Enrichment of Aerobic and Anaerobic Hydrocarbon-Degrading Bacteria from Multicontaminated Marine Sediment in Mar Piccolo Site (Taranto, Italy)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of the Marine Sediment
2.2. Microbial Enrichment Cultivation
- (i)
- Aerobic biostimulation with biphenyl to initiate PCB aerobic transformation (Ae);
- (ii)
- Anaerobic incubation without the addition of external electron donors or biostimulating compounds (Ana);
- (iii)
- Anaerobic incubation with periodic spikes of PCE (0.8 mM) as an electron acceptor and H2 (5 mL) as an electron donor, to stimulate PCB reductive dechlorination (Ana*), and to facilitate the growth of dechlorinating bacteria and enhance the reductive PCB dechlorination, as previously documented [12].
2.3. DNA Extraction
2.4. 16S rRNA Gene Amplicon Sequencing
2.5. Biomarker Genes Quantification
2.6. PCBs and Chlorinated Ethenes Determination
2.7. Calculations
3. Results
3.1. PCB Contamination
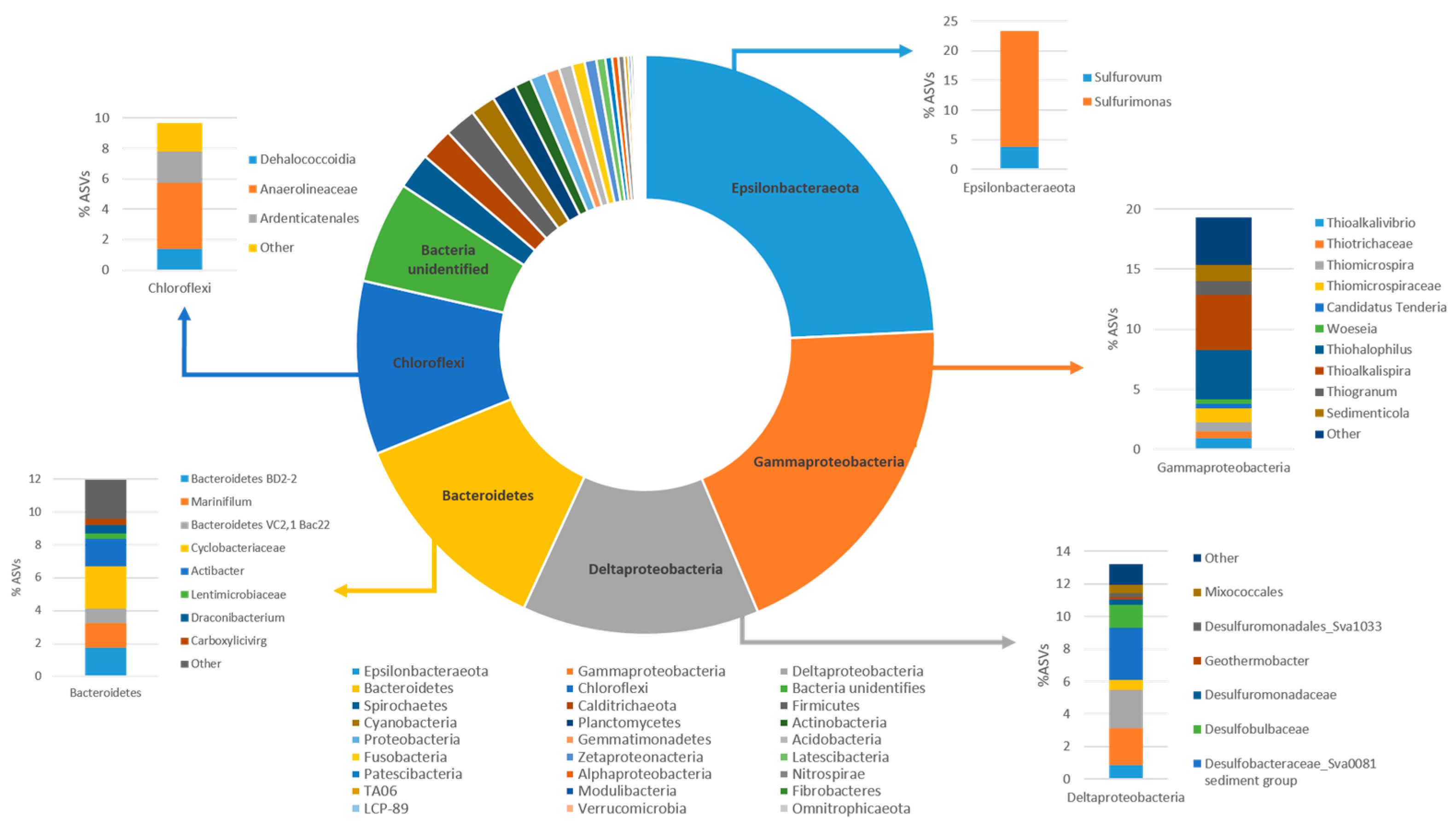
3.2. Composition of the Indigenous Bacterial Community of the Multicontaminated Marine Sediment
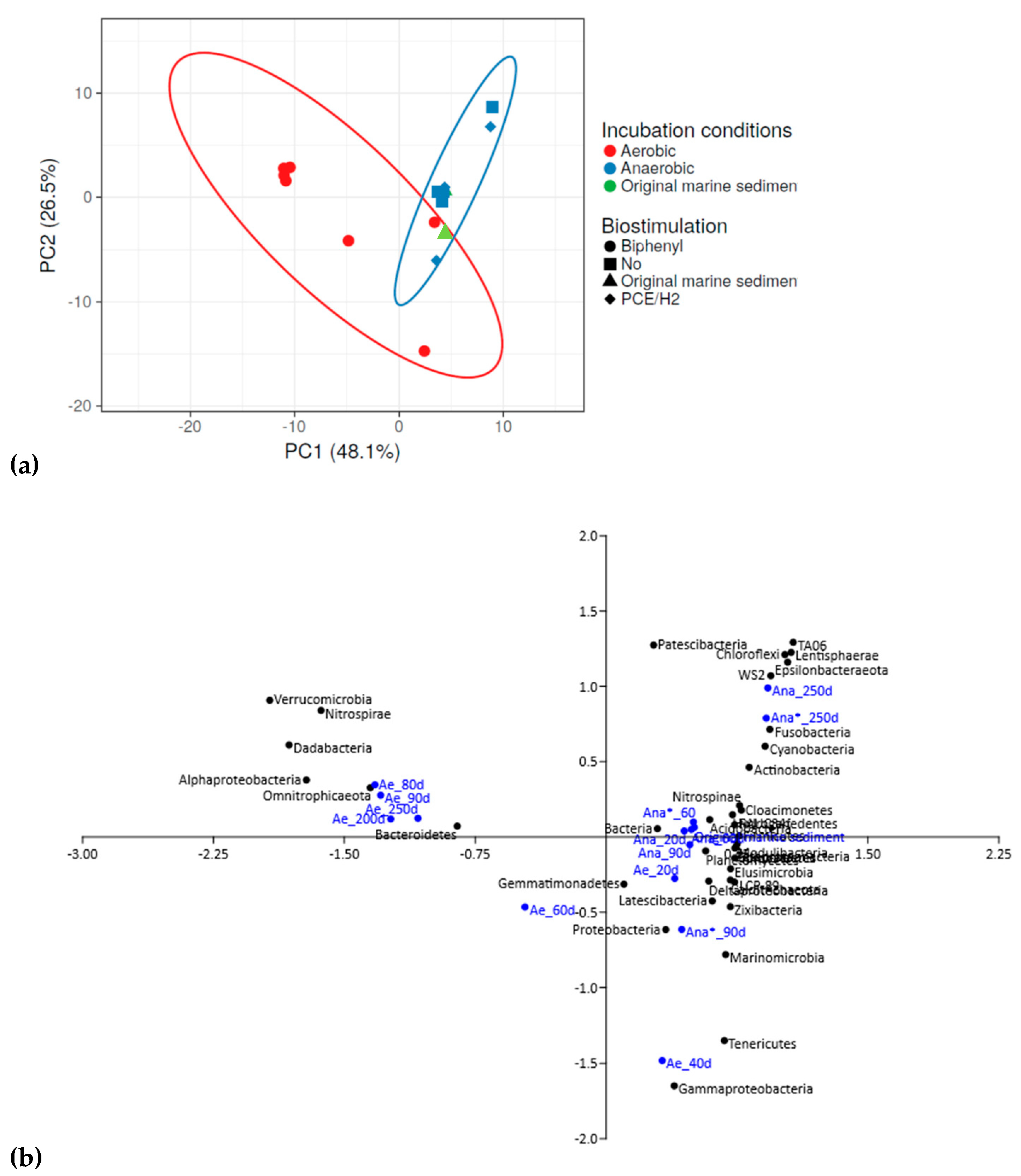
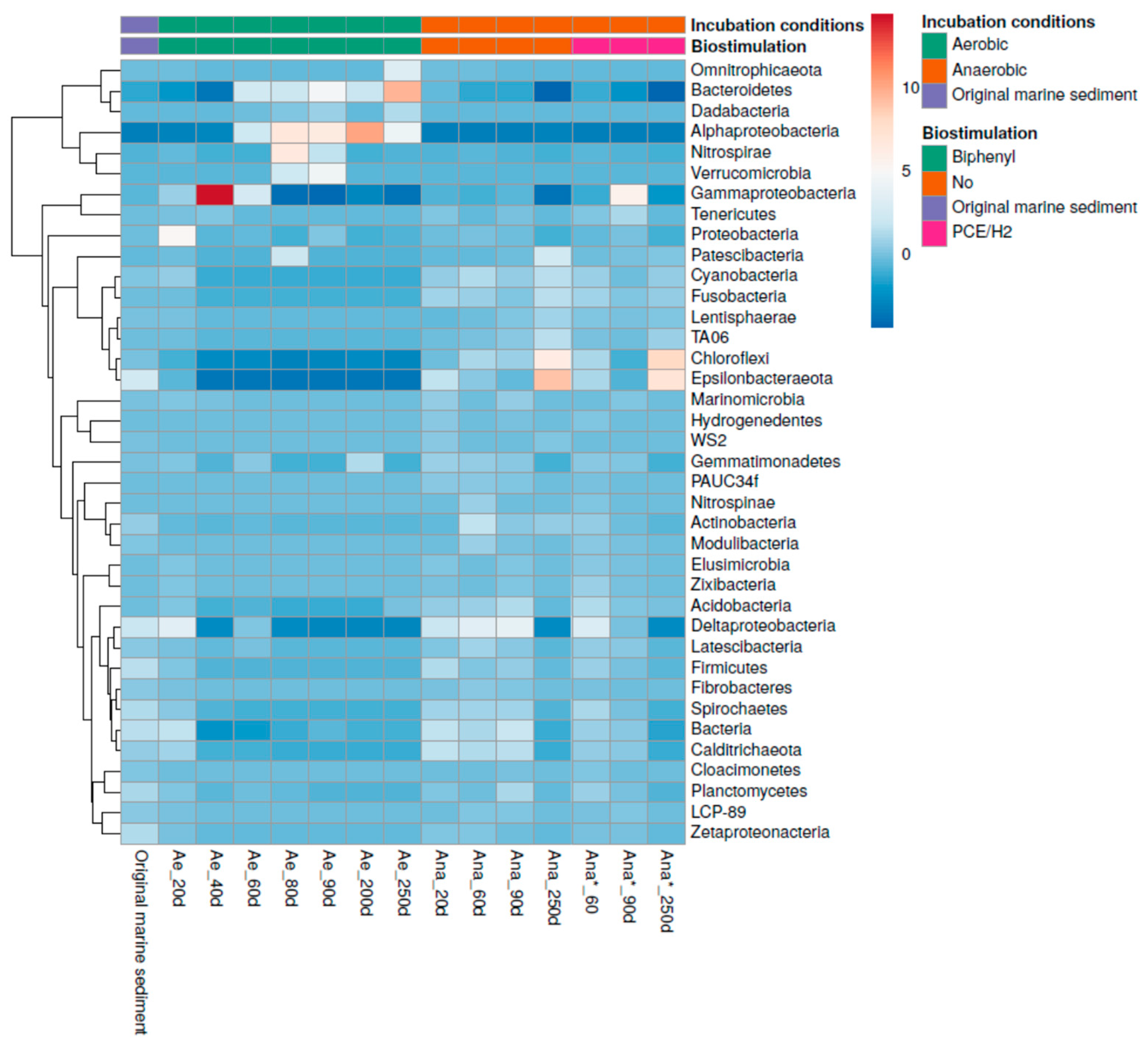
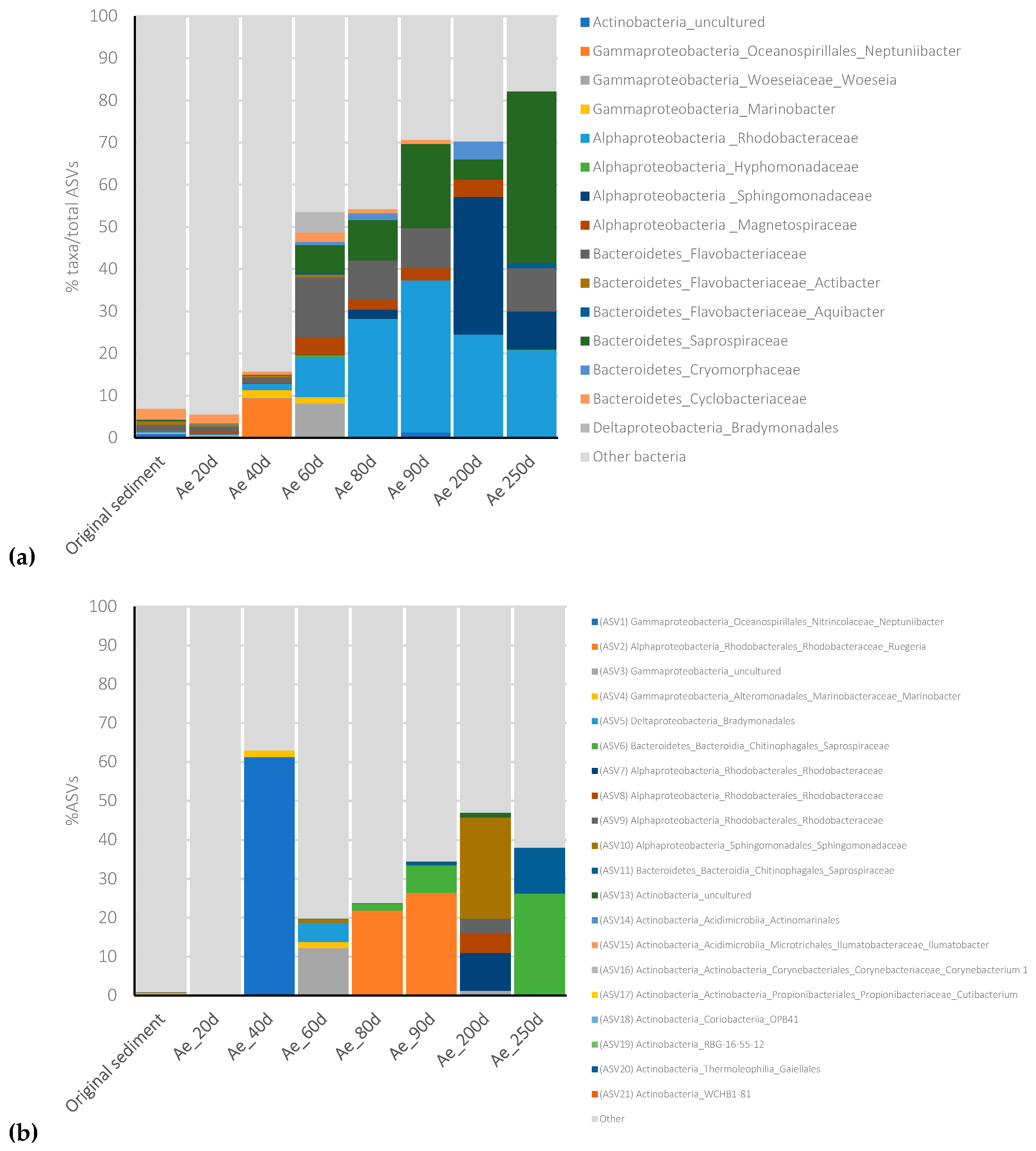
3.3. Dynamic Changes in the Bacterial Community during Aerobic and Anaerobic Incubation of Contaminated Marine Sediment
