Unusual Oligomeric Laccase-like Oxidases from Ascomycete Curvularia geniculata VKM F-3561 Polymerizing Phenylpropanoids and Phenolic Compounds under Neutral Environmental Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cultivation Procedure
2.3. Laccase Activity Assay
2.4. Purification of Laccase-like Oxidases of C. geniculata VKM F-3561
2.5. Enzyme Characterization
2.6. Isolation and Identification of Phenylpropanoid and Phenolic Compound Intermediates Produced by the Oxidases of C. geniculata VKM F-3561
2.7. Identification of Nucleotide Sequence of the Laccase Gene Expressed at the Peak of Alkaliphilic Laccase Activity of the Fungus C. geniculata VKM F-3561
2.8. Analysis of the Obtained Sequences
2.9. Nucleotide Sequence of the C. geniculata VKM F-3561 Laccase Gene
2.10. Homology Modelling and Structural Analysis
2.11. Phylogenetic Analysis
3. Results and Discussion
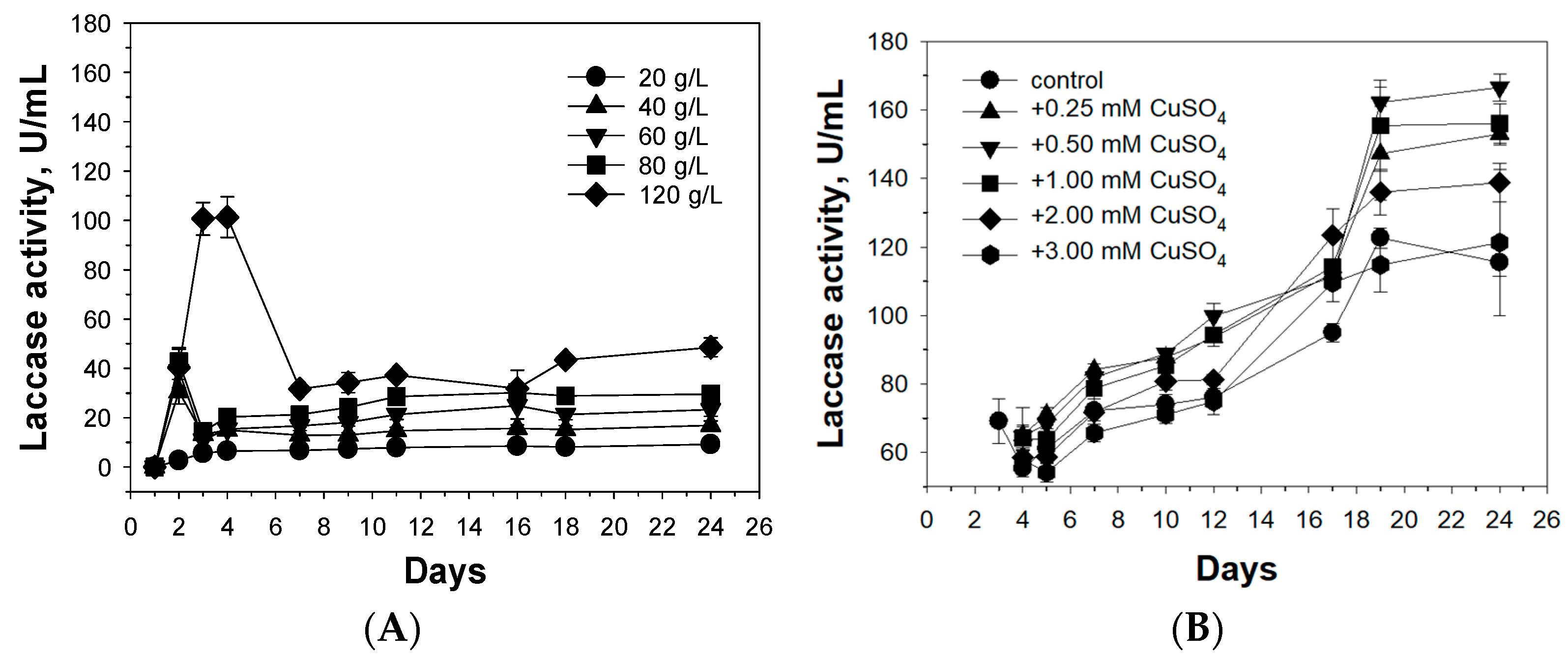
3.1. Optimization of Cultivation Conditions for Alkaliphilic Laccase Production by the Fungus C. geniculata VKM F-3561
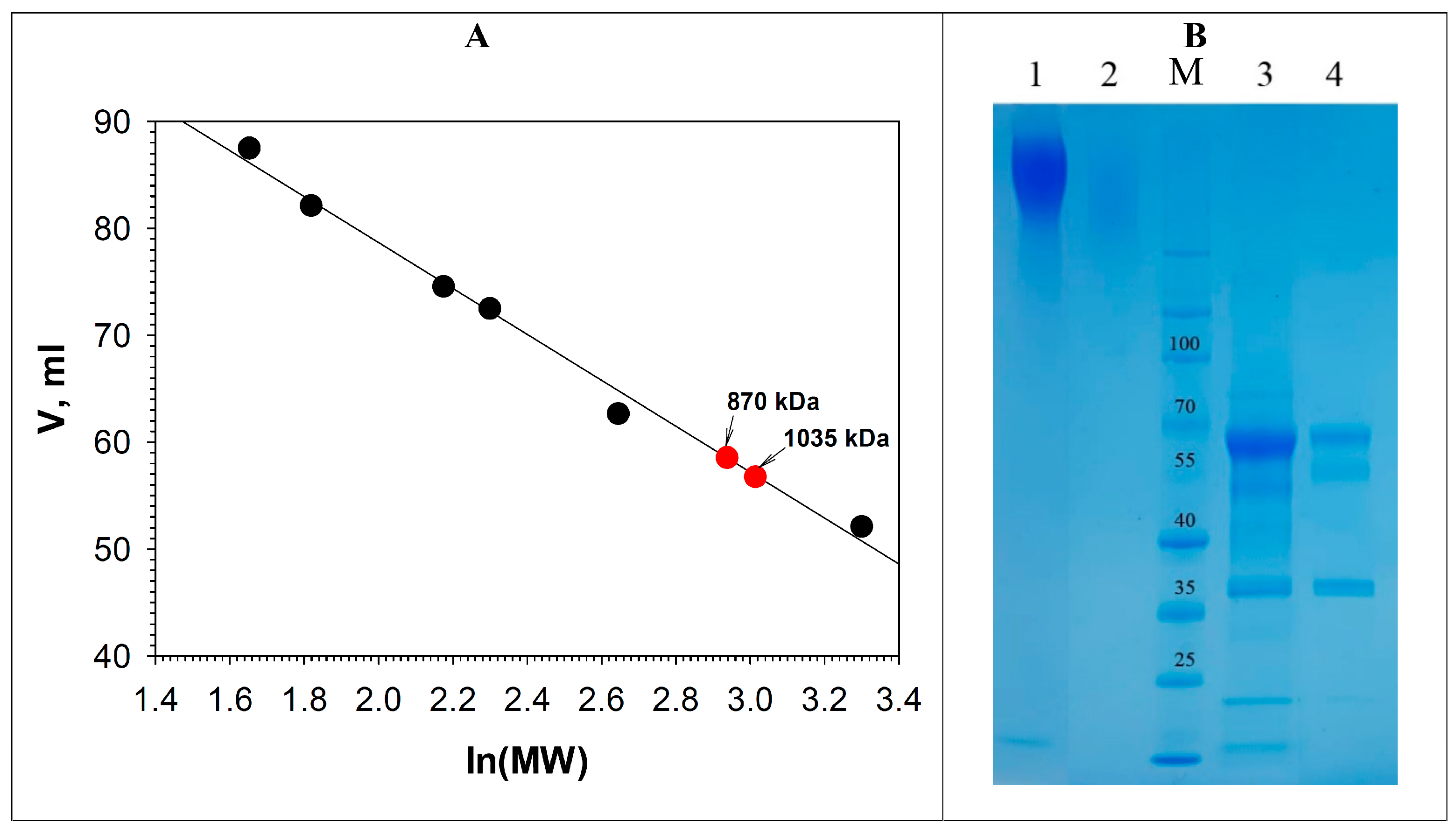
3.2. Purification of the Alkaliphilic Laccase-like Oxidases of C. geniculata VKM F-3561
3.3. T-, pH Optima, and Stability of Isolated Oxidases
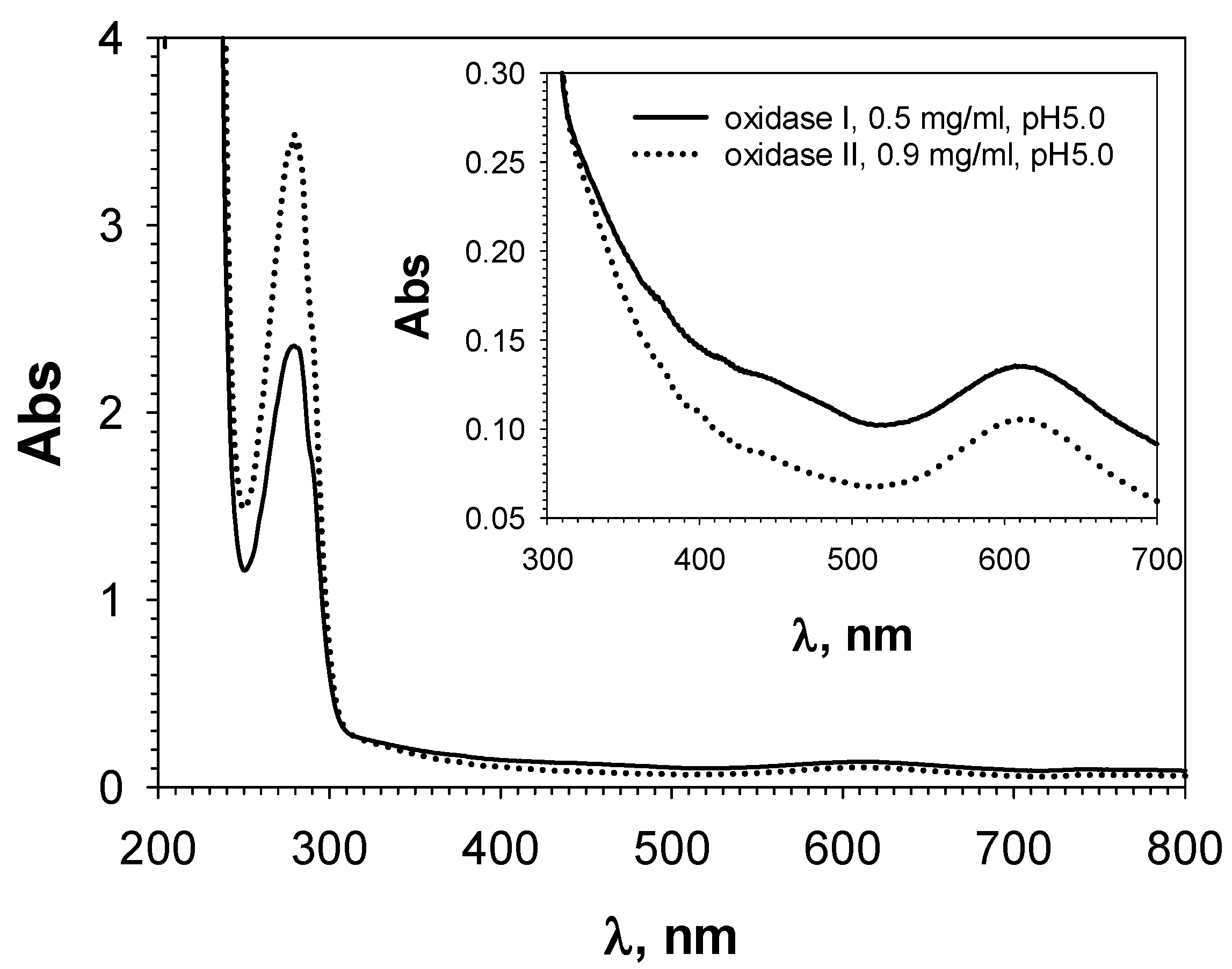
3.4. Spectral Properties of Isolated Laccase-like Oxidases
| Organisms | pHopt with SG | T3-Peak, 330 nm | Modified T3-Peak, nm | T1- Peak, nm | References |
|---|---|---|---|---|---|
| Coprinus cinereus | 7.0 | no | no | 614 | [9] |
| Rhizoctonia praticola | 7.4 | smooth | 370, 420 | 610 | [11] |
| Rhizoctonia solani RS22 | 7.0 | no | no | 602 | [12] |
| Rhus vernicifera | 7.0, 8.5 * | no | 370, 420 | 614 | [77,78] |
| Melanocarpus albomyces | 6.0–7.0 | no | 360, 450 | 600 | [76] |
| Myceliophthora thermophila CBS 117.65 | 6.0–7.0 | no | no | 589 | [21] |
| Myrothecium roridum VKM F-3565 | 7.8 | no | 370, 440 | 587 | [28] |
| Curvularia geniculata VKM F-3561 | |||||
| oxidase I | 7.5 | no | 372, 411, 450 | 610 | the present work |
| oxidase II | 7.0 | smooth | 374, 409–414, 450 | 611 | the present work |
3.5. Kinetic Properties of the Isolated Laccase-like Oxidases
3.6. Transformation of Phenylpropanoids and Phenolic Compounds by Laccase-like Oxidases of C. geniculata VKM F-3561
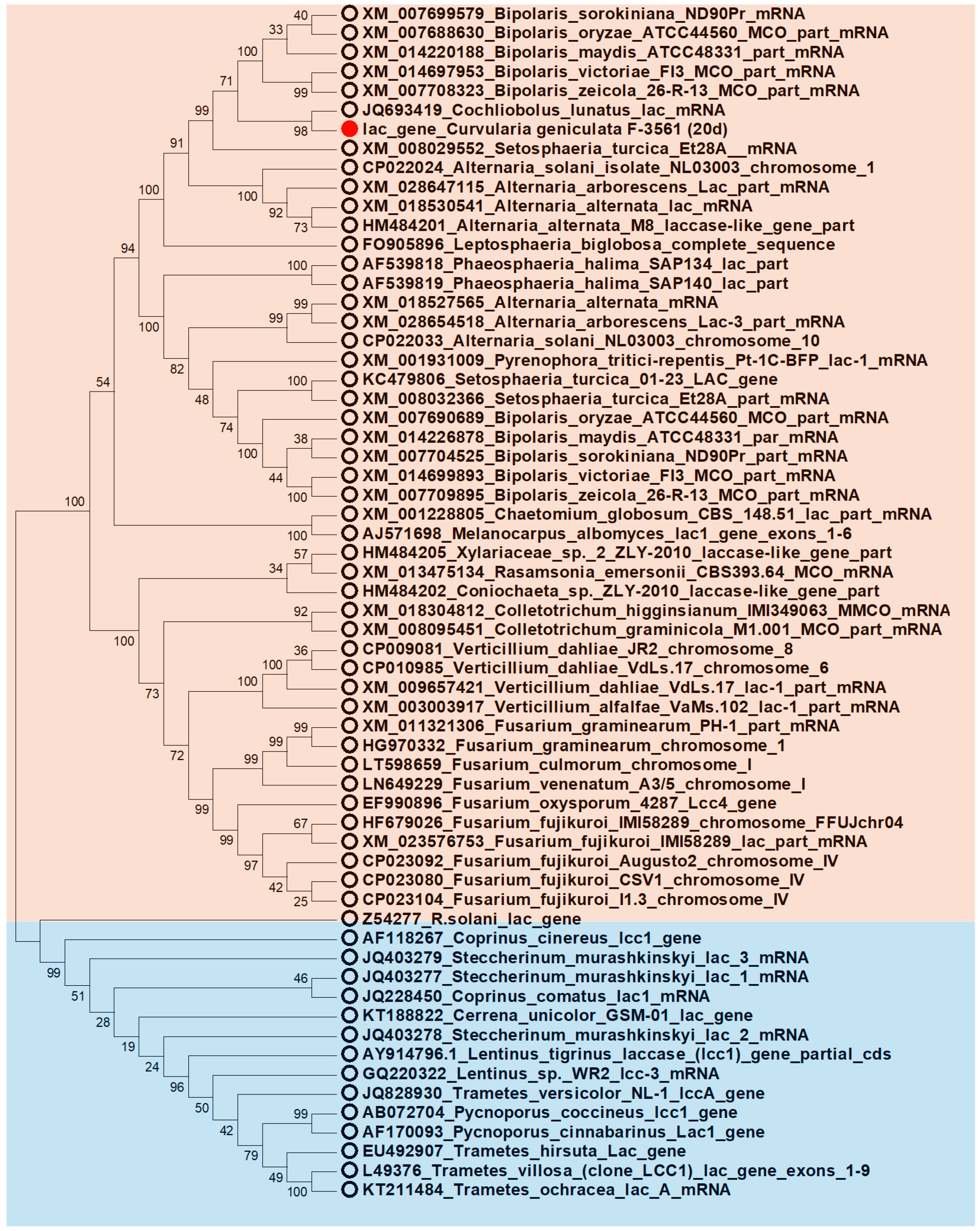
3.7. Identification and Phylogenetic Analysis of the Nucleotide Sequence of the Laccase Gene of C. geniculata VKM F-3561 Expressed at the Peak of Alkaliphilic Laccase Activity
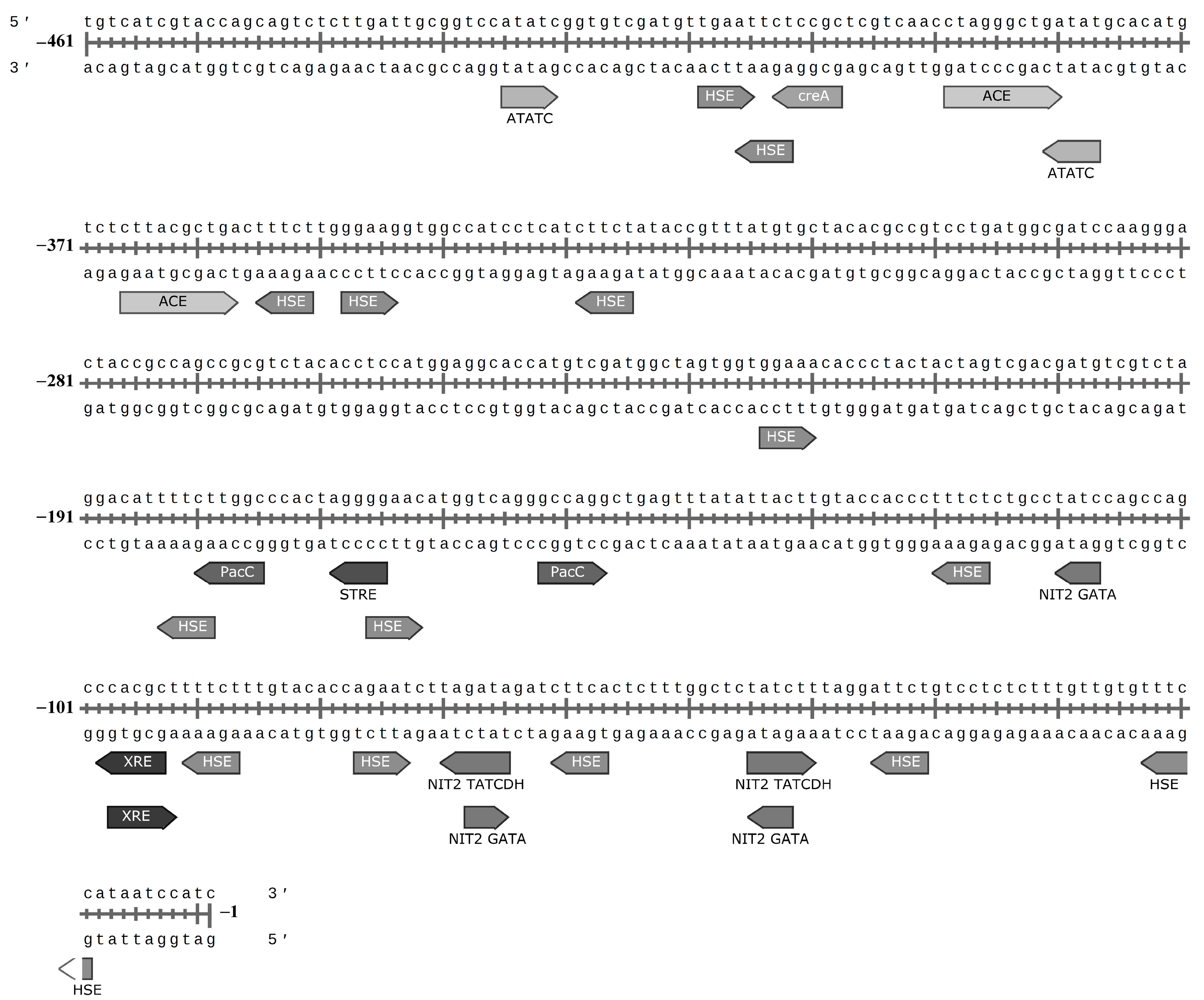
3.8. Regulatory Elements in Promoter Region of the Laccase Gene of C. geniculata VKM F-3561
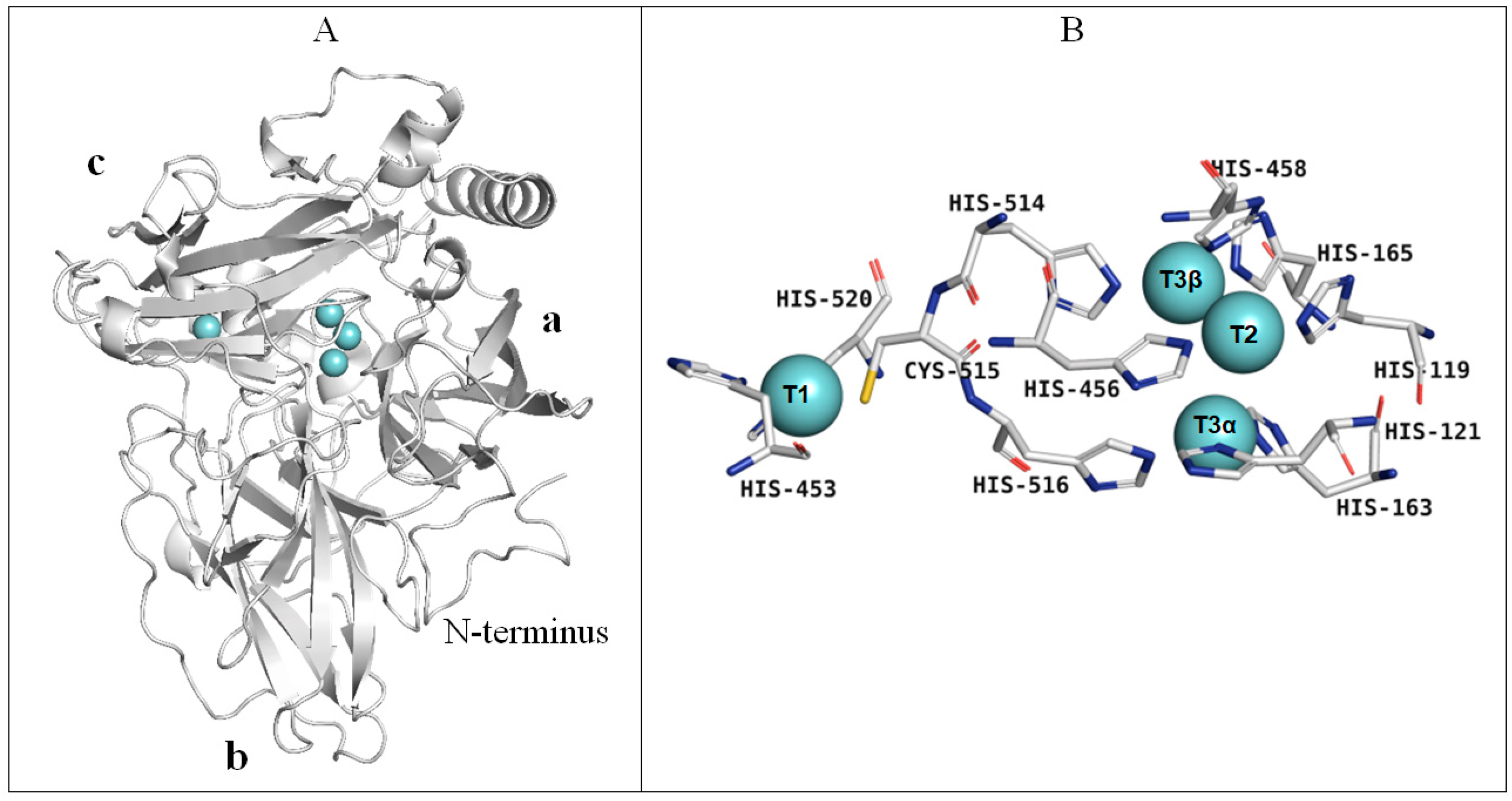
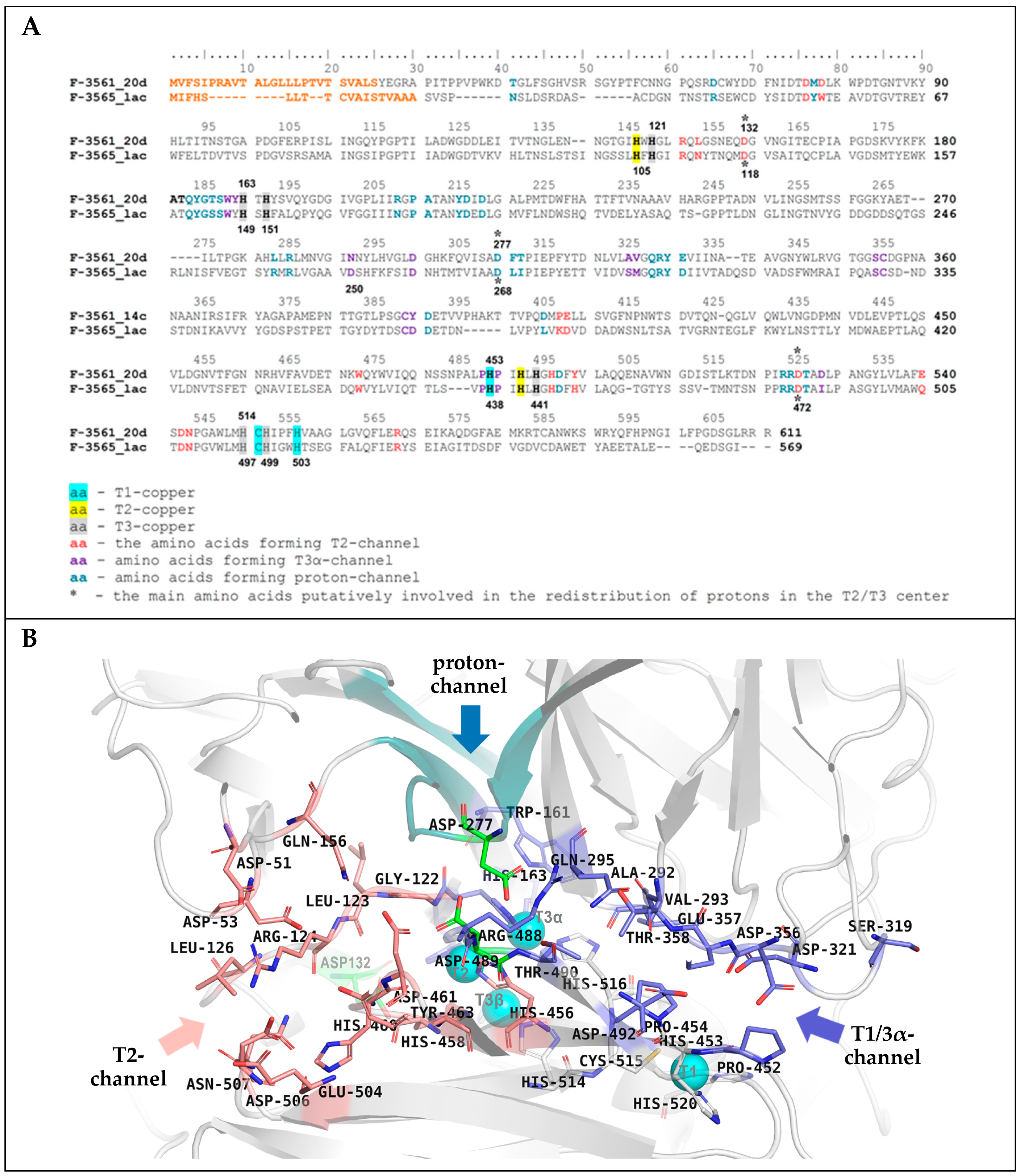
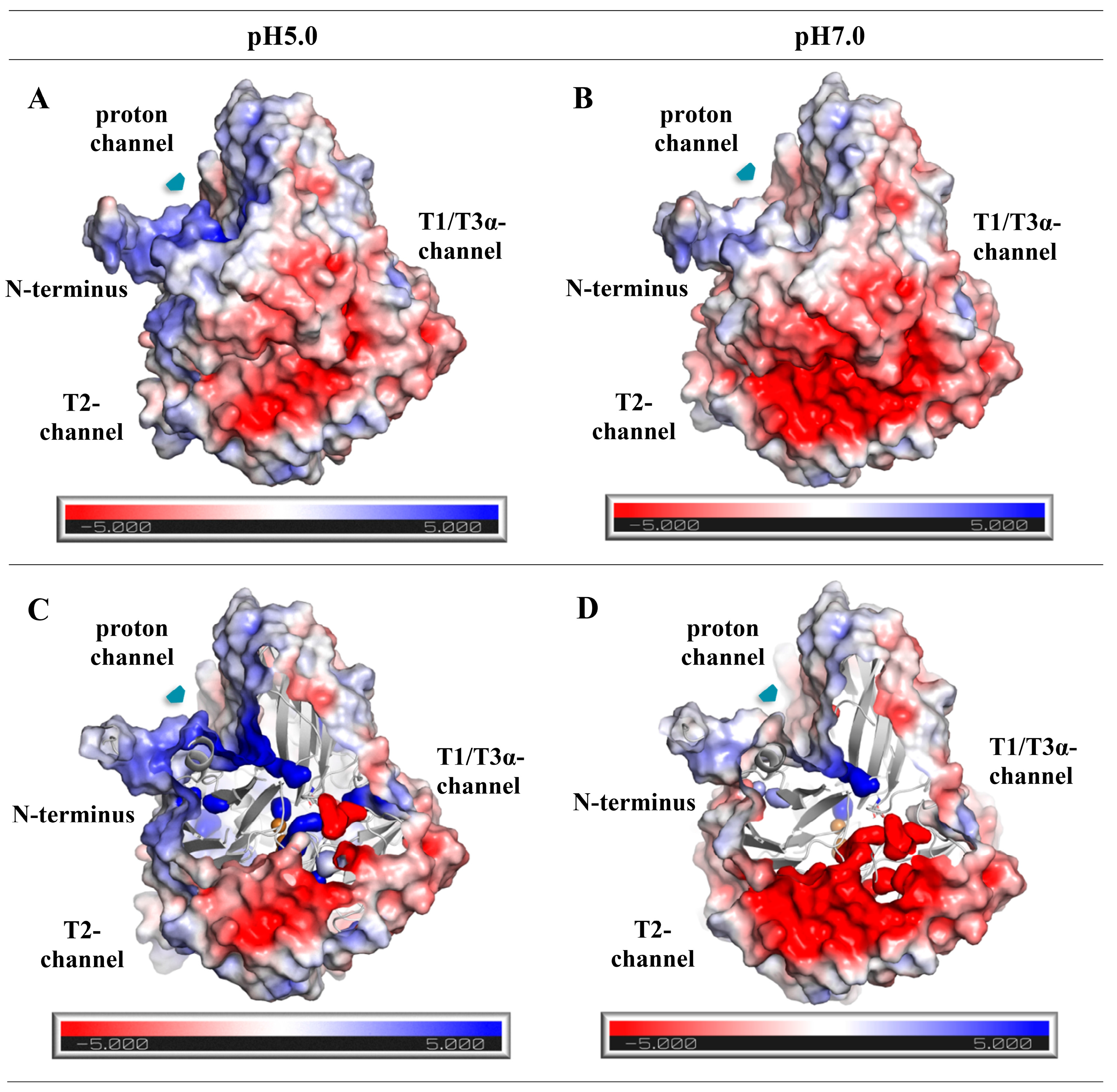
3.9. Homological Model of the Laccase of C. geniculata VKM F-3561 and Structural Analysis
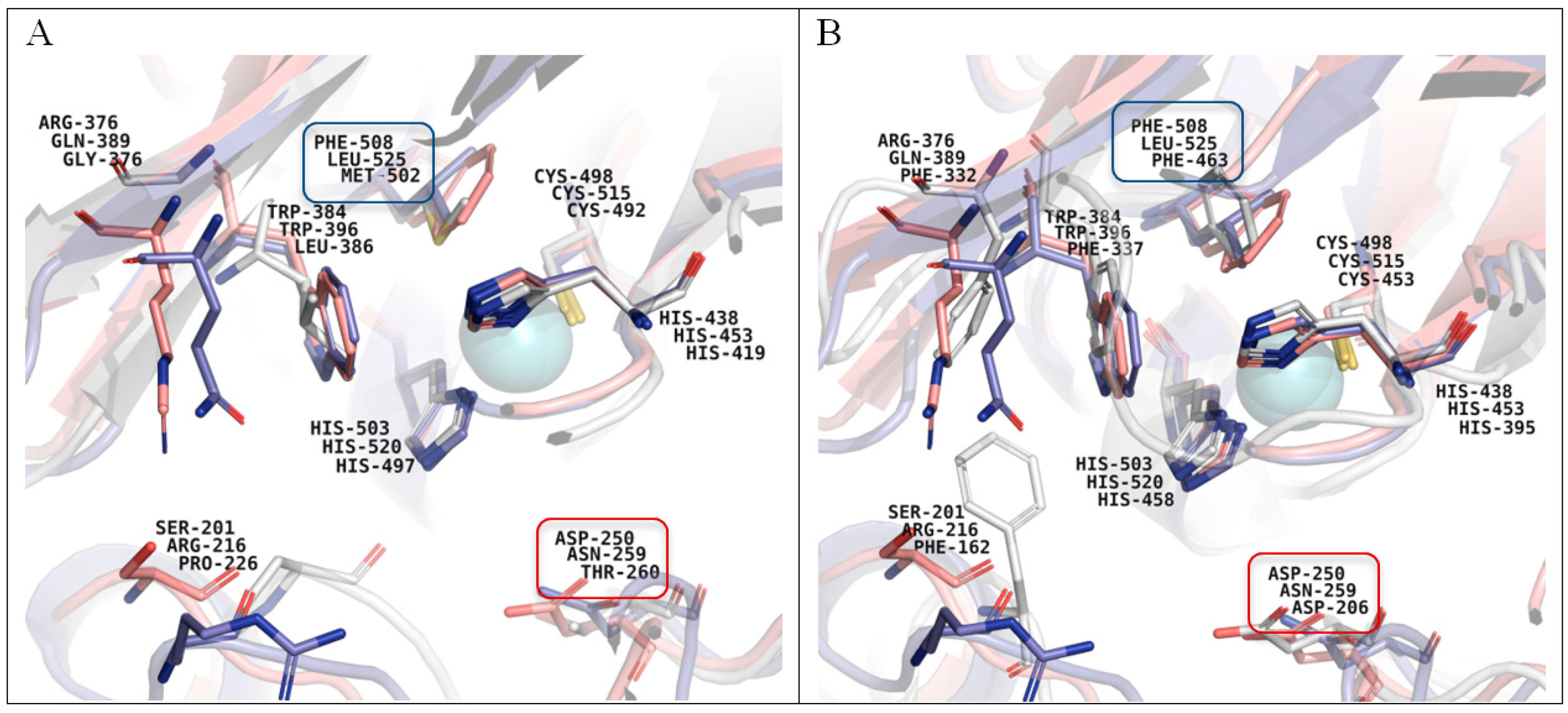
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferry, Y.; Leech, D. Amperometric detection of catecholamine neurotransmitters using electrocatalytic substrate recycling at a laccase electrode. Electroanalysis 2005, 17, 113–119. [Google Scholar] [CrossRef]
