A Novel Carotenoid-Producing Bacterium, Paenibacillus aurantius sp. nov., Isolated from Korean Marine Environment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation, Cultivation, and Growth Conditions
2.2. 16S rRNA Gene Phylogeny
2.3. Phenotypic Characterization
2.4. Chemotaxonomic Characterization
2.5. Whole-Genome Sequencing
2.6. Comparative Genomic Analysis
2.7. Genome Annotation and Secondary Metabolite Annotation
2.8. Carotenoid Extraction and HPLC Analysis
3. Results and Discussion
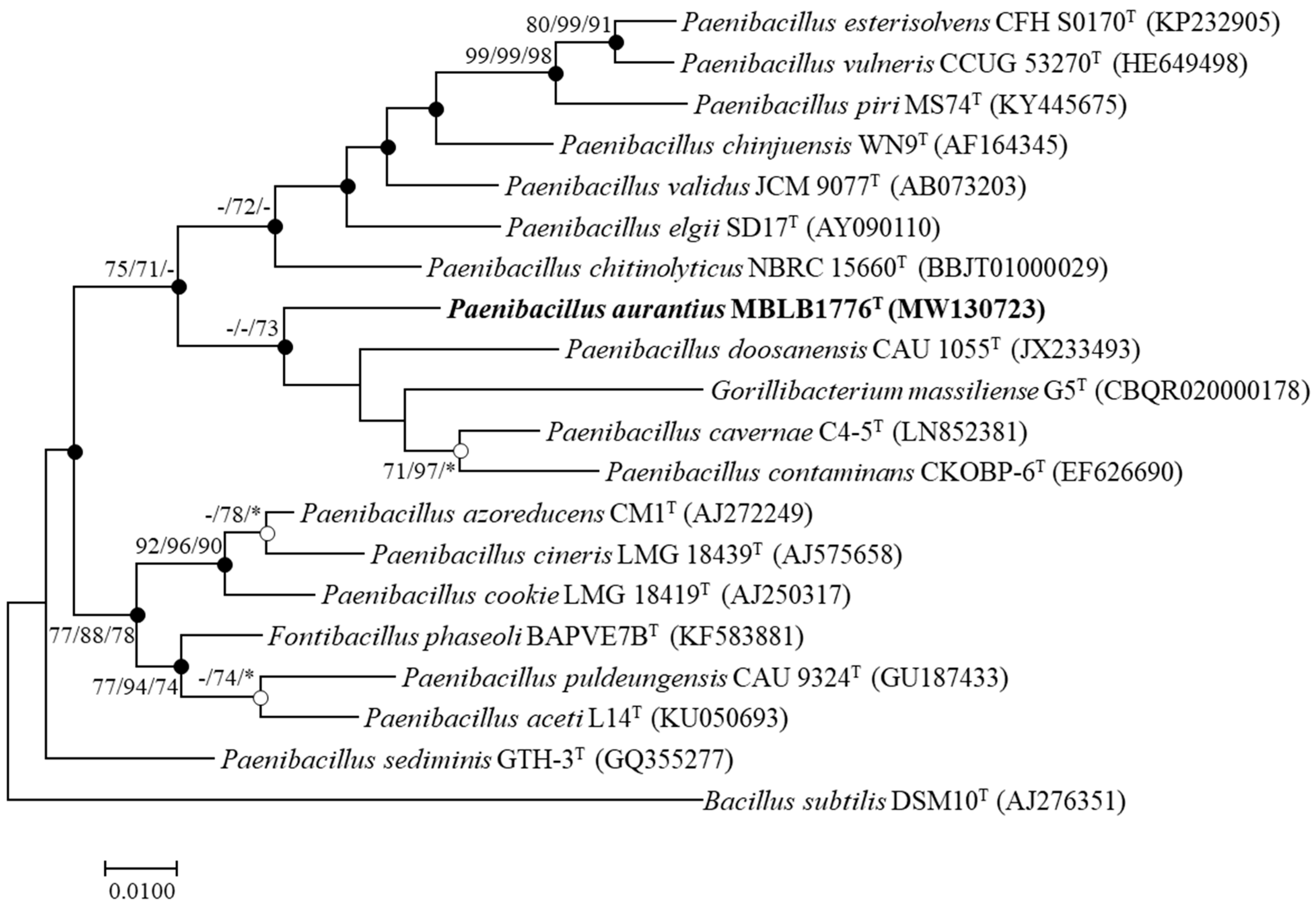
3.1. Phylogenetic Analysis
3.2. Morphological, Physiological, and Biochemical Characteristics
3.3. Chemotaxonomic Characteristics
3.4. Genome Information and Authenticity
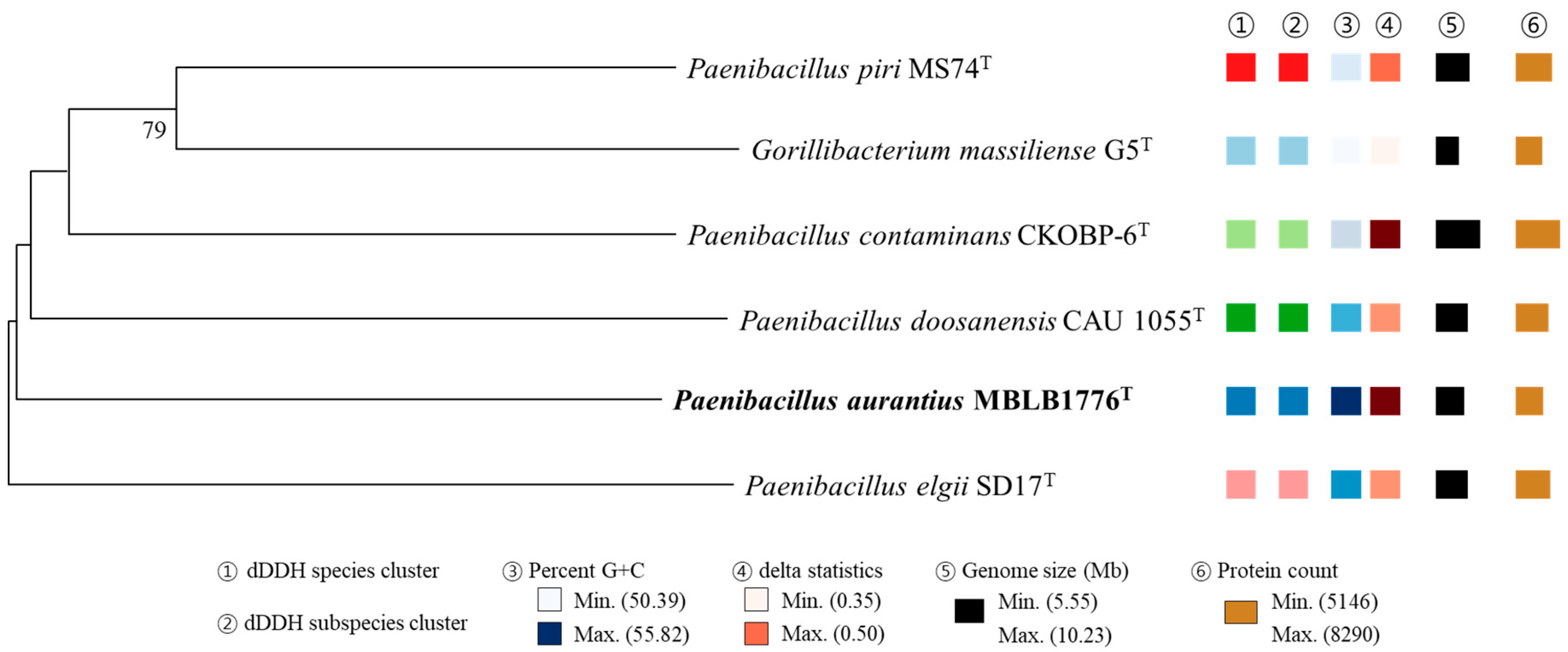
3.5. Phylogenomic Analysis
3.6. Genomic Features and Bio-Functional Potential
3.7. Identification of Carotenoid
4. Conclusions
Description of Paenibacillus aurantius sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ash, C.; Farrow, J.; Wallbanks, S.; Collins, M. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 1991, 13, 202–206. [Google Scholar] [CrossRef]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Oh, H.-M.; Yoon, B.-D.; Kang, K.H.; Park, Y.-H. Paenibacillus kribbensis sp. nov. and Paenibacillus terrae sp. nov., bioflocculants for efficient harvesting of algal cells. Int. J. Syst. Evol. Microbiol. 2003, 53, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Sukweenadhi, J.; Balusamy, S.R.; Kim, Y.-J.; Lee, C.H.; Kim, Y.-J.; Koh, S.C.; Yang, D.C. A growth-promoting bacteria, Paenibacillus yonginensis DCY84T enhanced salt stress tolerance by activating defense-related systems in Panax ginseng. Front. Plant Sci. 2003, 9, 813. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G.; Albrecht, M.; Schnurr, G.; Knörzer, O.; Böger, P. The biotechnological potential and design of novel carotenoids by gene combination in Escherichia coli. Trends Biotechnol. 1999, 17, 223–237. [Google Scholar] [CrossRef]
- Padhan, B.; Poddar, K.; Sarkar, D.; Sarkar, A. Production, purification, and process optimization of intracellular pigment from novel psychrotolerant Paenibacillus sp. BPW19. Biotechnol. Rep. 2021, 29, e00592. [Google Scholar] [CrossRef]
- Malik, K.; Tokkas, J.; Goyal, S. Microbial Pigments: A review. Int. J. Microb. Resour. Technol. 2012, 1, 361–365. [Google Scholar]
