Development of a Melting-Curve-Based Multiplex Real-Time PCR Assay for the Simultaneous Detection of Viruses Causing Respiratory Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oligonucleotide Primers and Positive Controls
2.2. Viral and Microbial Strains
2.3. Nucleic Acid Purification
2.4. PCR Design
2.5. Analytical Specificity, Sensitivity, and Performance in Virus-Spiked Swabs
2.6. Performance of the Multiplex Real-Time PCR Assay in Clinical Samples
3. Results and Discussion
3.1. Melting-Curve-Based Multiplex Real-Time PCR Assay
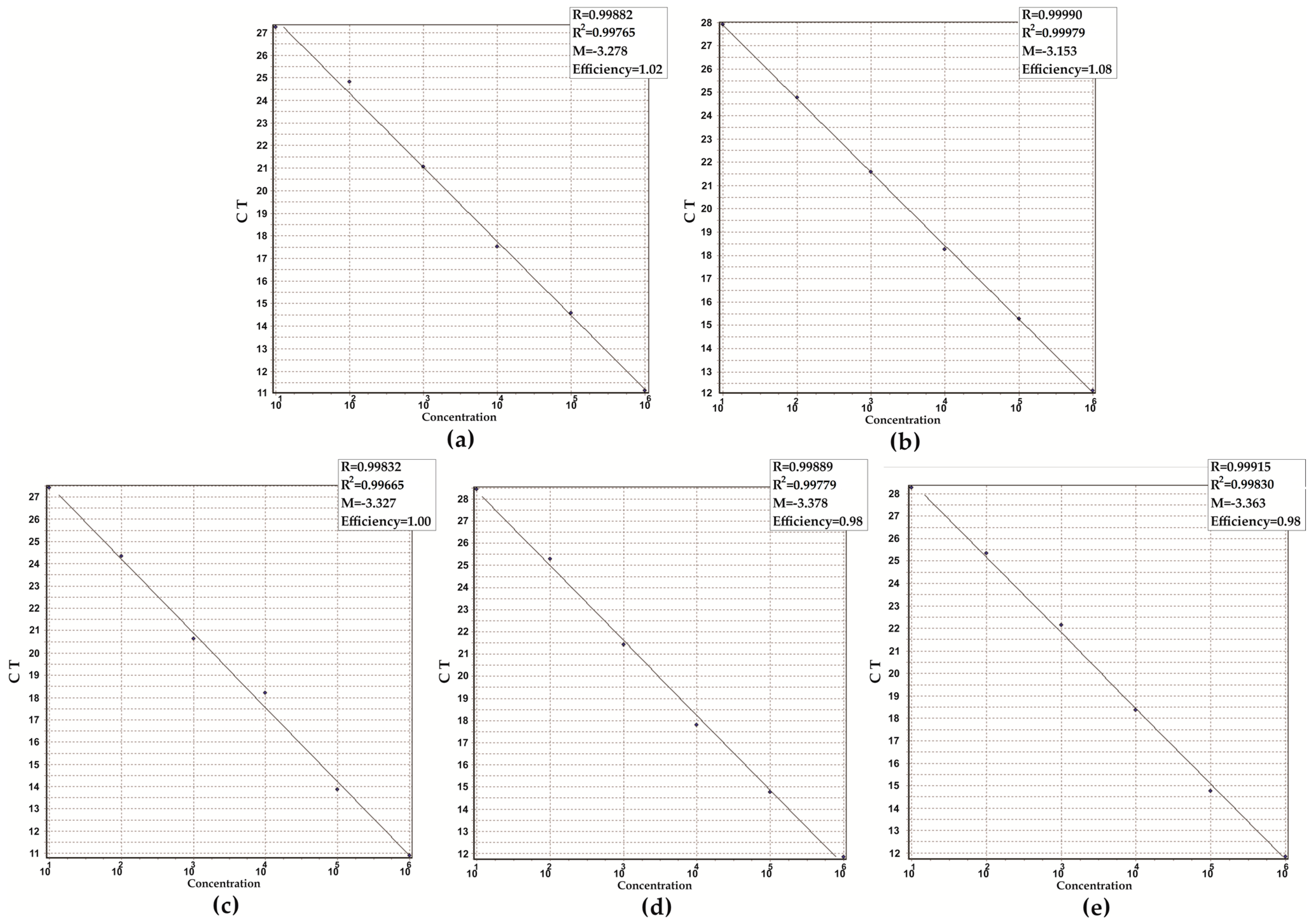
3.2. Analytical Performance of the Assay
3.3. Performance of the Assay in Clinical Samples
4. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cunha, B.A. Influenza: Historical aspects of epidemics and pandemics. Infect. Dis. Clin. N. Am. 2004, 18, 141–155. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus Disease (COVID-19). Available online: https://www.who.int/health-topics/coronavirus#tab=tab_1 (accessed on 19 October 2023).