- Szczupak, A.; Halámek, J.; Halámková, L.; Bocharova, V.; Alfonta, L.; Katz, E. Living battery—Biofuel cells operating in vivo in clams. Energy Environ. Sci. 2012, 5, 8891–8895. [Google Scholar] [CrossRef]
- Casero, E.; Petit-Domínguez, M.D.; Vázquez, L.; Ramírez-Asperilla, I.; Parra-Alfambra, A.M.; Pariente, F.; Lorenzo, E. Laccase biosensors based on different enzyme immobilization strategies for phenolic compounds determination. Talanta 2013, 115, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Reuillard, B.; Abreu, C.; Lalaoui, N.; Le Goff, A.; Holzinger, M.; Ondel, O.; Buret, F.; Cosnier, S. One-year stability for a glucose/oxygen biofuel cell combined with pH reactivation of the laccase/carbon nanotube biocathode. Bioelectrochemistry 2015, 106, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Vicente, A.I.; Viña-Gonzalez, J.; Santos-Moriano, P.; Marquez-Alvarez, C.; Ballesteros, A.O.; Alcalde, M. Evolved alkaline fungal laccase secreted by Saccharomyces cerevisiae as useful tool for the synthesis of C–N heteropolymeric dye. J. Mol. Catal. B Enzym. 2016, 134, 323–330. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase: A multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Cardullo, N.; Muccilli, V.; Tringali, C. Laccase-mediated synthesis of bioactive natural products and their analogues. RSC Chem. Biol. 2022, 3, 614–647. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Caspersen, M.B.; Mondorf, K.; Halkier, T.; Skov, L.K.; Østergaard, P.R.; Brown, K.M.; Brown, S.H.; Xu, F. Characterization of a Coprinus cinereus laccase. Enzym. Microb. Technol. 1999, 25, 502–508. [Google Scholar] [CrossRef]
- Yaver, D.S.; Overjero, M.D.; Xu, F.; Nelson, B.A.; Brown, K.M.; Halkier, T.; Bernauer, S.; Brown, S.H.; Kauppinen, S. Molecular characterization of laccase genes from the basidiomycete Coprinus cinereus and heterologous expression of the laccase lcc1. Appl. Environ. Microbiol. 1999, 65, 4943–4948. [Google Scholar] [CrossRef]
- Liu, H.; Tong, C.; Du, B.; Liang, S.; Lin, Y. Expression and characterization of LacMP, a novel fungal laccase of Moniliophthora perniciosa FA553. Biotechnol. Lett. 2015, 37, 1829–1835. [Google Scholar] [CrossRef]
- Rogalski, J.; Janutz, G.; Legiec, D.; Cho, N.S.; Shin, S.J.; Ohga, S. Purification of extracellular laccase from Rhizoctonia praticola. J. Fac. Agric. Kyushu Univ. 2011, 56, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wahleithner, J.A.; Xu, F.; Brown, K.M.; Brown, S.H.; Golightly, E.J.; Halkier, T.; Kauppinen, S.; Pederson, A.; Schneider, P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr. Genet. 1996, 29, 395–403. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.G.M.; Peralta, R.M. Purification and characterization of the main laccase produced by the white-rot fungus Pleurotus pulmonarius on wheat bran solid state sedium. J. Basic Microbiol. 2003, 43, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Heinzkill, M.; Bech, L.; Halkier, T.; Schneider, P.; Anke, T. Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl. Environ. Microbiol. 1998, 64, 1601–1606. [Google Scholar] [CrossRef]
