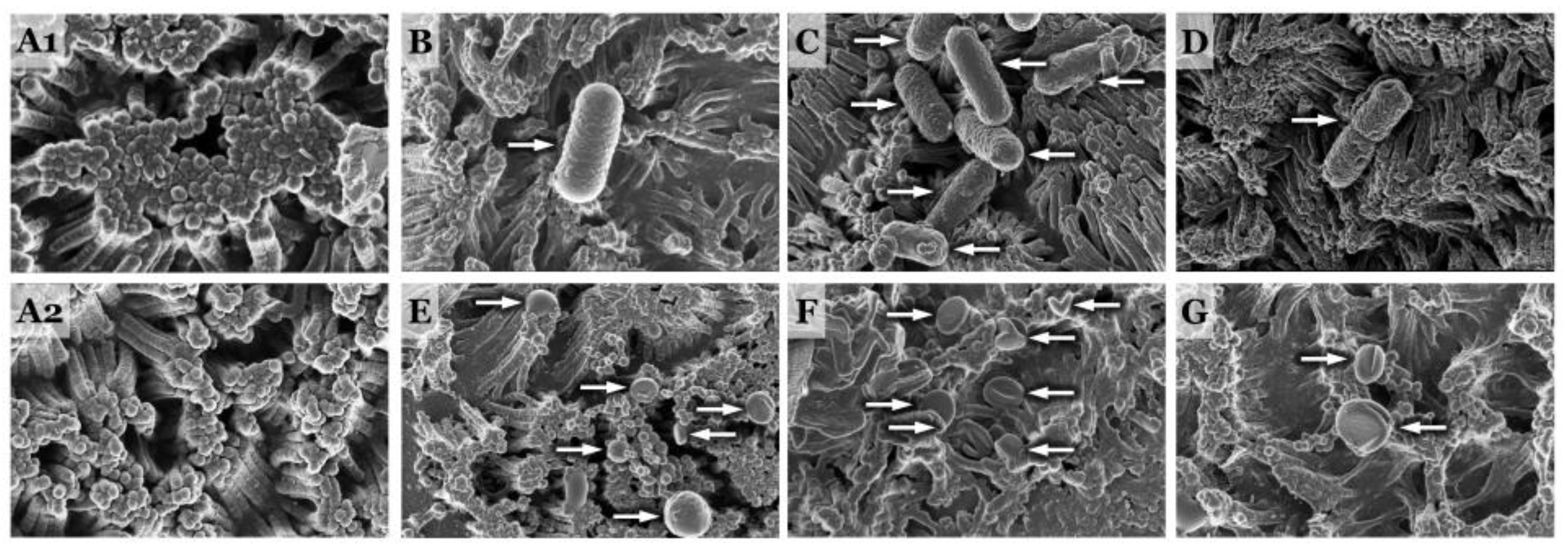

| Bacteria Strain | Characteristic | Source |
|---|---|---|
| EAEC, 3591-87 a | Clinical and diarrhoeagenic (positive reference strain) | NICD of NHLS c |
| EAEC, K2 a | Diarrhoeagenic | Unpasteurised fresh milk d |
| EAEC, N23 a | Non-Diarrhoeagenic | Unpasteurised fresh milk d |
| B. bifidum, ATCC 11863 b | Reference probiotic bacteria | ATTC Collections e |
| L. plantarum, FS2 b | Promising probiotic characteristics | Traditional fermented food (ogi) f |
| P. pentosaceus, D39 b | Promising probiotic characteristics | Traditional fermented food (ogi) f |
| LAB Strains | EAEC Strains | ||
|---|---|---|---|
| 3591-87 | K2 | N23 | |
| 11863 | 3591-87 + 11863 | K2 + 11863 | N23 + 11863 |
| FS2 | 3591-87 + FS2 | K2 + FS2 | N23 + FS2 |
| D39 | 3591-87 + D39 | K2 + D39 | N23 + D39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agbemavor, W.S.K.; Buys, E.M. Dynamic Interactions between Diarrhoeagenic Enteroaggregative Escherichia coli and Presumptive Probiotic Bacteria: Implications for Gastrointestinal Health. Microorganisms 2023, 11, 2942. https://doi.org/10.3390/microorganisms11122942
Agbemavor WSK, Buys EM. Dynamic Interactions between Diarrhoeagenic Enteroaggregative Escherichia coli and Presumptive Probiotic Bacteria: Implications for Gastrointestinal Health. Microorganisms. 2023; 11(12):2942. https://doi.org/10.3390/microorganisms11122942
Chicago/Turabian StyleAgbemavor, Wisdom Selorm Kofi, and Elna Maria Buys. 2023. "Dynamic Interactions between Diarrhoeagenic Enteroaggregative Escherichia coli and Presumptive Probiotic Bacteria: Implications for Gastrointestinal Health" Microorganisms 11, no. 12: 2942. https://doi.org/10.3390/microorganisms11122942
APA StyleAgbemavor, W. S. K., & Buys, E. M. (2023). Dynamic Interactions between Diarrhoeagenic Enteroaggregative Escherichia coli and Presumptive Probiotic Bacteria: Implications for Gastrointestinal Health. Microorganisms, 11(12), 2942. https://doi.org/10.3390/microorganisms11122942