- Guest, P.C.; Hawkins, S.F.C.; Rahmoune, H. Rapid Detection of SARS-CoV-2 Variants of Concern by Genomic Surveillance Techniques. Adv. Exp. Med. Biol. 2023, 1412, 491–509. [Google Scholar] [PubMed]
- Ramaswamy, K.; Rashid, M.; Ramasamy, S.; Jayavelu, T.; Venkataraman, S. Revisiting Viral RNA-Dependent RNA Polymerases: Insights from Recent Structural Studies. Viruses 2022, 14, 2200. [Google Scholar] [CrossRef] [PubMed]
- Langedijk, A.C.; Bont, L.J. Respiratory syncytial virus infection and novel interventions. Nat. Rev. Microbiol. 2023, 21, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y. Pathogenicity and virulence of influenza. Virulence 2023, 14, 2223057. [Google Scholar] [CrossRef] [PubMed]
- Ljubin-Sternak, S.; Meštrović, T. Rhinovirus-A True Respiratory Threat or a Common Inconvenience of Childhood? Viruses 2023, 15, 825. [Google Scholar] [CrossRef]
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, 7, 83–101. [Google Scholar] [CrossRef]
- Nikolai, L.A.; Meyer, C.G.; Kremsner, P.G.; Velavan, T.P. Asymptomatic SARS Coronavirus 2 infection: Invisible yet invincible. Int. J. Infect. Dis. 2020, 100, 112–116. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Costa, R.; Bueno, F.; Albert, E.; Torres, I.; Carbonell-Sahuquillo, S.; Barrés-Fernández, A.; Sánchez, D.; Padrón, C.; Colomina, J.; Lázaro Carreño, M.I.; et al. Upper respiratory tract SARS-CoV-2 RNA loads in symptomatic and asymptomatic children and adults. Clin. Microbiol. Infect. 2021, 27, 1858.e1–1858.e7. [Google Scholar] [CrossRef]
- Dorta-Gorrín, A.; Navas-Méndez, J.; Gozalo-Margüello, M.; Miralles, L.; García-Hevia, L. Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT). Int. J. Mol. Sci. 2023, 24, 10233. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Varela, F.H.; Scotta, M.C.; Polese-Bonatto, M.; Sartor, I.T.S.; Ferreira, C.F.; Fernandes, I.R.; Zavaglia, G.O.; de Almeida, W.A.F.; Arakaki-Sanchez, D.; Pinto, L.A.; et al. Absence of detection of RSV and influenza during the COVID-19 pandemic in a Brazilian cohort: Likely role of lower transmission in the community. J. Glob. Health 2021, 11, 05007. [Google Scholar] [CrossRef] [PubMed]
- Habbous, S.; Hota, S.; Allen, V.G.; Henry, M.; Hellsten, E. Changes in hospitalizations and emergency department respiratory viral diagnosis trends before and during the COVID-19 pandemic in Ontario, Canada. PLoS ONE 2023, 18, e0287395. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Autore, G.; Ramundo, G.; Esposito, S. Epidemiology of Respiratory Infections during the COVID-19 Pandemic. Viruses 2023, 15, 1160. [Google Scholar] [CrossRef] [PubMed]