- Xu, F.; Shin, W.; Brown, S.H.; Wahleithner, J.A.; Sundaram, U.M.; Solomon, E.I. A sudy of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta 1996, 1292, 303–311. [Google Scholar] [CrossRef]
- Gouka, R.J.; van der Heiden, M.; Swarthoff, T.; Verrips, C.T. Cloning of a phenol oxidase gene from Acremonium murorum and its expression in Aspergillus awamori. Appl. Environ. Microbiol. 2001, 67, 2610–2616. [Google Scholar] [CrossRef] [PubMed]
- Sulistyaningdyah, W.T.; Ogawa, J.; Tanaka, H.; Maeda, C.; Shimizu, S. Characterization of alkaliphilic laccase activity in the culture supernatant of Myrothecium verrucaria 24G-4 in comparison with bilirubin oxidase. FEMS Microbiol. Lett. 2004, 230, 209–214. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Yin, Q.; Fang, W.; Xiao, Y.; Fang, Z. Expression of a thermo-and alkali-philic fungal laccase in Pichia pastoris and its application. Prot. Expr. Purif. 2019, 154, 16–24. [Google Scholar] [CrossRef]
- Saito, K.O.; Ikeda, R.; Endo, K.; Tsujino, Y.; Takagi, M.; Tamiya, E. Isolation of a novel alkaline-induced laccase from Flammulina velutipes and its application for hair coloring. J. Biosci. Bioeng. 2012, 113, 575–579. [Google Scholar] [CrossRef]
- Bonomo, R.P.; Boudet, A.M.; Cozzolino, R.; Rizzarelli, E.; Santoro, A.M.; Sterjiades, R.; Zappalà, R. A comparative study of two isoforms of laccase secreted by the “white-rot” fungus Rigidoporus lignosus, exhibiting significant structural and functional differences. J. Inorg. Biochem. 1998, 71, 205–211. [Google Scholar] [CrossRef]
- Berka, R.M.; Schneider, P.; Golightly, E.J.; Brown, S.H.; Madden, M.; Brown, K.M.; Halkier, T.; Mondorfm, K.; Xu, F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 1997, 63, 3151–3157. [Google Scholar] [CrossRef]
- Kiiskinen, L.-L.; Viikari, L.; Kruus, K. Purification and characterisation of a novel laccase from the ascomycete Melanocarpus albomyces. Appl. Microbiol. Biotechnol. 2002, 59, 198–204. [Google Scholar] [CrossRef]
- Tamayo-Ramos, J.A.; van Berkel, W.J.H.; de Graaff, L.H. Biocatalytic potential of laccase-like multicopper oxidases from Aspergillus niger. Microbial Cell Fact. 2012, 11, 165. [Google Scholar] [CrossRef]
- Ferraroni, M.; Westphal, A.H.; Borsari, M.; Tamayo-Ramos, J.A.; van Berkel, W.J.H. Structure and function of Aspergillus niger laccase McoG. Biocatalysis 2017, 3, 1–21. [Google Scholar] [CrossRef]
- Yang, X.; Gu, C.; Lin, Y. A novel fungal laccase from Sordaria macrospora K-Hell: Expression, characterization, and application for lignin degradation. Bioprocess Biosyst. Eng. 2020, 43, 1133–1139. [Google Scholar] [CrossRef]
- Kolomytseva, M.; Myasoedova, N.; Samoilova, A.; Chernykh, A.; Podieiablonskaia, E.; Classen, T.; Pietruszka, J.; Golovleva, L. Rapid identification of fungal laccases/oxidases with different pH-optimum. Process Biochem. 2017, 62, 174–183. [Google Scholar] [CrossRef]
- Podieiablonskaia, E.V.; Kolomytseva, M.P.; Myasoedova, N.M.; Baskunov, B.P.; Chernykh, A.M.; Classen, T.; Pietruszka, J.; Golovleva, L.A. Myrothecium verrucaria F-3851, a producer of laccases transforming phenolic compounds at neutral and alkaline conditions. Microbiology 2017, 86, 370–376. [Google Scholar] [CrossRef]
