First Report on Leptospira Species Isolated from Patients in Slovenia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Leptospira Clinical Isolates
2.2. Leptospira Serogroup Determination
2.3. Leptospira Serovar Identification
2.4. Leptospira Identification Using MALDI-TOF MS
2.5. DNA Extraction
2.6. Leptospira Species Identification Using PCR-Tm
2.7. Leptospira Sequence Typing
2.8. Whole-Genome Sequencing
2.9. Phylogenetic Analysis
2.10. Data Availability
3. Results
3.1. Leptospira Serogroup Determination
3.2. Leptospira Serovar Identification
3.3. Leptospira Identification Using MALDI-TOF MS
3.4. Leptospira Species Identification Using PCR-Tm
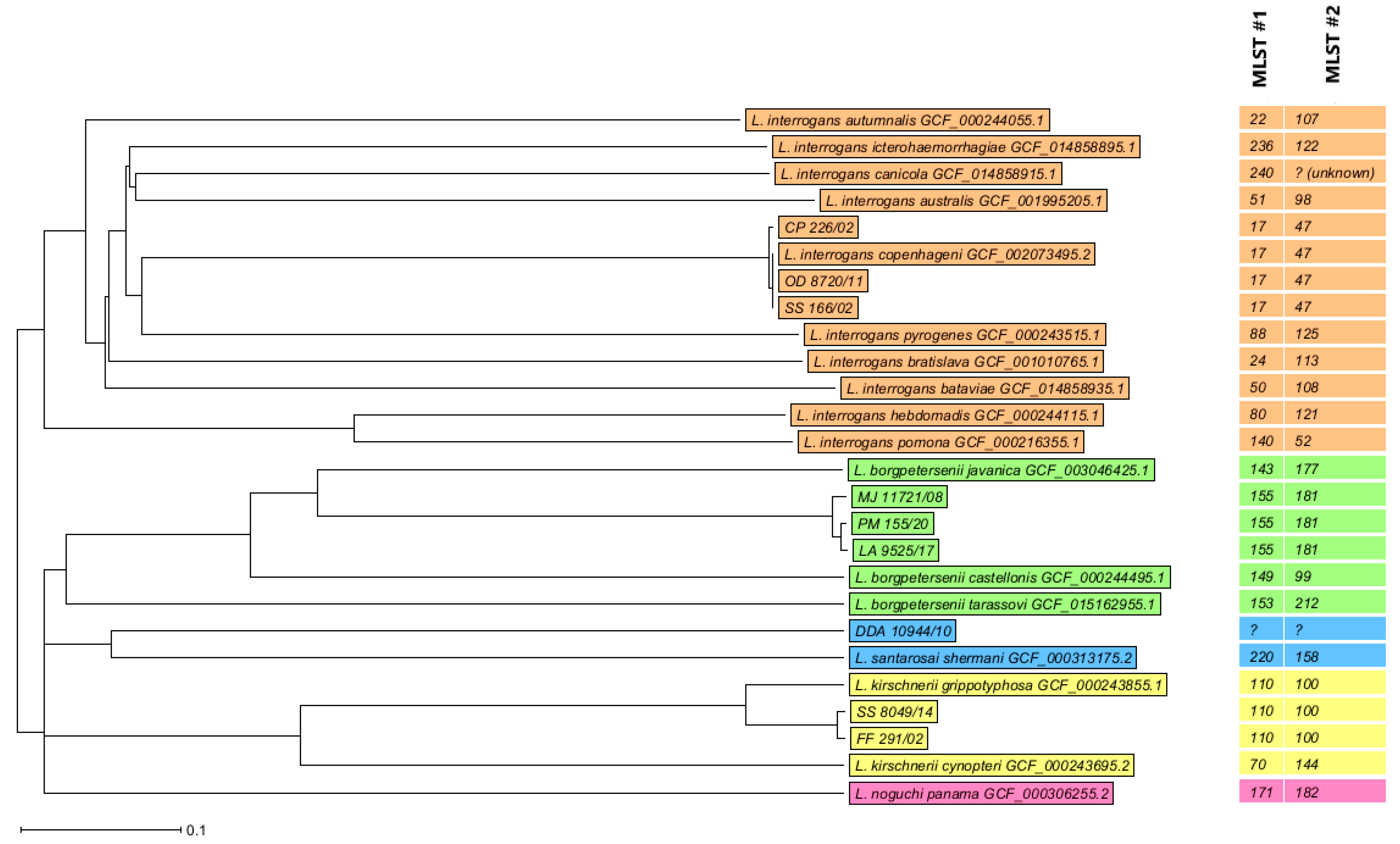
3.5. Leptospira Sequence Typing
3.6. WGS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bedernjak, J. Leptospiroze Pri Nas in v Svetu; Pomurska založba: Murska Sobota, Slovenia, 1993. [Google Scholar]
- Podgorsek, D.; Ruzic-Sabljic, E.; Logar, M.; Pavlovic, A.; Remec, T.; Baklan, Z.; Pal, E.; Cerar, T. Evaluation of Real-Time PCR Targeting the LipL32 Gene for Diagnosis of Leptospira Infection. BMC Microbiol. 2020, 20, 59. [Google Scholar] [CrossRef]
- Zele-Vengust, D.; Lindtner-Knific, R.; Mlakar-Hrzenjak, N.; Jerina, K.; Vengust, G. Exposure of Free-Ranging Wild Animals to Zoonotic Leptospira Interrogans Sensu Stricto in Slovenia. Animals 2021, 11, 2722. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the Taxonomy and Evolution of Pathogenicity of the Genus Leptospira through the Prism of Genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.A.; Lockaby, G. Leptospirosis and the Environment: A Review and Future Directions. Pathogens 2023, 12, 1167. [Google Scholar] [CrossRef]
- Picardeau, M. Leptospira and Leptospirosis. Methods Mol. Biol. 2020, 2134, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Nacionalni Inštitut za Javno Zdravje. Ljublijana. 2022. Available online: https://Nijz.Si/Wp-Content/Uploads/2021/05/Epidemiolosko_spremljanje_nalezljivih_bolezni_v_sloveniji_v_letu_2019_in_2020_0.pdf (accessed on 2 October 2023).
- Rajapakse, S. Leptospirosis: Clinical Aspects. Clin. Med. 2022, 22, 14–17. [Google Scholar] [CrossRef]
- Goarant, C.; Girault, D.; Thibeaux, R.; Soupe-Gilbert, M.E. Isolation and Culture of Leptospira from Clinical and Environmental Samples. In Leptospira spp. Methods and Protocols; Springer US: New York, NY, USA, 2020; Volume 2134, pp. 1–9. [Google Scholar] [CrossRef]
- Gorman, M.; Xu, R.; Prakoso, D.; Salvador, L.C.M.; Rajeev, S. Leptospira Enrichment Culture Followed by ONT Metagenomic Sequencing Allows Better Detection of Leptospira Presence and Diversity in Water and Soil Samples. PLoS Negl. Trop. Dis. 2022, 16, e0010589. [Google Scholar] [CrossRef]