- Guido, G.; Lalle, E.; Mosti, S.; Mencarini, P.; Lapa, D.; Libertone, R.; Ianniello, S.; Ricciuto, G.M.; Vaia, F.; Maggi, F.; et al. Recovery from Triple Infection with SARS-CoV-2, RSV and Influenza virus: A case report. J. Infect. Public Health 2023, 16, 1045–1047. [Google Scholar] [CrossRef]
- Malveste Ito, C.R.; Moreira, A.L.E.; Silva, P.A.N.D.; Santos, M.O.; Santos, A.P.D.; Rézio, G.S.; Brito, P.N.; Rezende, A.P.C.; Fonseca, J.G.; Peixoto, F.A.O.; et al. Viral Coinfection of Children Hospitalized with Severe Acute Respiratory Infections during COVID-19 Pandemic. Biomedicines 2023, 11, 1402. [Google Scholar] [CrossRef]
- Maltezou, H.C.; Papanikolopoulou, A.; Vassiliu, S.; Theodoridou, K.; Nikolopoulou, G.; Sipsas, N.V. COVID-19 and Respiratory Virus Co-Infections: A Systematic Review of the Literature. Viruses 2023, 15, 865. [Google Scholar] [CrossRef]
- WHO Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 20 September 2023).
- Luo, J.; Zhang, Z.; Zhao, S.; Gao, R. A Comparison of Etiology, Pathogenesis, Vaccinal and Antiviral Drug Development between Influenza and COVID-19. Int. J. Mol. Sci. 2023, 24, 6369. [Google Scholar] [CrossRef]
- Pillai, T.K.; Johnson, K.E.; Song, T.; Gregianini, T.S.; Tatiana, G.; Wang, G.; Medina, R.A.; Van Bakel, H.; García-Sastre, A.; Nelson, M.I.; et al. Tracking the emergence of antigenic variants in influenza A virus epidemics in Brazil. Virus Evol. 2023, 9, vead027. [Google Scholar] [CrossRef]
- Fontes, V.; Ferreira, H.; Ribeiro, M.; Pinheiro, A.; Maramaldo, C.; Pereira, E.; Batista, L.; Júnior, A.; Lobato, L.; Silva, F.; et al. High Incidence of Respiratory Syncytial Virus in Children with Community-Acquired Pneumonia from a City in the Brazilian Pre-Amazon Region. Viruses 2023, 15, 1306. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Van-Tam, J.S.; O’Leary, M.; Martin, E.T.; Heijnen, E.; Callendret, B.; Fleischhackl, R.; Comeaux, C.; Tran, T.M.P.; Weber, K. Burden of respiratory syncytial virus infection in older and high-risk adults: A systematic review and meta-analysis of the evidence from developed countries. Eur. Respir. Rev. 2022, 31, 220105. [Google Scholar] [CrossRef] [PubMed]