3.4. Microbial Selection in the Aerobic Consortium
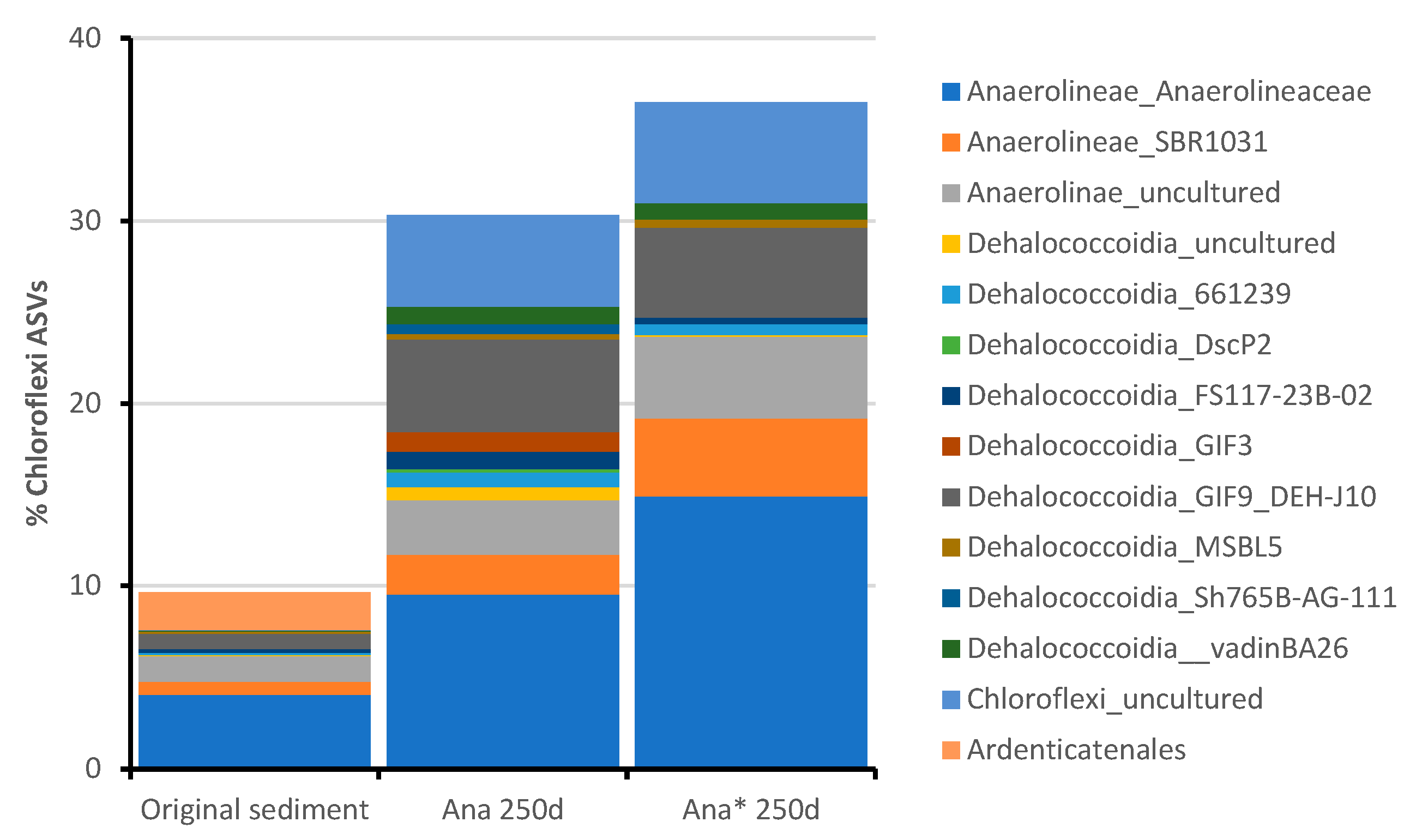
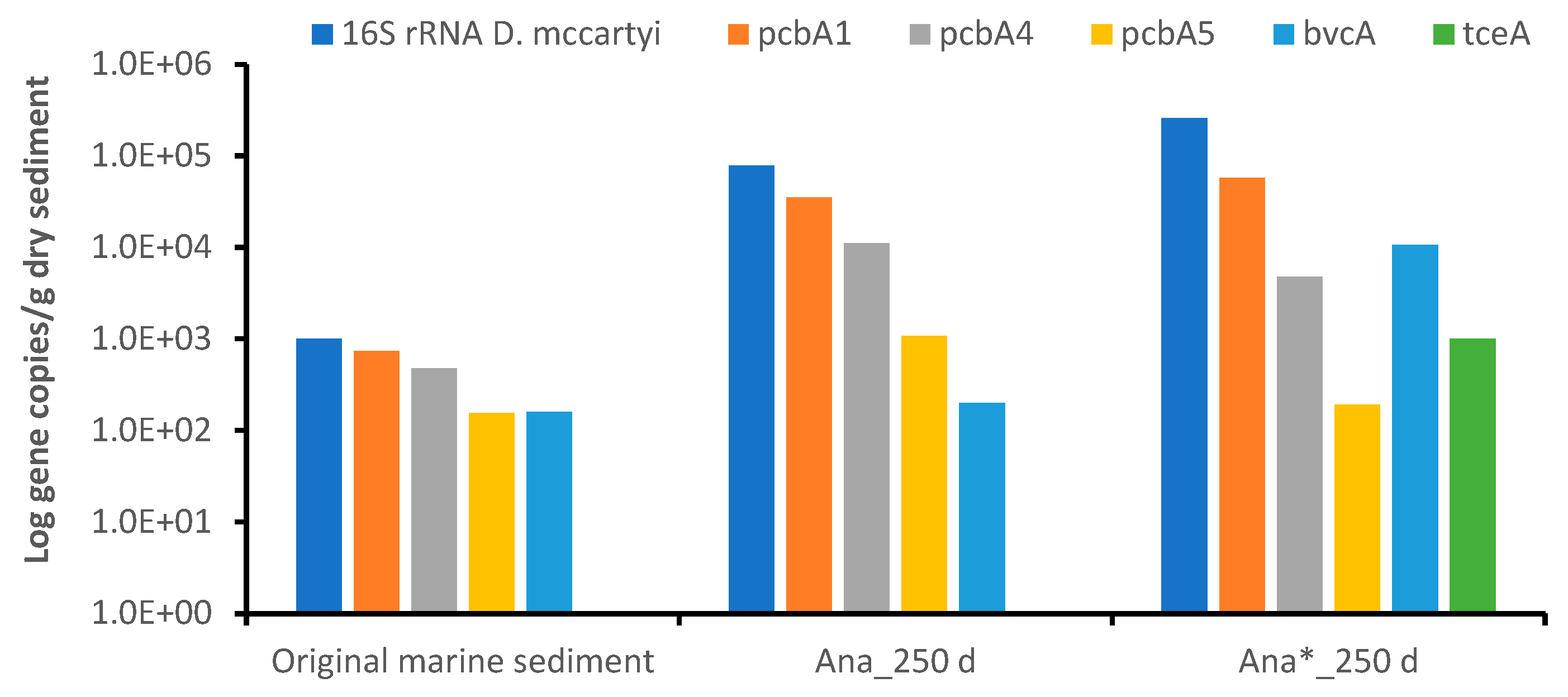
3.5. Microbial Community Selection in the Anaerobic Consortia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kronenberg, M.; Trably, E.; Bernet, N.; Patureau, D. Biodegradation of Polycyclic Aromatic Hydrocarbons: Using Microbial Bioelectrochemical Systems to Overcome an Impasse. Environ. Pollut. 2017, 231, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Gong, Y.; Reilly, S.E.O.; Zhao, D. Effects of Oil Dispersant on Solubilization, Sorption and Desorption of Polycyclic Aromatic Hydrocarbons in Sediment—Seawater Systems. Mar. Pollut. Bull. 2015, 92, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Environmental Protection Agency EPA. Alternate PCB Extraction Methods and Amendments to PCB Cleanup and Disposal Regulations. 2023. Available online: https://www.federalregister.gov/documents/2023/08/29/2023-17708/alternate-pcb-extraction-methods-and-amendments-to-pcb-cleanup-and-disposal-regulations (accessed on 20 October 2023).
- Agency for Toxic Substances and Disease Registry; US Department of Health and Human Services; Public Health Service. ATSDR: Toxicological Profile for Polychlorinat–Ed Biphenyls (PCBs); AISDR: San Francisco, CA, USA, 2000.