- Herrmann, J.L.; Bellenger, E.; Perolat, P.; Baranton, G.; Saint Girons, I. Pulsed-Field Gel Electrophoresis of NotI Digests of Leptospiral DNA: A New Rapid Method of Serovar Identification. J. Clin. Microbiol. 1992, 30, 1696–1702. [Google Scholar] [CrossRef]
- Turk, N.; Milas, Z.; Mojcec, V.; Ruzic-Sabljic, E.; Staresina, V.; Stritof, Z.; Habus, J.; Postic, D. Molecular Analysis of Leptospira Spp. Isolated from Humans by Restriction Fragment Length Polymorphism, Real-Time PCR and Pulsed-Field Gel Electrophoresis. FEMS Microbiol. Lett. 2009, 300, 174–179. [Google Scholar] [CrossRef]
- Mende, K.; Galloway, R.L.; Becker, S.J.; Beckius, M.L.; Murray, C.K.; Hospenthal, D.R. Interlaboratory Agreement of Pulsed-Field Gel Electrophoresis Identification of Leptospira Serovars. Am. J. Trop. Med. Hyg. 2013, 89, 380–384. [Google Scholar] [CrossRef]
- Djelouadji, Z.; Roux, V.; Raoult, D.; Kodjo, A.; Drancourt, M. Rapid MALDI-TOF Mass Spectrometry Identification of Leptospira Organisms. Vet. Microbiol. 2012, 158, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Piccolo, G.; Gorrini, C.; Montecchini, S.; Buttrini, M.; Rossi, S.; Piergianni, M.; De Conto, F.; Arcangeletti, M.C.; Chezzi, C.; et al. Leptospira Species and Serovars Identified by MALDI-TOF Mass Spectrometry after Database Implementation. BMC Res. Notes 2014, 7, 330. [Google Scholar] [CrossRef] [PubMed]
- Sonthayanon, P.; Jaresitthikunchai, J.; Mangmee, S.; Thiangtrongjit, T.; Wuthiekanun, V.; Amornchai, P.; Newton, P.; Phetsouvanh, R.; Day, N.P.; Roytrakul, S. Whole Cell Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) for Identification of Leptospira Spp. in Thailand and Lao PDR. PLoS Negl. Trop. Dis. 2019, 13, e0007232. [Google Scholar] [CrossRef] [PubMed]
- Karcher, D.; Grenfell, R.C.; Moreno, A.M.; Moreno, L.Z.; Vasconcellos, S.A.; Heinemann, M.B.; de Almeida, J.N., Jr.; Juliano, L.; Juliano, M.A. Identification of Pathogenic and Nonpathogenic Leptospira Species of Brazilian Isolates by Matrix Assisted Laser Desorption/Ionization and Time Flight Mass Spectrometry. Braz. J. Microbiol. 2018, 49, 900–908. [Google Scholar] [CrossRef]
- Merien, F.; Portnoi, D.; Bourhy, P.; Charavay, F.; Berlioz-Arthaud, A.; Baranton, G. A Rapid and Quantitative Method for the Detection of Leptospira Species in Human Leptospirosis. FEMS Microbiol. Lett. 2005, 249, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 October 2023).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatic 2020, 70, e102. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid Whole-Genome Sequencing for Detection and Characterization of Microorganisms Directly from Clinical Samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome Analysis Using the Kraken Software Suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-Wide Leptospira Core Genome Multilocus Sequence Typing for Strain Taxonomy and Global Surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef]