- Renfeld, Z.V.; Chernykh, A.M.; Egorova (Shebanova), A.D.; Baskunov, B.P.; Gaidina, A.S.; Myasoedova, N.M.; Kolomytseva, M.P. The laccase of Myrothecium roridum VKM F-3565: A new look at the fungal laccase tolerance to neutral alkaline conditions. ChemBioChem 2023, 24, e202200600. [Google Scholar] [CrossRef]
- Myasoedova, N.M.; Gasanov, N.B.; Chernykh, A.M.; Kolomytseva, M.P.; Golovleva, L.A. Selective regulation of laccase isoform production by the Lentinus strigosus 1566 fungus. Appl. Biochem. Microbiol. 2015, 51, 222–229. [Google Scholar] [CrossRef]
- Perez, J.; Jeffries, T.W. Mineralization of C-ring-labeled synthetic lignin correlates with the production of lignin peroxidase, not of manganese peroxidase or laccase. Appl. Environ. Microbiol. 1990, 56, 1806. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Diezel, W.; Kopperschläger, G.; Hofmann, E. An improved procedure for protein staining in polyacrylamide gels with a new type of Coomassie Brilliant Blue. Anal. Biochem. 1972, 48, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Xu, F.; Kulys, J.J.; Duke, K.; Li, K.C.; Krikstopaitis, K.; Deussen, H.-J.W.; Abbate, E.; Galinyte, V.; Schneider, P. Redox chemistry in laccase-catalyzed oxidation of N-hydroxy compounds. Appl. Environ. Microbiol. 2000, 66, 2052–2056. [Google Scholar] [CrossRef]
- Kolomytseva, M.P.; Myasoedova, N.M.; Chernykh, A.M.; Gaidina (Samoilova), A.S.; Shebanova, A.D.; Baskunov, B.P.; Aschenbrenner, J.; Rosengarten, J.F.; Renfeld, Z.V.; Gasanov, N.B.; et al. Laccase isoform diversity in basidiomycete Lentinus strigosus 1566: Potential for phenylpropanoid polymerization. Int. J. Biol. Macromol. 2019, 137, 1199–1210. [Google Scholar] [CrossRef]
- Leneva, N.A.; Kolomytseva, M.P.; Baskunov, B.P.; Golovleva, L.A. Phenanthrene and anthracene degradation by microorganisms of the genus Rhodococcus. Appl. Biochem. Microbiol. 2009, 45, 188–194. [Google Scholar] [CrossRef]
- Lyons, J.I.; Newell, S.Y.; Buchan, A.; Moran, M.A. Diversity of ascomycete laccase gene seguences in a southeastern US salt marsh. Microb. Ecol. 2003, 45, 270–281. [Google Scholar] [CrossRef]
- Antal, Z.; Rascle, C.; Fèvre, M.; Bruel, C. Single oligonucleotide nested PCR: A rapid method for the isolation of genes and their flanking regions from expressed sequence tags. Curr. Genet. 2004, 46, 240–246. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10.1–S10.12. [Google Scholar] [CrossRef]
- Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Cañero, D.C.; Roncero, M.I.G. Functional analyses of laccase genes from Fusarium oxysporum. Phytopathology 2008, 98, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Kucharzyk, K.H.; Pawlik, A.; Staszczak, M.; Paszczynski, A.J. Fungal laccase, manganese peroxidase and lignin peroxidase: Gene expression and regulation. Enzym. Microb. Technol. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Janusz, G.; Mazur, A.; Checinsks, A.; Malek, W. Cloning and characterization of a lacease gene from biotechnologically important basidiomycete Cerrerna unicolor. J. Fac. Agric. Kyushu Univ. 2012, 57, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins 2005, 61, 704–721. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kolomytseva, M.P.; Myasoedova, N.M.; Podieiablonskaya, E.V.; Renfeld, Z.V.; Chernykh, A.M.; Classen, T.; Pietruszka, J.; Golovleva, L.A. Method for Producing Curvularia geniculata VKM F-3561 Oxidases, Active with Phenolic Compounds in Neutral Environmental Conditions. Patent No. RU2664483C2, 17 August 2018. [Google Scholar]