- Venil, C.K.; Zakaria, Z.A.; Ahmad, W.A. Bacterial pigments and their applications. Process Biochem. 2013, 48, 1065–1079. [Google Scholar] [CrossRef]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef]
- Baker, B.J.; Appler, K.E.; Gong, X. New Microbial Biodiversity in Marine Sediments. Annu. Rev. Mar. Sci. 2021, 13, 161–175. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kluge, A.G.; Farris, J.S. Quantitative phyletics and the evolution of anurans. Syst. Biol. 1969, 18, 1–32. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tittsler, R.P.; Sandholzer, L.A. The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 1936, 31, 575–580. [Google Scholar] [CrossRef]
- Benson, H.J. Microbiological Applications: A Laboratory Manual in General Microbiology; McGraw-Hill Company Inc.: Bedford, MA, USA, 2002; p. 432. [Google Scholar]
- Bauer, A.W.; Kirby, M.M.; Sherris, J.C.; Truck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Miller, L.T. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J. Clin. Microbiol. 1982, 16, 584–586. [Google Scholar] [CrossRef]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl. 1990, 20, 1–6. [Google Scholar]
- Collins, M.D.; Jones, D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Micro-biol. Rev. 1981, 45, 316–354. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference pro-gram. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2009, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Choi, S.R.; Lim, S.-H.; Yeo, Y.; Kweon, S.J.; Bae, Y.-S.; Kim, K.W.; Im, K.-H.; Ahn, S.K.; Ha, S.-H.; et al. Identification and quantification of carotenoids in paprika fruits and cabbage, kale, and lettuce leaves. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 355–358. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; de Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Lee, S.D. Paenibacillus cavernae sp. nov., isolated from soil of a natural cave. Int. J. Syst. Evol. Microbiol. 2016, 66, 598–603. [Google Scholar] [CrossRef]
- Chou, J.-H.; Lee, J.-H.; Lin, M.-C.; Chang, P.-S.; Arun, A.B.; Young, C.-C.; Chen, Y.-M. Paenibacillus contaminans sp. nov., isolated from a contaminated laboratory plate. Int. J. Syst. Evol. Microbiol. 2009, 59, 125–129. [Google Scholar] [CrossRef]
- Kim, J.-H.; Kang, H.; Kim, W. Paenibacillus doosanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 1271–1277. [Google Scholar] [CrossRef]