- Robertson, L.W.; Hansen, L.G. PCBs: Recent Advances in Environmental Toxicology and Health Effects; University Press of Kentucky: Lexington, KY, USA, 2001; Volume 8. [Google Scholar]
- Beyer, A.; Biziuk, M. Environmental Fate and Global Distribution of Polychlorinated Biphenyls; Whitacre, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 201, ISBN 9781441900326. [Google Scholar]
- Furukawa, K.; Fujihara, H. Microbial Degradation of Polychlorinated Biphenyls: Biochemical and Molecular Features. J. Biosci. Bioingineering 2008, 105, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Laroe, S.L.; Fricker, A.D.; Bedard, D.L. Dehalococcoides Mccartyi Strain JNA in Pure Culture Extensively Dechlorinates Aroclor 1260 According to Polychlorinated Biphenyl (PCB) Dechlorination Process N. Environ. Sci. Technol. 2014, 48, 9187–9196. [Google Scholar] [CrossRef]
- Chun, C.L.; Payne, R.B.; Sowers, K.R.; May, H.D. Electrical Stimulation of Microbial PCB Degradation in Sediment. Water Res. 2013, 47, 141–152. [Google Scholar] [CrossRef]
- Matturro, B.; Frascadore, E.; Rossetti, S. High-Throughput Sequencing Revealed Novel Dehalococcoidia in Dechlorinating Microbial Enrichments from PCB-Contaminated Marine Sediments. FEMS Microbiol. Ecol. 2017, 93, fix134. [Google Scholar] [CrossRef]
- Bedard, D.L.; Pohl, E.A.; Bailey, J.J.; Murphy, A.A. Characterization of the PCB Substrate Range of Microbial Dechlorination Process LP. Environ. Sci. Technol. 2005, 39, 6831–6838. [Google Scholar] [CrossRef]
- Wang, S.; Rei, K.; Wilm, A.; Zhao, S.; Yang, K.; Nagarajan, N. Genomic Characterization of Three Unique Dehalococcoides That Respire on Persistent Polychlorinated Biphenyls. Proc. Natl. Acad. Sci. USA 2014, 111, 12103–12108. [Google Scholar] [CrossRef]
- Petrić, I.; Hršak, D.; Fingler, S.; Udiković-Kolić, N.; Bru, D.; Martin-Laurent, F. Insight in the PCB-Degrading Functional Community in Long-Term Contaminated Soil under Bioremediation. J. Soil Sediments 2011, 11, 290–300. [Google Scholar] [CrossRef]
- Matturro, B.; Presta, E.; Rossetti, S. Reductive Dechlorination of Tetrachloroethene in Marine Sediments: Biodiversity and Dehalorespiring Capabilities of the Indigenous Microbes. Sci. Total Environ. 2016, 545–546, 445–452. [Google Scholar] [CrossRef]
- Bedard, D.L. A Case Study for Microbial Biodegradation: Anaerobic Bacterial Reductive Dechlorination of Polychlorinated Biphenyls—From Sediment to Defined Medium. Annu. Rev. Microbiol. 2008, 62, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; He, J. Phylogenetically Distinct Bacteria Involve Extensive Dechlorination of Aroclor 1260 in Sediment-Free Cultures. PLoS ONE 2013, 8, e59178. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chng, K.R.; Chen, C.; Bedard, D.L.; He, J. Genomic Characterization of Dehalococcoides Mccartyi Strain JNA That Reductively Dechlorinates Tetrachloroethene and Polychlorinated Biphenyls. Environ. Sci. Technol. 2015, 49, 14319–14325. [Google Scholar] [CrossRef]
- Matturro, B.; Ubaldi, C.; Grenni, P.; Caracciolo, A.B.; Rossetti, S. Polychlorinated Biphenyl (PCB) Anaerobic Degradation in Marine Sediments: Microcosm Study and Role of Autochthonous Microbial Communities. Environ. Sci. Pollut. Res. 2016, 23, 12613–12623. [Google Scholar] [CrossRef] [PubMed]
- May, H.D.; Sowers, K.R. “Dehalobium chlorocoercia” DF-1—From Discovery to Application. In Organohalide-Respiring Bacteria; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9783662498750. [Google Scholar]
- Zanaroli, G.; Balloi, A.; Negroni, A.; Borruso, L.; Daffonchio, D.; Fava, F. A Chloroflexi Bacterium Dechlorinates Polychlorinated Biphenyls in Marine Sediments under in Situ -like Biogeochemical Conditions. J. Hazard. Mater. 2012, 209–210, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Bedard, D.L.; Unterman, R.; Bopp, L.H.; Brennan, M.J.; Haberl, M.L.; Johnson, C. Rapid Assay for Screening and Characterizing Microorganisms for the Ability to Degrade Polychlorinated Biphenyls. Appl. Environ. Microbiol. 1986, 51, 761–768. [Google Scholar] [CrossRef]
- Gibson, D.T.; Parales, R.E. Aromatic Hydrocarbon Dioxygenases in Environmental Biotechnology. Curr. Opin. Biotechnol. 2000, 11, 236–243. [Google Scholar] [CrossRef]
- Mackova, M.; Uhlik, O.; Lovecka, P.; Viktorova, J.; Novakova, M.; Demnerova, K.; Sylvestre, M.; Macek, T. Bacterial Degradation of Polychlorinated Biphenyls. In Geomicrobiology: Molecular and Environmental Perspective; Barton, L., Mandl, M., Loy, A., Eds.; Springer: Dordrecht, The Netherland, 2010. [Google Scholar]
- Pavlů, L.; Vosàhlova, J.; Klierova, H.; Prouza, M.; Demnerova, K.; Brenner, V. Characterization of Chlorobenzoate Degraders Isolated from Polychlorinated Biphenyl-Contaminated Soil and Sediment in the Czech Republic. J. Appl. Microbiol. 1999, 87, 381–386. [Google Scholar] [CrossRef]
- Field, J.A.; Sierra-alvarez, R. Microbial Transformation and Degradation of Polychlorinated Biphenyls. Environ. Microbiol. 2008, 155, 1–12. [Google Scholar] [CrossRef]