- Baldrian, P. Fungal laccases—Occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Durrens, P. The phenoloxidases of the ascomycete Podospora anserina: The three forms of the major laccase activity. Arch. Microbiol. 1981, 130, 121–124. [Google Scholar] [CrossRef]
- Lisov, A.V.; Trubitsina, L.I.; Lisova, Z.A.; Trubitsin, I.V.; Zavarzina, A.S.G.; Leontievsky, A.A. Transformation of humic acids by two-domain laccase from Streptomyces anulatus. Process Biochem. 2019, 76, 128–135. [Google Scholar] [CrossRef]
- Otto, B.; Schlosser, D.; Reisser, W. First description of a laccase-like enzyme in soil algae. Arch. Microbiol. 2010, 192, 759–768. [Google Scholar] [CrossRef]
- Otto, B.; Schlosser, D. First laccase in green algae: Purification and characterization of an extracellular phenol oxidase from Tetracystis aeria. Planta 2014, 240, 1225–1236. [Google Scholar] [CrossRef]
- Choy, H.A.; Jones, G.H. Phenoxazinone synthase from Streptomyces antibioticus: Purification of the large and small enzyme forms. Arch. Biochem. Biophys. 1981, 211, 55–65. [Google Scholar] [CrossRef]
- Smith, A.W.; Camara-Artigas, A.; Wang, M.; Allen, J.P.; Francisco, W.A. Structure of phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. Biochemistry 2006, 45, 4378–4387. [Google Scholar] [CrossRef]
- Paavola, J.L.; Battistin, U.; Ogata, C.M.; Georgiadis, M.M. Crystal structures of a dodecameric multicopper oxidase from Marinithermus hydrothermalis. Acta Cryst. 2021, D77, 1336–1345. [Google Scholar] [CrossRef]
- Hildén, K.; Hakala, T.K.; Lundell, T. Thermotolerant and thermostable laccases. Biotechnol. Lett. 2009, 31, 1117–1128. [Google Scholar] [CrossRef]
- Xu, F. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 1997, 272, 924–928. [Google Scholar] [CrossRef]
- Branchi, B.; Galli, C.; Gentili, P. Kinetics of oxidation of benzyl alcohols by the dication and radical cation of ABTS. Comparison with laccase–ABTS oxidations: An apparent paradox. Org. Biomol. Chem. 2005, 3, 2604–2614. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Sahay, R. Fungal laccase as a green catalyst. Int. J. Environ. Agricult. Biotechnol. 2021, 6, 45–58. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; de los Santos, M.H.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell. Fact. 2019, 18, 2–33. [Google Scholar] [CrossRef]
- Loi, M.; Glazunova, O.; Fedorova, T.; Logrieco, A.F.; Mulè, G. Fungal laccases: The forefront of enzymes for sustainability. J. Fungi 2021, 7, 1048. [Google Scholar] [CrossRef] [PubMed]
- Curran, L.M.C.L.K.; Pham, L.T.M.; Sale, K.L.; Simmons, B.A. Review of advances in the development of laccases for the valorization of lignin to enable the production of lignocellulosic biofuels and bioproducts. Biotechnol. Adv. 2022, 54, 107809. [Google Scholar] [CrossRef]