- Ningsih, F.; Sari, D.C.A.F.; Yabe, S.; Yokota, A.; Sjamsuridzal, W. Potential secondary metabolite biosynthetic gene clusters and antibacterial activity of novel taxa Gandjariella. Biodiversitas 2020, 21, 5674–5684. [Google Scholar] [CrossRef]
- Zhu, S.; Hegemann, J.D.; Fage, C.D.; Zimmermann, M.; Xie, X.; Linne, U.; Marahiel, M.A. Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem. 2016, 291, 13662–13678. [Google Scholar] [CrossRef] [PubMed]
- Köcher, S.; Breitenbach, J.; Müller, V.; Sandmann, G. Structure, function and biosynthesis of carotenoids in the moderately halophilic bacterium Halobacillus halophilus. Arch. Microbiol. 2009, 191, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Duc, L.H.; Fraser, P.; Cutting, S.M. Carotenoids present in halotolerant Bacillus spore formers. FEMS Microbiol. Lett. 2006, 255, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Khaneja, R.; Perez-Fons, L.; Fakhry, S.; Baccigalupi, L.; Steiger, S.; To, E.; Sandmann, G.; Dong, T.C.; Ricca, E.; Fraser, P.D.; et al. Carotenoids found in Bacillus. J. Appl. Microbiol. 2010, 108, 1889–1902. [Google Scholar]
- Perez-Fons, L.; Steiger, S.; Khaneja, R.; Bramley, P.M.; Cutting, S.M.; Sandmann, G.; Fraser, P.D. Identification and the developmental formation of carotenoid pigments in the yellow/orange Bacillus spore-formers. Biochem. Biophys. Acta 2011, 1811, 177–185. [Google Scholar] [CrossRef]
- Zang, L.-Y.; Sommerburg, O.; van Kuijk, F.J.G.M. Absorbance Changes of Carotenoids in Different Solvents. Free Radic. Biol. Med. 1997, 23, 1086–1089. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, P.C. Functional Expression and Extension of Staphylococcal Staphyloxanthin Biosynthetic Pathway in Escherichia coli. J. Biol. Chem. 2012, 287, 21575–21583. [Google Scholar] [CrossRef]


| Characteristic | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Temp. range (°C) (optimum) | 10–45 (30) * | 20–42 (30) a | 10–37 (30) b | 4–45 (30) c |
| NaCl range (%, w/v) (optimum) | 0–4 (1) * | 0–1 a | 0–2 (0.5) b | 0–4 (2) c |
| pH range for growth (optimum) | 6.0–8.0 (7.0) * | 6.0–10.0 (7.0) a | 6.5–8.0 (7.0) b | 4.5–7.5 (6.5) c |
| Catalase * | − | + | + | + |
| Oxidase * | − | + | + | + |
| API 20NE * | ||||
| Protease (gelatin) | − | + | + | − |
| Mannose | − | − | − | + |
| Mannitol | − | − | + | − |
| N-acetyl-glucosamine | − | + | + | − |