- Pieper, D.H.; Seeger, M. Bacterial Metabolism of Polychlorinated. J. Mol. Microbiol. Biotechnol. 2008, 15, 121–138. [Google Scholar] [CrossRef]
- Sharma, J.K.; Gautam, R.K.; Nanekar, S.V.; Weber, R.; Singh, B.K.; Singh, S.K.; Juwarkar, A.A. Advances and Perspective in Bioremediation of Polychlorinated Biphenyl-Contaminated Soils. Environ. Sci. Pollut. Res. 2018, 25, 16355–16375. [Google Scholar] [CrossRef] [PubMed]
- Hoostal, M.J.; Bouzat, J.L. Spatial Patterns of BphA Gene Diversity Reveal Local Adaptation of Microbial Communities to PCB and PAH Contaminants. Microb. Ecol. 2016, 72, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Quero, G.M.; Cassin, D.; Botter, M.; Perini, L.; Luna, G.M. Patterns of Benthic Bacterial Diversity in Coastal Areas Contaminated by Heavy Metals, Polycyclic Aromatic Hydrocarbons (PAHs) and Polychlorinated Biphenyls (PCBs). Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Matturro, B.; Mascolo, G.; Rossetti, S. Microbiome Changes and Oxidative Capability of an Anaerobic PCB Dechlorinating Enrichment Culture after Oxygen Exposure. N. Biotechnol. 2020, 56, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Firrincieli, A.; Negroni, A.; Zanaroli, G.; Cappelletti, M. Unraveling the Metabolic Potential of Asgardarchaeota in a Sediment from the Mediterranean Hydrocarbon-Contaminated Water Basin Mar Piccolo (Taranto, Italy). Microorganisms 2021, 9, 859. [Google Scholar] [CrossRef]
- Botti, A.; Musmeci, E.; Negroni, A.; Capuozzo, R.; Fava, F.; Biagi, E.; Zanaroli, G. Site-Specific Response of Sediment Microbial Community to Supplementation of Polyhydroxyalkanoates as Biostimulants for PCB Reductive Dechlorination. Sci. Total Environ. 2023, 898, 165485. [Google Scholar] [CrossRef]
- Cardellicchio, N.; Covelli, S.; Cibic, T. Integrated Environmental Characterization of the Contaminated Marine Coastal Area of Taranto, Ionian Sea (Southern Italy). Environ. Sci. Pollut. Res. 2016, 23, 12491–12494. [Google Scholar] [CrossRef][Green Version]
- Cotecchia, F.; Vitone, C.; Sollecito, F.; Mali, M.; Miccoli, D.; Petti, R.; Milella, D.; Ruggieri, G.; Bottiglieri, O.; Santaloia, F.; et al. A Geo-chemo-mechanical Study of a Highly Polluted Marine System (Taranto, Italy) for the Enhancement of the Conceptual Site Model. Sci. Rep. 2021, 11, 4017. [Google Scholar] [CrossRef]
- Xiang, Y.; Xing, Z.; Liu, J.; Qin, W.; Huang, X. Recent Advances in the Biodegradation of Polychlorinated Biphenyls. World J. Microbiol. Biotechnol. 2020, 36, 145. [Google Scholar] [CrossRef]
- Dyksterhouse, S.E.; Gray, J.P.; Herwig, R.P.; Lara, J.C.A.N.; Staley, J.T. Cycloclasticus Pugetii Gen. Nov., Sp. Nov., an Aromatic Hydrocarbon-Degrading Bacterium from Marine Sediments. Int. J. Syst. Bacteriol. 1995, 45, 116–123. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Gabriel, A.; Ghalith, A.; Alexander, H.; Alm, E.J.; Arumugam, M.; et al. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ Prepr. 2018. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Jonson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Glo, F.O.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef]
- Mattes, T.E.; Ewald, J.M.; Liang, Y.; Martinez, A.; Awad, A.M.; Hornbuckle, K.C.; Schnoor, J.L. Microbial Communities in Polychlorinated Biphenyl (PCB)-Contaminated Wastewater Lagoon Sediments: PCB Congener, Quantitative PCR, and 16S RRNA Gene Amplicon Sequencing Datasets. Data Br. 2021, 39, 107546. [Google Scholar] [CrossRef] [PubMed]
- Aulenta, F.; Majone, M.; Verbo, P.; Tandoi, V. Complete Dechlorination of Tetrachloroethene to Ethene in Presence of Methanogenesis and Acetogenesis by an Anaerobic Sediment Microcosm. Biodegradation 2002, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Lu, Q.; Yu, L.; Wang, S. Tetrachloroethene Primes Reductive Dechlorination of Polychlorinated Biphenyls in a River Sediment Microcosm. Water Res. 2019, 152, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Perner, M. The Globally Widespread Genus Sulfurimonas: Versatile Energy Metabolisms and Adaptations to Redox Clines. Front. Microbiol. 2015, 6, 989. [Google Scholar] [CrossRef]
- Inagaki, F.; Takai, K.; Kobayashi, H.; Nealson, K.H.; Horikoshi, K. Sulfurimonas Autotrophica Gen. Nov., Sp. Nov., a Novel Sulfur-Oxidizing e -Proteobacterium Isolated from Hydrothermal Sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2003, 53, 1801–1805. [Google Scholar] [CrossRef]
- Dyksma, S.; Pjevac, P.; Ovanesov, K.; Mussmann, M. Evidence for H 2 Consumption by Uncultured Desulfobacterales in Coastal Sediments. Environ. Microbiol. 2018, 20, 450–461. [Google Scholar] [CrossRef]