- Gałązka, A.; Jankiewicz, U.; Szczepkowski, A. Biochemical characteristics of laccases and their practical application in the removal of xenobiotics from water. Appl. Sci. 2023, 13, 4394. [Google Scholar] [CrossRef]
- Banerjee, U.C.; Vohra, R.M. Production of laccase by Curvularia sp. Folia Microbiol. 1991, 36, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ren, J.; Yu, Y.; Wang, B. Purification and characterization of laccase from Curvularia trifold. Adv. Mater. Res. 2010, 113, 2215–2219. [Google Scholar] [CrossRef]
- Chernykh, A.M.; Myasoedova, N.M.; Kolomytseva, M.P.; Ferraroni, M.; Scozzafava, A.; Briganti, F.; Golovleva, L.A. Laccase isoforms with unusual properties from the basidiomycete Steccherinum ochraceum strain 1833. J. Appl. Microbiol. 2008, 105, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Kiiskinen, L.-L.; Kruus, K.; Saloheimo, M.; Paananen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Mol. Biol. 2002, 9, 601–605. [Google Scholar] [CrossRef]
- Huang, H.-W.; Zoppellaro, G.; Sakurai, T. Spectroscopic and kinetic studies on the oxygen-centered radical formed during the four-electron reduction process of dioxygen by Rhus vernicifera laccase. J. Biol. Chem. 1999, 274, 32718–32724. [Google Scholar] [CrossRef]
- Lu, R.; Miyakoshi, T. Studies on acetone powder and purified Rhus laccase immobilized on zirconium chloride for oxidation of phenols. Enzym. Res. 2012, 2012, 375309. [Google Scholar] [CrossRef]
- Bourrier, O.; Butlin, J.; Hourani, R.; Kakkar, A.K. Aggregation of 3,5-dihydroxybenzyl alcohol based dendrimers and hyperbranched polymers, and encapsulation of DR1 in such dendritic aggregates. Inorg. Chim. Acta 2004, 357, 3836–3846. [Google Scholar] [CrossRef]
- Bourrier, O.; Kakkar, A.K. Dendritic polymers containing a dimethylsilyl linked dihydroxybenzyl alcohol backbone: Divergent synthesis, aggregation, functionalization, and an evaluation of their applications in catalysis. J. Mater. Chem. 2003, 13, 1306–1315. [Google Scholar] [CrossRef]
- Robles-Almazan, M.; Pulido-Moran, M.; Moreno-Fernandez, J.; Ramirez-Tortosa, C.; Rodriguez-Garcia, C.; Quiles, J.L.; Ramirez-Tortosa, M.C. Hydroxytyrosol: Bioavailability, toxicity, and clinical applications. Food Res. Int. 2017, 105, 654–667. [Google Scholar] [CrossRef]
- Morozova, O.V.; Shumakovich, G.P.; Gorbacheva, M.A.; Shleev, S.V.; Yaropolov, A.I. “Blue” laccases. Biochemistry 2007, 72, 1136–1150. [Google Scholar] [CrossRef]
- Margot, J.; Bennati-Granier, C.; Maillard, J.; Blánquez, P.; Barry, D.A.; Holliger, C. Bacterial versus fungal laccase: Potential for micropollutant degradation. AMB Express 2013, 3, 63. [Google Scholar] [CrossRef] [PubMed]
- Valles, M.; Kamaruddin, A.F.; Wong, L.S.; Blanford, C.F. Inhibition in multicopper oxidases: A critical review. Catal. Sci. Technol. R. Soc. Chem. 2020, 10, 5386–5410. [Google Scholar] [CrossRef]