| Gluconate | + | − | + | − |
| Malate | + | − | + | − |
| API® ZYM * | ||||
| Esterase lipase (C8) | + | − | + | + |
| Leucine arylamidase | − | − | − | + |
| α-chymotrypsin | − | + | − | − |
| Acid phosphatase | + | + | − | + |
| α-galactosidase | + | − | − | − |
| α-glucosidase | − | + | + | + |
| β-glucosidase | − | − | + | + |
| Attribute | Characteristics |
|---|---|
| Sequencing platforms | PacBio |
| Assembler | FLYE v. 2.7 |
| Genome coverage | 111.98× |
| Assembly status | Complete |
| Assembly size (bp) | 6,303,770 |
| G + C content (mol%) | 55.8 |
| Total contigs | 1 |
| Total CDS | 5588 |
| RNAs | 112 |
| -rRNA genes (5S, 16S, 23S) | 30 |
| -tRNAs | 82 |
| Query Genome | Reference Genome | isDDH Value (%) | Model Confidence Interval (%) | G+C Content Difference (%) |
|---|---|---|---|---|
| MBLB1776T | Paenibacillus piri MS74T | 25.2 | [22.9–27.7] | 4.78 |
| Paenibacillus contaminans CKOBP-6T | 23.4 | [21.1–25.9] | 4.31 | |
| Paenibacillus doosanensis CAU 1055T | 22.7 | [20.4–25.1] | 2.53 | |
| Gorillibacterium massiliense G5T | 22.4 | [20.1–24.8] | 5.44 | |
| Paenibacillus elgii SD17T | 21.2 | [18.9–23.6] | 2.42 |
| Gene Type | Product | Span (nt) | Most Similar Biosynthetic Gene Cluster | BGC Similarity (%) |
|---|---|---|---|---|
| Ectoine | Ectoine | 414,375–424,767 | Streptomyces chrysomallus ATCC 11523T | 75.0 |
| Lassopeptide | Paeninodin | 1,318,636–1,342,527 | Paenibacillus dendritiformis C454 | 80.0 |
| LAP | 2,211,773–2,235,324 | - | - | |
| T3PKS | 2,799,722–2,840,831 | - | - | |
| Terpene | 3,482,565–3,500,973 | - | - | |
| Terpene | Carotenoid | 4,537,895–4,558,767 | Halobacillus halophilus DSM 2266T | 33.0 |

| Name | Top Hit | Description | E-Value | Identity (%) |
|---|---|---|---|---|
| ORF1 | WP_138193179 | Sulfatase (Paenibacillus antri SYSU K30003T) | 0.0 | 77.2 |
| AraC (ORF2) | WJH34801 | AraC family transcriptional regulator (Paenibacillus sp. CC-CFT747) | 0.0 | 98.6 |
| IDI (ORF3) | WJH34802 | Type 2 isopentenyl-diphosphate delta-isomerase (Paenibacillus sp. CC-CFT747) | 0.0 | 97.7 |
| CrtNb (ORF4) | WP_307396138 | Phytoene desaturase family protein (Bacillus horti) | 0.0 | 59.1 |
| GT (ORF5) | WP_130609653 | Glycosyltransferase family 2 protein (Cohnella abietis) | 5 × 10−147 | 56.4 |
| LPAT (ORF6) | WJH34805 | Lysophospholipid acyltransferase family protein (Paenibacillus sp. CC-CFT747) | 3 × 10−118 | 98.3 |