- Altman, J.D.; Rouse, B.T. The Long-Awaited Respiratory Syncytial Virus Vaccine. J. Interferon Cytokine Res. 2023, 43, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.L.D.S.; Costa, K.L.P.; Cariolano, M.S.; Oliveira, G.S.; Felipe, K.K.P.; Silva, E.S.A.; Alves, M.S.; Maramaldo, C.E.C.; de Sousa, E.M.; Rego, J.S.; et al. High incidence of rhinovirus infection in children with community-acquired pneumonia from a city in the Brazilian pre-Amazon region. J. Med. Virol. 2019, 91, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.L.E.; da Silva, P.A.N.; Assunção, L.D.P.; Santos, M.O.; Ito, C.R.M.; de Araújo, K.M.; Cunha, M.O.; Rabelo, V.D.C.; de Souza, P.P.; Maia, S.B.S.; et al. Profile analysis of emerging respiratory virus in children. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 873–882. [Google Scholar] [CrossRef]
- Nakagome, K.; Nagata, M. Innate Immune Responses by Respiratory Viruses, Including Rhinovirus, During Asthma Exacerbation. Front. Immunol. 2022, 13, 865973. [Google Scholar] [CrossRef] [PubMed]
- Brendish, N.J.; Malachira, A.K.; Beard, K.R.; Ewings, S.; Clark, T.W. Impact of turnaround time on outcome with point-of-care testing for respiratory viruses: A post hoc analysis from a randomised controlled trial. Eur. Respir. J. 2018, 52, 1800555. [Google Scholar] [CrossRef]
- Hodinka, R.L. Point: Is the era of viral culture over in the clinical microbiology laboratory? J. Clin. Microbiol. 2013, 51, 2–4. [Google Scholar] [CrossRef]
- Otaguiri, E.S.; Morguette, A.E.B.; Morey, A.T.; Tavares, E.R.; Kerbauy, G.; de Almeida Torres, R.S.L.; Chaves Júnior, M.; Tognim, M.C.B.; Góes, V.M.; Krieger, M.A.; et al. Development of a melting-curve based multiplex real-time PCR assay for simultaneous detection of Streptococcus agalactiae and genes encoding resistance to macrolides and lincosamides. BMC Pregnancy Childbirth 2018, 18, 126. [Google Scholar] [CrossRef]
- Gunson, R.N.; Collins, T.C.; Carman, W.F. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J. Clin. Virol. 2005, 33, 341–344. [Google Scholar] [CrossRef]
- Paulino, R.D.S.; Benega, M.A.; Santos, K.C.; Silva, D.B.; Pereira, J.C.; Sasaki, N.A.; Silva, P.E.; Curti, S.P.; Oliveira, M.I.; Carvalhanas, T.R.; et al. Differential diagnosis of respiratory viruses by using real time RT-PCR methodology. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 432. [Google Scholar] [CrossRef] [PubMed]