- Singha, A.; Sekretareva, A.; Tao, L.; Lim, H.; Ha, Y.; Braun, A.; Jones, S.M.; Hedman, B.; Hodgson, K.O.; Britt, R.D.; et al. Tuning the type 1 reduction potential of multicopper oxidases: Uncoupling the effects of electrostatics and H-bonding to histidine ligands. J. Am. Chem. Soc. 2023, 145, 13284–13301. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Madzak, C.; Vadal, R.; Jolivalt, C.; Gentili, P. Concerted electron/proton transfer mechanism in the oxidation of phenols by laccase. ChemBioChem 2013, 14, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Zovo, K.; Pupart, H.; Wieren, A.V.; Gillilan, R.E.; Huang, Q.; Majumdar, S.; Lukk, T. Substitution of the methionine axial ligand of the T1 copper for the fungal-like phenylalanine ligand (M298F) causes local structural perturbations that lead to thermal instability and reduced catalytic efficiency of the small laccase from Streptomyces coelicolor A3(2). ACS Omega 2022, 7, 6184–6194. [Google Scholar] [CrossRef]

| Substrates | Oxidase I | Oxidase II | ||||||
|---|---|---|---|---|---|---|---|---|
| pH- Optimum | Km, µM | kcat, min−1 | kcat/Km, min−1 µM−1 | pH- Optimum | Km, µM | kcat, min−1 | kcat/Km, min−1 µM−1 | |
| ABTS | 2.0 | 280.0 | 10758.0 | 38.4 | 2.0 | 152.0 | 8400.0 | 55.3 |
| syringaldazine | 7.5 | 2.3 | 12364.0 | 5375.7 | 7.0 | 46.0 | 41040.0 | 892.2 |
| 2,6-dimethoxyphenol | 7.5–8.0 | 26.0 | 858.0 | 33.0 | 7.5–8.5 | 108.0 | 840.0 | 7.8 |
| ferulic acid | 7.0 | 37.6 | 48.6 | 1.3 | 6.8 | 51.0 | 885.0 | 17.4 |
| coniferyl alcohol | 7.0–8.0 | 130.0 | 1793.0 | 13.8 | 7.5 | 125.0 | 1472.0 | 11.8 |
| № | Coloring | TLC (11 cm), Rf | Mass Spectrometric Analysis (Probable Structure), The Intensity of Fragment Ions is Indicated in Parentheses, % | |
|---|---|---|---|---|
| By Benzidine Reagent | In Visible Light | |||
| Ferulic acid | ||||
| 1 | light brown | light orange | 0.0 | [M+H]+637(50%), 581(100%)—a condensation product of at least 3 molecules of ferulic acid, with successive elimination during ionization of two fragments with m/z = 56 (CH2=C+–CH=O) and formation of a fragment ion m/z 525. ([M+H]+581(90%), 525(100%)—daughter ion (resistant to ionization), also stable as m/z 525) |
| 2 | brown | light brown | 0.0 | [M+H]+736(100%), 574(42%), 556(90%), 538(23%)—condensation product of 4 molecules of ferulic acid, with successive elimination of m/z = 162 and m/z = 18 or simultaneously m/z = 180 (ferulic acid) during ionization |
| Coniferyl alcohol | ||||
| 3 | no | orange | 0.0 | [M+H]+341(100%), 323(42%), 311(17%), 208(76%)—a polymer |
| 4 | no | no | 0.07 | [M+H]+341(58%), 323(98%), 311(27%), 175(16%), 161(100%), 137(25%)—a polymer |
| Caffeic acid | ||||
| 5 | dark brown | rufous | 0.0 | [M+H]+141(100%), 123(80%), 95(85%), 77(20%)—hydroxybenzyl alcohol with m/z = 140—(HO)2C6H3CH2OH |
| 6 | dark brown | rufous | 0.0 | [M-H]−153(%), 137(7%), 120(10%), 108(40%), 88(100%)—hydroxytyrosol with m/z = 154 (HO)2C6H3CH2CH2OH |
| Guaiacol | ||||
| 7 | no | light cherry | 0.29 | [M-H]−455(90%), 325(100%), 294(40%)—presumably a heterotrimer with m/z = 456 |
| 8 | no | light cherry | 0.29 | [M-H]−473(60%), 458(10%), 413(100%), 365(15%)—presumably heterotri(tetra)mer with m/z = 474 |
| 9 | light orange | light yellow | 0.41 | [M+H]+369(45%), 337(100%), 305(40%), 245(20%)—a product of polymerization of three molecules of guaiacol with elimination of 4 protons, m/z = 368. Sequential elimination of two methoxyl groups is observed. The same metabolite is detected in negative ions [M-H]−367(90%), 352(100%) |