- World Health Organization. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control; World Health Organization: Geneva, Switzerland, 2003; ISBN 92-4-154589-5. [Google Scholar]
- Galloway, R.L.; Levett, P.N. Evaluation of a Modified Pulsed-Field Gel Electrophoresis Approach for the Identification of Leptospira Serovars. Am. J. Trop. Med. Hyg. 2008, 78, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Naigowit, P.; Charoenchai, S.; Biaklang, M.; Seena, U.; Wangroongsarb, P.; Sawanpanyalert, P.; Warachit, P. Identification of Clinical Isolates of Leptospira Spp by Pulsed Field Gel-Electrophoresis and Microscopic Agglutination Test. Southeast Asian J. Trop. Med. Public. Health 2007, 38, 97–103. [Google Scholar]
- Postic, D.M. Diagnostic Biologique Leptospirose—Borreliose de Lyme, 2nd ed.; Lamassoure, M.A., Ed.; Institut Pasteur: Paris, France, 2000; ISBN 2-901320-30-9. [Google Scholar]
- Almasian-Tehrani, N.; Alebouyeh, M.; Armin, S.; Soleimani, N.; Azimi, L.; Shaker-Darabad, R. Overview of Typing Techniques as Molecular Epidemiology Tools for Bacterial Characterization. Cell. Mol. Biomed. Rep. 2021, 1, 69–77. [Google Scholar] [CrossRef]
- van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the Validation and Application of Typing Methods for Use in Bacterial Epidemiology. Clin. Microbiol. Infect. 2007, 13 (Suppl. S3), 1–46. [Google Scholar] [CrossRef] [PubMed]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chierakul, W.; Smythe, L.D.; Petkanchanapong, W.; Limpaiboon, R.; Apiwatanaporn, A.; Slack, A.T.; Suputtamongkol, Y.; White, N.J.; et al. A Dominant Clone of Leptospira Interrogans Associated with an Outbreak of Human Leptospirosis in Thailand. PLoS Negl. Trop. Dis. 2007, 1, e56. [Google Scholar] [CrossRef] [PubMed]
- Leon, A.; Pronost, S.; Fortier, G.; Andre-Fontaine, G.; Leclercq, R. Multilocus Sequence Analysis for Typing Leptospira Interrogans and Leptospira Kirschneri. J. Clin. Microbiol. 2010, 48, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Devi, S.M.; Valverde Mde, L.; Vijayachari, P.; Machang’u, R.S.; Ellis, W.A.; Hartskeerl, R.A. Multilocus Sequence Typing Method for Identification and Genotypic Classification of Pathogenic Leptospira Species. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 28. [Google Scholar] [CrossRef]
- Sykes, J.E.; Gamage, C.D.; Haake, D.A.; Nally, J.E. Understanding Leptospirosis: Application of State-of-the-Art Molecular Typing Tools with a One Health Lens. Am. J. Vet. Res. 2022, 83, ajvr.22.06.0104. [Google Scholar] [CrossRef]