| ORF7 | WJH34807 | Carotenoid biosynthetic protein (Paenibacillus sp. CC-CFT747) | 4 × 10−103 | 95.3 |
| CrtM (ORF8) | WP_189012146 | Phytoene/squalene synthase family protein (Paenibacillus marchantiophytorum R55T) | 3 × 10−142 | 68.5 |
| CrtNc (ORF9) | WJH34808 | FAD-dependent oxidoreductase (Paenibacillus sp. CC-CFT747) | 0.0 | 96.9 |
| CrtNa (ORF10) | WP_076265663 | Phytoene desaturase family protein (Paenibacillus sp. FSL A5-0031) | 0.0 | 66.8 |
| ORF11 | WJH34810 | Cobalamin B12-binding domain-containing protein (Paenibacillus sp. CC-CFT747) | 8 × 10−122 | 84.3 |
| CrtO (ORF12) | WP_082882581 | Glycosyl-4,4’-diaponeurosporenoate acyltransferase (Pseudalkalibacillus sp. FJAT-53715) | 5 × 10−47 | 47.3 |
| GT2 (ORF13) | WJH34812 | Glycosyltransferase (Paenibacillus sp. CC-CFT747) | 0.0 | 92.8 |
| Aldedh (ORF14) | WP_099520946 | Aldehyde dehydrogenase family protein (Paenibacillus sp. BIHB 4019) | 4 × 10−176 | 55.1 |
| ORF15 | WJH34813 | Phytoene desaturase family protein (Paenibacillus sp. CC-CFT747) | 0.0 | 96.8 |
| ORF16 | WJH34814 | Dabb family protein (Paenibacillus sp. CC-CFT747) | 4 × 10−63 | 98.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, C.Y.; Seo, S.M.; Cho, E.-S.; Nam, Y.-D.; Park, S.-L.; Lim, S.-I.; Seo, M.-J. A Novel Carotenoid-Producing Bacterium, Paenibacillus aurantius sp. nov., Isolated from Korean Marine Environment. Microorganisms 2023, 11, 2719. https://doi.org/10.3390/microorganisms11112719
Hwang CY, Seo SM, Cho E-S, Nam Y-D, Park S-L, Lim S-I, Seo M-J. A Novel Carotenoid-Producing Bacterium, Paenibacillus aurantius sp. nov., Isolated from Korean Marine Environment. Microorganisms. 2023; 11(11):2719. https://doi.org/10.3390/microorganisms11112719
Chicago/Turabian StyleHwang, Chi Young, Sung Man Seo, Eui-Sang Cho, Young-Do Nam, So-Lim Park, Seong-Il Lim, and Myung-Ji Seo. 2023. "A Novel Carotenoid-Producing Bacterium, Paenibacillus aurantius sp. nov., Isolated from Korean Marine Environment" Microorganisms 11, no. 11: 2719. https://doi.org/10.3390/microorganisms11112719
APA StyleHwang, C. Y., Seo, S. M., Cho, E.-S., Nam, Y.-D., Park, S.-L., Lim, S.-I., & Seo, M.-J. (2023). A Novel Carotenoid-Producing Bacterium, Paenibacillus aurantius sp. nov., Isolated from Korean Marine Environment. Microorganisms, 11(11), 2719. https://doi.org/10.3390/microorganisms11112719