| 10 | light orange | light yellow | 0.41 | [M+H]+247(95%), 215(100%), 183(35%)—a product of polymerization of two molecules of guaiacol with elimination of 2 protons, m/z = 246. The spectrum shows the sequential elimination of two methoxyl groups. The same metabolite is detected in negative ions—[M-H]−245(72%), 230(100%) |
| 11 | light orange | red-brown | 0.45 | [M+H]+489(100%), 457(80%), 425(50%), 366(10%), 259(100%), 213(40%), 185(10%)—presumably the condensation product of 4 molecules of guaiacol with the elimination of 8 protons, m/z = 488 |
| 12 | light orange | red-brown | 0.45 | [M+H]+261(95%), 229(100%)—presumably a condensation product of 2 molecules of guaiacol with monohydroxylation and concomitant elimination of 4 protons (m/z = 260) |
| 13 | light orange | red-brown | 0.45 | [M-H]−275(80%), 260(100%), 247(20%)—a product of condensation of 2 molecules of guaiacol with a dihydroxyl derivative and the formation of two bonds |
| 14 | light orange | light orange | 0.51 | [M-H]−271(100%), 253(45%), 243(20%), 192(30%)—presumably methylated guaiacol dimer with a double bond (CH2)2), m/z = 272 |
| 15 | light orange | light orange | 0.51 | [M-H]−245(100%), 244(51%), 230(90%), 229(65%)—a product of guaiacol condensation with the formation of one bond (124 × 2 = 248-H2) = 246 |
| 3-methoxycatechol | ||||
| 16 | dark brown | red-brown | 0.0 | [M+H]+317(70%), 259(100%) or [M-H]−315(100%), 297(35%), 271(70%)—presumably a dimer with m/z = 316 |
| 17 | dark brown | red-brown | 0.0 | [M-H]−289(100%), 245(90%), 226(15%)—presumably a dimer of the parent compound, m/z = 290 |
| 18 | dark brown | red-brown | 0.0 | [M-H]−181(55%), 139(100%)—putative methylation product ((CH2)3) of parent compound, m/z = 182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renfeld, Z.V.; Chernykh, A.M.; Baskunov, B.P.; Gaidina, A.S.; Myasoedova, N.M.; Egorova, A.D.; Moiseeva, O.V.; Gorina, S.Y.; Kolomytseva, M.P. Unusual Oligomeric Laccase-like Oxidases from Ascomycete Curvularia geniculata VKM F-3561 Polymerizing Phenylpropanoids and Phenolic Compounds under Neutral Environmental Conditions. Microorganisms 2023, 11, 2698. https://doi.org/10.3390/microorganisms11112698
Renfeld ZV, Chernykh AM, Baskunov BP, Gaidina AS, Myasoedova NM, Egorova AD, Moiseeva OV, Gorina SY, Kolomytseva MP. Unusual Oligomeric Laccase-like Oxidases from Ascomycete Curvularia geniculata VKM F-3561 Polymerizing Phenylpropanoids and Phenolic Compounds under Neutral Environmental Conditions. Microorganisms. 2023; 11(11):2698. https://doi.org/10.3390/microorganisms11112698
Chicago/Turabian StyleRenfeld, Zhanna V., Alexey M. Chernykh, Boris P. Baskunov, Anastasya S. Gaidina, Nina M. Myasoedova, Anna D. Egorova, Olga V. Moiseeva, Sophya Yu Gorina, and Marina P. Kolomytseva. 2023. "Unusual Oligomeric Laccase-like Oxidases from Ascomycete Curvularia geniculata VKM F-3561 Polymerizing Phenylpropanoids and Phenolic Compounds under Neutral Environmental Conditions" Microorganisms 11, no. 11: 2698. https://doi.org/10.3390/microorganisms11112698