- Koizumi, N.; Picardeau, M. (Eds.) Leptospira spp.: Methods and Protocols; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; Volume 2134, ISBN 978-1-07-160458-8. [Google Scholar]



| No. | Isolate Identification | Year of Isolation |
|---|---|---|
| 1 | SS 166/02 | 2002 |
| 2 | KB 185/02 | 2002 |
| 3 | VV 206/02 | 2002 |
| 4 | FF 291/02 | 2002 |
| 5 | ČP 226/02 | 2002 |
| 6 | PZ 9474/06 | 2006 |
| 7 | MJ 112721/08 | 2008 |
| 8 | DDA 10944/10 | 2010 |
| 9 | OD 8720/11 | 2011 |
| 10 | SS 8049/14 | 2014 |
| 11 | LA 9525/17 | 2017 |
| 12 | PM 155/20 | 2020 |
| No. | Species | Serological Group | Serovar | Strain Name | Origin |
|---|---|---|---|---|---|
| 1 | L. interrogans | Canicola | Canicola | Hond Utrecht IV 1 | A |
| 2 | L. interrogans | Pomona | Pomona | Pomona 1 | A |
| 3 | L. interrogans | Icterohaemorrhagiae | Copenhageni | Wijnberg 1 | LJ |
| 4 | L. interrogans | Australis | Australis | Ballico 1 | A |
| 5 | L. interrogans | Autumnalis | Autumnalis | Akiyami A 1 | A |
| 6 | L. interrogans | Pyrogenes | Pyrogenes | Salinem 1 | A |
| 7 | L. interrogans | Bataviae | Bataviae | Van Tienen 1 | P |
| 8 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | RGA | A |
| 9 | L. interrogans | Australis | Bratislava | Jez Bratislava | P |
| 10 | L. interrogans | Hebdomadis | Hebdomadis | Hebdomadis | LJ |
| 11 | L. borgpetersenii | Sejroe | Sejroe | M84 1 | P |
| 12 | L. borgpetersenii | Tarrassovi | Tarassovi | Mitis Johnson 1 | LJ |
| 13 | L. borgpetersenii | Ballum | Castellanis | Castellon 3 1 | A |
| 14 | L. borgpetersenii | Javanica | Javanica | Veldart Batavia 46 1 | A |
| 15 | L. kirschneri | Cynopteri | Cynopteri | 3522C 1 | A |
| 16 | L. kirschneri | Grippothyphosa | Grippotyphosa | Moskva V 1 | A |
| 17 | L. santarosai | Shermani | Shermani | 1342K | A |
| 18 | L. noguchii | Panama | Panama | CZ 214 K 1 | A |
| 19 | L. noguchii | Pomona | Proechiniys | 1161 U | P |
| 20 | L. alexanderi | Hebdomadis | Manzhuang | A23 | P |
| 21 | L. biflexa | Semaranga | Patoc | Patoc 1 1 | A, P, ZG, LJ |
| WGS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Isolate No. | PCR-Tm Identification | Serotyping Identification | NotI I RFLP | MALDI-TOF Identification | MLST 1 8 | MLST 1 9 | Kmer Finder | secY | lfb1 |
| 1 | SS 166/02 | L. interrogans | Icterohaemorrhagiae | 87.5% match to ICW 6 | Leptospira sp. (L. interrogans) 4 | 17 | 17 | L. interrogans serovar Copenhageni str. FDAARGOS | L. interrogans | L. interrogans |
| 2 | KB 185/02 | L. kirschneri | Grippotyphosa | 90.4% match to GGMV 7 | ND 5 | 110 | ND 5 | ND 5 | ND 5 | ND 5 |
| 3 | VV 206/02 | L. kirschneri | Grippotyphosa | 90.4% match to GGMV 7 | ND 5 | 110 | ND 5 | ND 5 | ND 5 | ND 5 |