- Watanabe, M.; Higashioka, Y.; Kojima, H.; Fukui, M. Desulfosarcina Widdelii Sp. Nov. and Desulfosarcina Alkanivorans Isolated from Marine Sediment and Emended Description of the Genus Desulfosarcina. Int. J. Syst. Evol. Microbiol. 2017, 67, 2994–2997. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, X.; Sun, K.; Lin, C.; Ma, J.; He, M.; Ouyang, W. Persulfate-Based Advanced Oxidation Processes (AOPs) for Organic- Contaminated Soil Remediation: A Review. Chem. Eng. J. 2019, 372, 836–851. [Google Scholar] [CrossRef]
- Machado, L.F.; Leite, D.C.A.; Rachid, C.T.C.C.; Paes, J.E.; Martins, E.F.; Peixoto, R.S.; Rosado, A.S. Tracking Mangrove Oil Bioremediation Approaches and Bacterial Diversity at Different Depths in an in Situ Mesocosms System. Front. Microbiol. 2019, 10, 2107. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, T.; Berry, D.; Teske, A.; Aitken, M.D. Enrichment of Fusobacteria in Sea Surface Oil Slicks from the Deepwater Horizon Oil Spill. Microorganisms 2016, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Roy, A.; Kazy, S.K. Exploring Microbial Diversity and Function in Petroleum Hydrocarbon Associated Environments through Omics Approaches; Elsevier: Amsterdam, The Netherlands, 2019; ISBN 9780128148495. [Google Scholar]
- Rani, S.; Jin, W.; Koh, H.; Kim, Y.; Kang, M.; Park, S. Marine Genomics Genomic Potential of Marinobacter Salinus Hb8 T as Sulfur Oxidizing and Aromatic Hydrocarbon Degrading Bacterium. Mar. Genom. 2017, 34, 19–21. [Google Scholar] [CrossRef]
- Fathepure, B.Z.; Coolen, M.J.L.; Hole, W. Recent Studies in Microbial Degradation of Petroleum Hydrocarbons in Hypersaline Environments. Front. Microbiol. 2014, 5, 173. [Google Scholar] [CrossRef]
- Du, Z.-J.; Wang, Z.-J.; Zhao, J.-X.; Chen, G. Woeseia Oceani Gen. Nov., Sp. Nov., a Chemoheterotrophic Member of the Order Chromatiales, and Proposal of Woeseiaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 107–112. [Google Scholar] [CrossRef]
- Mußmann, M.; Pjevac, P.; Krüger, K.; Dyksma, S. Genomic Repertoire of the Woeseiaceae / JTB255, Cosmopolitan and Abundant Core Members of Microbial Communities in Marine Sediments. ISME J. 2017, 11, 1276–1281. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Erdner, D.L.; Rosenheim, B.E.; Shetty, P.; Seitz, K.W.; Baker, B.J.; Liu, Z. Hydrocarbon Degradation and Response of Sea Fl Oor Sediment Bacterial Community in the Northern Gulf of Mexico to Light Louisiana Sweet Crude Oil. ISME J. 2018, 12, 2532–2543. [Google Scholar] [CrossRef]
- Molari, M.; Janssen, F.; Vonnahme, T.R.; Wenzhöfer, F.; Boetius, A. The Contribution of Microbial Communities in Polymetallic Nodules to the Diversity of the Deep-Sea Microbiome of the Peru Basin (4130—4198 m Depth). Biogeosciences 2020, 17, 3203–3222. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Kawasaki, A.; Stolz, A. Aerobic Hydrocarbon-Degrading Alphaproteobacteria: Sphingomonadales. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. Handbook of Hydrocarbon and Lipid Microbiology; McGenity, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 9783030147952. [Google Scholar]
- Ribicic, D.; Mcfarlin, K.M.; Netzer, R.; Brakstad, O.G.; Winkler, A.; Throne-Holst, M.; Størseth, T.R. Oil Type and Temperature Dependent Biodegradation Dynamics—Combining Chemical and Microbial Community Data through Multivariate Analysis. BMC Microbiol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Thompson, H.F.; Summers, S.; Yuecel, R.; Gutierrez, T. Hydrocarbon-Degrading Bacteria Found Tightly Associated with the 50–70 Μm Cell-Size Population of Eukaryotic Phytoplankton in Surface Waters of a Northeast Atlantic Region. Microorganisms 2020, 8, 1955. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mu, D.; Du, Z. Persicimonas Caeni Gen. Nov., Sp. Nov., the Representative of a Novel Wide-Ranging Predatory Taxon in Bradymonadales. Front. Environ. Sci. 2020, 11, 698. [Google Scholar] [CrossRef]
- Chen, S.; Budhraja, R.; Adrian, L.; Calabrese, F.; Stryhanyuk, H.; Musat, N.; Richnow, H.-H.; Duan, G.-L.; Zhu, Y.-G.; Musat, F. Novel Clades of Soil Biphenyl Degraders Revealed by Integrating Isotope Probing, Multi-Omics, and Single-Cell Analyses. ISME J. 2021, 15, 3508–3521. [Google Scholar] [CrossRef] [PubMed]
- Meier, D.V.; Pjevac, P.; Bach, W.; Markert, S.; Schweder, T.; Jamieson, J.; Petersen, S.; Amann, R.; Meyerdierks, A. Microbial Metal-Sulfide Oxidation in Inactive Hydrothermal Vent Chimneys Suggested by Metagenomic and Metaproteomic Analyses. Environ. Microbiol. 2019, 21, 682–701. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Perner, M. The Role of Hydrogen for Sulfurimonas Denitrificans’ Metabolism. PLoS ONE 2014, 9, e106218. [Google Scholar] [CrossRef]
- Graw, M.F.; Angelo, G.D.; Borchers, M.; Thurber, A.R.; Johnson, J.E.; Zhang, C.; Liu, H.; Colwell, F.S. Energy Gradients Structure Microbial Communities Across Sediment Horizons in Deep Marine Sediments of the South China Sea. Front. Microbiol. 2018, 9, 729. [Google Scholar] [CrossRef]
- Yang, Y.; Sanford, R.; Yan, J.; Chen, G.; Càpiro, N.L.; Li, X.; Löffler, F.E. Roles of Organohalide-Respiring Dehalococcoidia in Carbon Cycling. mSystems 2020, 5, e00757-19. [Google Scholar] [CrossRef]
- Loffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Muller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides Mccartyi Gen. Nov., Sp. Nov., Obligately Organohalide-Respiring Anaerobic Bacteria Relevant to Halogen Cycling and Bioremediation, Belong to a Novel Bacterial Class, Dehalococcoidia Classis Nov., Order Dehalococcoidales Ord. Nov. and Famil. Int. J. Syst. Evol. Microbiol. 2013, 63, 625–635. [Google Scholar] [CrossRef]
- Key, T.A.; Bowman, K.S.; Lee, I.; Chun, J.; Albuquerque, L.; Costa, M.S.; Rainey, F.A.; Moe, W.M. Dehalogenimonas Formicexedens Sp. Nov., a Chlorinated Alkane- Respiring Bacterium Isolated from Contaminated Groundwater. Int. J. Syst. Evol. Microbiol. 2017, 67, 1366–1373. [Google Scholar] [CrossRef]
- Zhao, S.; Rogers, M.J.; Ding, C.; He, J. Reductive Debromination of Polybrominated Diphenyl Ethers—Microbes, Processes and Dehalogenases. Front. Microbiol. 2018, 9, 1292. [Google Scholar] [CrossRef]
- Krzmarzick, M.J.; Crary, B.B.; Harding, J.J.; Oyerinde, O.O.; Leri, A.C.; Myneni, S.C.B.; Novak, P.J. Natural Niche for Organohalide-Respiring Chloroflexi. Appl. Environ. Microbiol. 2011, 78, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Krzmarzick, M.J.; Mcnamara, P.J.; Crary, B.B.; Novak, P.J. Abundance and Diversity of Organohalide-Respiring Bacteria in Lake Sediments across a Geographical Sulfur Gradient. FEMS Microbiol. Ecol. 2013, 84, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Kaster, A.; Mayer-blackwell, K.; Pasarelli, B.; Spormann, A.M. Single Cell Genomic Study of Dehalococcoidetes Species from Deep-Sea Sediments of the Peruvian Margin. ISME J. 2014, 8, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Wasmund, K.; Schreiber, L.; Lloyd, K.G.; Petersen, D.G.; Schramm, A.; Stepanauskas, R.; Jørgensen, B.B.; Adrian, L. Genome Sequencing of a Single Cell of the Widely Distributed Marine Subsurface Dehalococcoidia, Phylum Chloroflexi. ISME J. 2014, 8, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, H.; Moyer, C.L. Comparative Single-Cell Genomics of Chloroflexi from the Okinawa Trough Deep-Subsurface Biosphere. Appl. Environ. Microbiol. 2016, 82, 3000–3008. [Google Scholar] [CrossRef] [PubMed]
- Sewell, H.L.; Kaster, A.-K.; Spormann, A.M. Homoacetogenesis in Deep-Sea Chloroflexi, as Inferred by Single-Cell Genomics, Provides a Link to Reductive Dehalogenation in Terrestrial Dehalococcoidetes. MBio 2017, 8, e02022-17. [Google Scholar] [CrossRef]
- Selesi, D.; Jehmlich, N.; Von Bergen, M.; Schmidt, F.; Rattei, T.; Tischler, P.; Lueders, T.; Meckenstock, R.U. Combined Genomic and Proteomic Approaches Identify Gene Clusters Involved in Anaerobic 2-Methylnaphthalene Degradation in the Sulfate-Reducing Enrichment Culture N47. J. Biacteriology 2010, 192, 295–306. [Google Scholar] [CrossRef]
- Leahy, J.G.; Colwell, R.R. Microbial Degradation of Hydrocarbons in the Environment. Microbiol. Rev. 1990, 54, 305–315. [Google Scholar] [CrossRef]
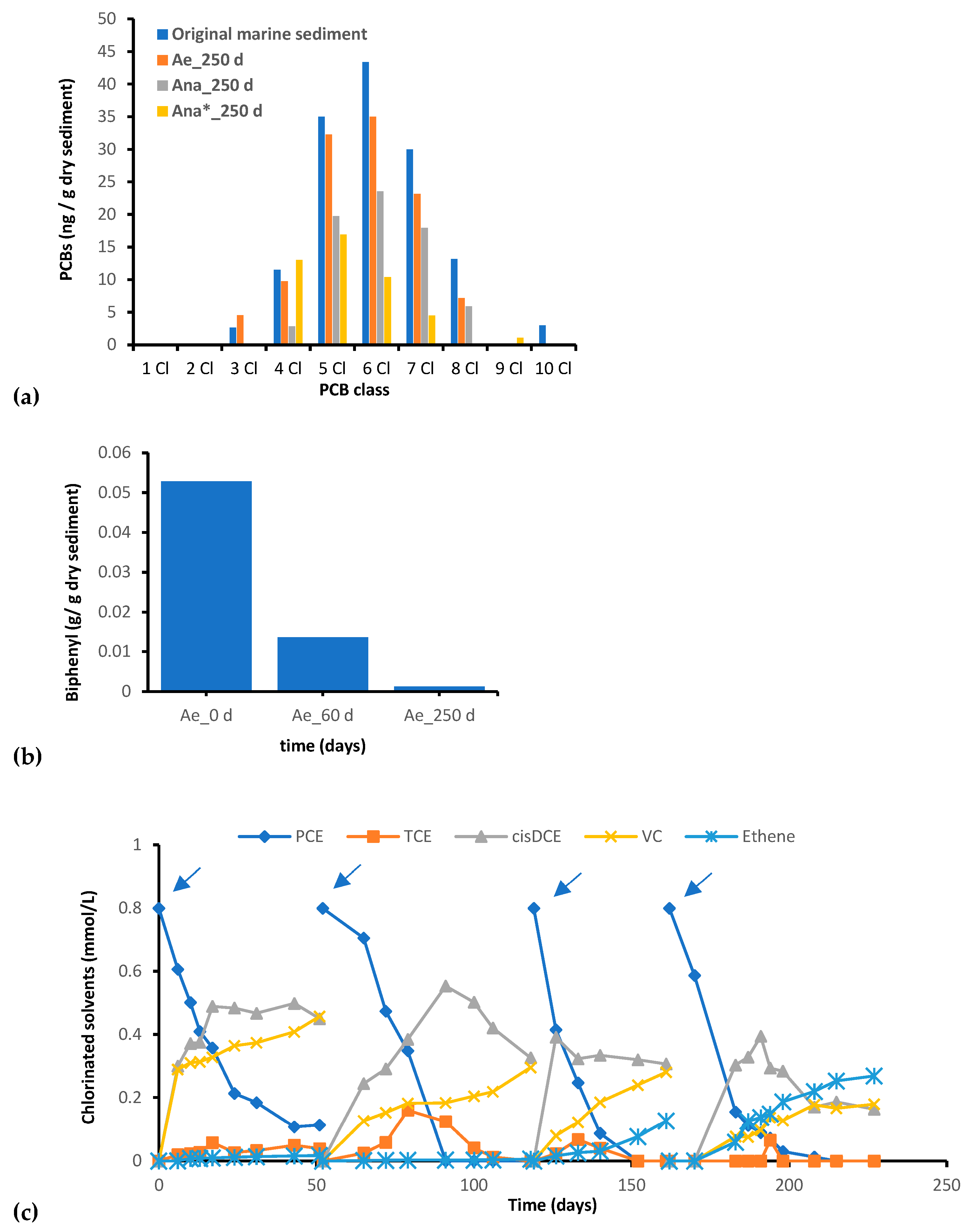
| Cl | m/z | Time Range (min) |
|---|---|---|
| - * | 154.1 | 5–10 |
| 1 | 188.1–190.1 | 10–13 |
| 2 | 222.0–224.0 | 12.2–18 |
| 3 | 256.0–258.0 | 15–23 |
| 4 | 291.9–293.9 | 18–29 |
| 5 | 325.9–327.9 | 21–32.5 |
| 6 | 359.9–361.9 | 24.5–34.5 |
| 7 | 393.9–397.8 | 29–35.5 |
| 8 | 429.8–431.8 | 32–36.5 |
| 9 | 463.8–465.8 | 34.5–37 |
| 10 | 497.7–499.7 | 36–41.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matturro, B.; Di Franca, M.L.; Tonanzi, B.; Cruz Viggi, C.; Aulenta, F.; Di Leo, M.; Giandomenico, S.; Rossetti, S. Enrichment of Aerobic and Anaerobic Hydrocarbon-Degrading Bacteria from Multicontaminated Marine Sediment in Mar Piccolo Site (Taranto, Italy). Microorganisms 2023, 11, 2782. https://doi.org/10.3390/microorganisms11112782
Matturro B, Di Franca ML, Tonanzi B, Cruz Viggi C, Aulenta F, Di Leo M, Giandomenico S, Rossetti S. Enrichment of Aerobic and Anaerobic Hydrocarbon-Degrading Bacteria from Multicontaminated Marine Sediment in Mar Piccolo Site (Taranto, Italy). Microorganisms. 2023; 11(11):2782. https://doi.org/10.3390/microorganisms11112782
Chicago/Turabian StyleMatturro, Bruna, Maria Letizia Di Franca, Barbara Tonanzi, Carolina Cruz Viggi, Federico Aulenta, Magda Di Leo, Santina Giandomenico, and Simona Rossetti. 2023. "Enrichment of Aerobic and Anaerobic Hydrocarbon-Degrading Bacteria from Multicontaminated Marine Sediment in Mar Piccolo Site (Taranto, Italy)" Microorganisms 11, no. 11: 2782. https://doi.org/10.3390/microorganisms11112782
APA StyleMatturro, B., Di Franca, M. L., Tonanzi, B., Cruz Viggi, C., Aulenta, F., Di Leo, M., Giandomenico, S., & Rossetti, S. (2023). Enrichment of Aerobic and Anaerobic Hydrocarbon-Degrading Bacteria from Multicontaminated Marine Sediment in Mar Piccolo Site (Taranto, Italy). Microorganisms, 11(11), 2782. https://doi.org/10.3390/microorganisms11112782