| 4 | FF 291/02 | L. kirschneri | Grippotyphosa | 90.4% match to GGMV 7 | Leptospira sp. (L. interrogans) 4 | 110 | 110 | L. kirschneri strain FMAS_PN5 | L. kirschneri | L. kirschneri |
| 5 | ČP 226/02 | L. interrogans | Icterohaemorrhagiae | 87.5% match to ICW 6 | L. interrogans 3 | 17 | 17 | L. interrogans serovar Copenhageni str. FDAARGOS | L. interrogans | L. interrogans |
| 6 | PZ 9474/06 | L. kirschneri | Grippotyphosa | 90.4% match to GGMV 7 | ND 5 | 110 | ND 5 | ND 5 | ND 5 | ND 5 |
| 7 | MJ 11721/08 | L. borgpetersenii | Sejroe | Distinct pattern | L. borgpetersenii 2 | - | 155 | L. borgpetersenii FMAS_AP4 | L. borgpetersenii | L. borgpetersenii |
| 8 | DDA 10944/10 1 | L. borgpetersenii | Bataviae | Distinct pattern | Leptospira sp. (L. santarosai) 4 | ND | ND | L. santarosai serovar Shermani str. LT 821 | L. santarosai | L. santarosai |
| 9 | OD 8720/11 | L. interrogans | Icterohaemorrhagiae | 87.5% match to ICW 6 | L. interrogans 1 | 17 | 17 | L. interrogans serovar Copenhageni str. FDAARGOS | L. interrogans | L. interrogans |
| 10 | SS 8049/14 | L. kirschneri | Grippotyphosa | 90.4% match to GGMV 7 | Leptospira sp. (L. kirschneri) 4 | 110 | 110 | L. kirschneri strain FMAS_PN5 | L. kirschneri | L. kirschneri |
| 11 | LA 9525/17 | ND | Sejroe | ND | ND | ND | 155 | L. borgpetersenii FMAS_AP4 | L. borgpetersenii | L. borgpetersenii |
| 12 | PM 155/20 | ND | Sejroe | ND | ND | ND | 155 | L. borgpetersenii FMAS_AP4 | L. borgpetersenii | L. borgpetersenii |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ružić-Sabljić, E.; Podgoršek, D.; Strašek Smrdel, K.; Celar Šturm, A.; Logar, M.; Pavlović, A.; Remec, T.; Baklan, Z.; Pal, E.; Cerar Kišek, T. First Report on Leptospira Species Isolated from Patients in Slovenia. Microorganisms 2023, 11, 2739. https://doi.org/10.3390/microorganisms11112739
Ružić-Sabljić E, Podgoršek D, Strašek Smrdel K, Celar Šturm A, Logar M, Pavlović A, Remec T, Baklan Z, Pal E, Cerar Kišek T. First Report on Leptospira Species Isolated from Patients in Slovenia. Microorganisms. 2023; 11(11):2739. https://doi.org/10.3390/microorganisms11112739
Chicago/Turabian StyleRužić-Sabljić, Eva, Daša Podgoršek, Katja Strašek Smrdel, Andraž Celar Šturm, Mateja Logar, Andrea Pavlović, Tatjana Remec, Zvonko Baklan, Emil Pal, and Tjaša Cerar Kišek. 2023. "First Report on Leptospira Species Isolated from Patients in Slovenia" Microorganisms 11, no. 11: 2739. https://doi.org/10.3390/microorganisms11112739
APA StyleRužić-Sabljić, E., Podgoršek, D., Strašek Smrdel, K., Celar Šturm, A., Logar, M., Pavlović, A., Remec, T., Baklan, Z., Pal, E., & Cerar Kišek, T. (2023). First Report on Leptospira Species Isolated from Patients in Slovenia. Microorganisms, 11(11), 2739. https://doi.org/10.3390/microorganisms11112739