- WHO Information for the Molecular Detection of Influenza Viruses. Available online: https://cdn.who.int/media/docs/default-source/influenza/molecular-detention-of-influenza-viruses/protocols_influenza_virus_detection_feb_2021.pdf?sfvrsn=df7d268a_5 (accessed on 20 September 2023).
- Hayes, E.K.; Gouthro, M.T.; LeBlanc, J.J.; Gagnon, G.A. Simultaneous detection of SARS-CoV-2, influenza A, respiratory syncytial virus, and measles in wastewater by multiplex RT-qPCR. Sci. Total Environ. 2023, 889, 164261. [Google Scholar] [CrossRef] [PubMed]
- Altman, S. The road to RNase P. Nat. Struct. Biol. 2000, 7, 827–828. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Killington, R.A. Virology Methods Manual; Mahy, B., Kangro, H., Eds.; Academic Press: Amsterdam, The Netherlands, 1996; Chapter 2; pp. 25–46. [Google Scholar]
- Ghezzi, C.E.; Hartigan, D.R.; Hardick, J.P.; Gore, R.; Adelfio, M.; Diaz, A.R.; McGuinness, P.D.; Robinson, M.L.; Buchholz, B.O.; Manabe, Y.C. Preclinical Validation of a Novel Injection-Molded Swab for the Molecular Assay Detection of SARS-CoV-2. Diagnostics 2022, 12, 206. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Bauer, L.; Lyoo, H.; van der Schaar, H.M.; Strating, J.R.; van Kuppeveld, F.J. Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr. Opin. Virol. 2017, 24, 1–8. [Google Scholar] [CrossRef]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef]
- Royster, A.; Ren, S.; Ma, Y.; Pintado, M.; Kahng, E.; Rowan, S.; Mir, S.; Mir, M. SARS-CoV-2 Nucleocapsid Protein Is a Potential Therapeutic Target for Anticoronavirus Drug Discovery. Microbiol. Spectr. 2023, 11, e0118623. [Google Scholar] [CrossRef]
- Sutto-Ortiz, P.; Eléouët, J.F.; Ferron, F.; Decroly, E. Biochemistry of the Respiratory Syncytial Virus L Protein Embedding RNA Polymerase and Capping Activities. Viruses 2023, 15, 341. [Google Scholar] [CrossRef]
- Burns, B.L.; Moody, D.; Tu, Z.J.; Nakitandwe, J.; Brock, J.E.; Bosler, D.; Mitchell, S.L.; Loeffelholz, M.J.; Rhoads, D.D. Design and Implementation of Improved SARS-CoV-2 Diagnostic Assays to Mitigate the Impact of Genomic Mutations on Target Failure: The Xpert Xpress SARS-CoV-2 Experience. Microbiol. Spectr. 2022, 10, e0135522. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Lan, L.; Ramalingam, M.; He, J.; Yang, Y.; Gao, M.; Shi, Z. A review of current effective COVID-19 testing methods and quality control. Arch. Microbiol. 2023, 205, 239. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, Y.; Zhou, H.; Sun, C.; Zhang, S. The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J. 2023, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Mönttinen, H.A.M.; Ravantti, J.J.; Poranen, M.M. Structure Unveils Relationships between RNA Virus Polymerases. Viruses 2021, 13, 313. [Google Scholar] [CrossRef] [PubMed]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser-Liggett, C.M.; Liggett, S.B. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 2009, 324, 55–59. [Google Scholar] [CrossRef]
- Pasqualotto, A.C.; Seus, A.L. COVID-19 PCR: Frequency of internal control inhibition in clinical practice. Access Microbiol. 2023, 5, acmi000478.v3. [Google Scholar] [CrossRef]
- Lei, C.; Yang, J.; Hu, J.; Sun, X. On the calculation of TCID50 for quantitation of virus infectivity. Virol. Sin. 2021, 36, 141–144. [Google Scholar] [CrossRef]
- Tavares, E.R.; Azevedo, C.S.; Panagio, L.A.; Pelisson, M.; Pinge-Filho, P.; Venancio, E.J.; Barros, T.F.; Yamada-Ogatta, S.F.; Yamauchi, L.M. Accurate and sensitive real-time PCR assays using intergenic spacer 1 region to differentiate Cryptococcus gattii sensu lato and Cryptococcus neoformans sensu lato. Med. Mycol. 2016, 54, 89–96. [Google Scholar]
- Sun, L.; Wang, L.; Zhang, C.; Xiao, Y.; Zhang, L.; Zhao, Z.; Ren, L.; Peng, J. Rapid Detection of Predominant SARS-CoV-2 Variants Using Multiplex High-Resolution Melting Analysis. Microbiol. Spectr. 2023, 11, e0005523. [Google Scholar] [CrossRef]
- Biancolella, M.; Colona, V.L.; Mehrian-Shai, R.; Watt, J.L.; Luzzatto, L.; Novelli, G.; Reichardt, J.K.V. COVID-19 2022 update: Transition of the pandemic to the endemic phase. Hum. Genom. 2022, 16, 19. [Google Scholar] [CrossRef]
- Biancolella, M.; Colona, V.L.; Luzzatto, L.; Watt, J.L.; Mattiuz, G.; Conticello, S.G.; Kaminski, N.; Mehrian-Shai, R.; Ko, A.I.; Gonsalves, G.S.; et al. COVID-19 annual update: A narrative review. Hum. Genom. 2023, 17, 68. [Google Scholar] [CrossRef] [PubMed]
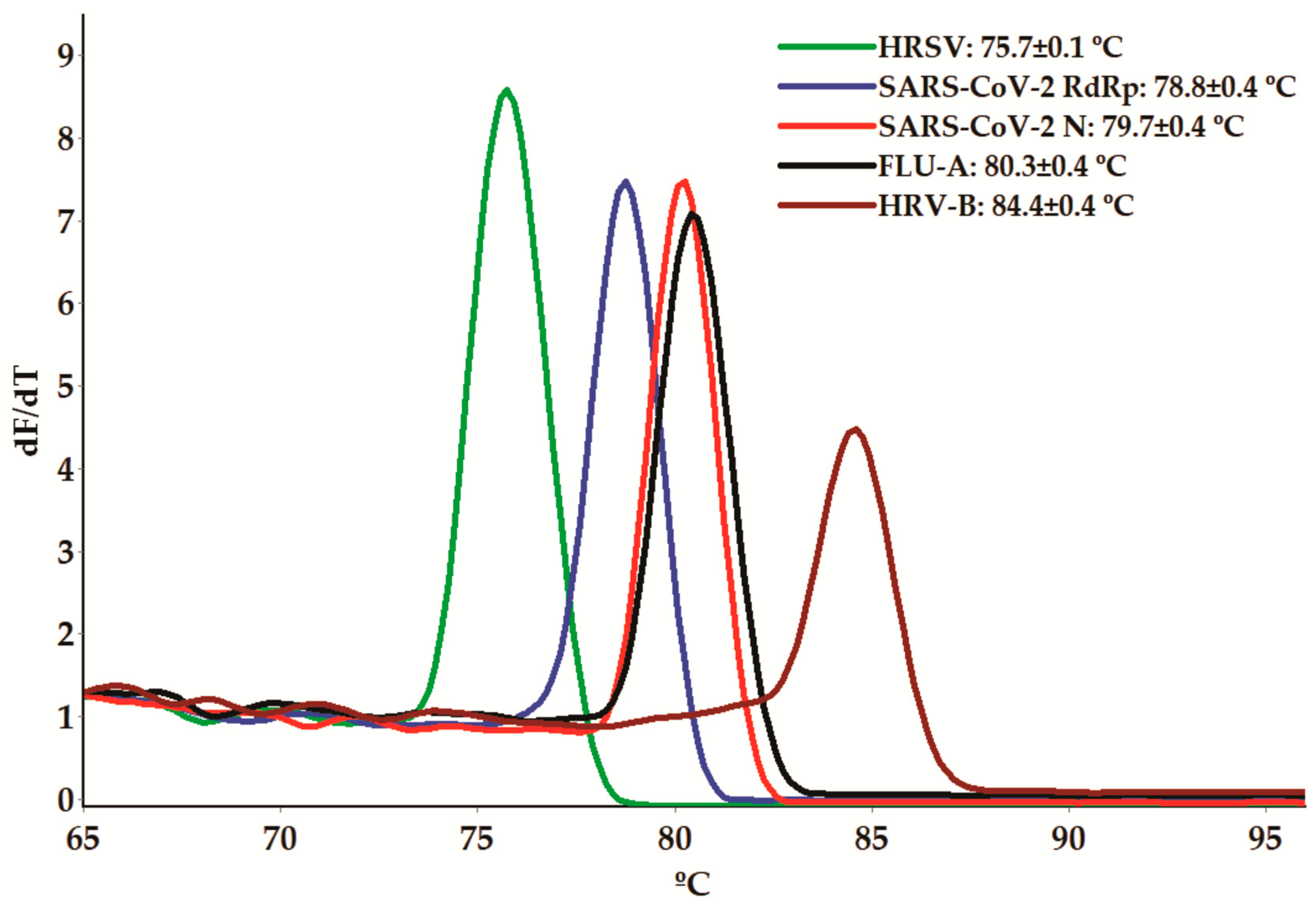
| Virus | Gene | Primers | Amplicon Size (pb) | Annealing Temperature (°C) | |
|---|---|---|---|---|---|
| SARS-CoV-2 | ORF1ab | RdRp-SARS-CoV-2-F | AGCTTGTCACACCGTTTCT | 282 | 61 |
| RdRp-SARS-CoV-2-R | AGTCTGTGTCAACATCTCTATTTCT | ||||
| N | NC-SARS-CoV-2-F | ACACCAATAGCAGTCCAGATG | 197 | 61 | |
| NC-SARS-CoV-2-R | ATTCAAGGCTCCCTCAGTTG | ||||
| FLU-A | M2 | M-IAV-F | TCTAACCGAGGTCGAAACGTA | 142 | 61 |
| M-IAV-R | TGGTCTTGTCTTTAGCCATTCC | ||||
| HRSV | L | RdRp-HRSV- F | AGAGAGGACCCACTAAACCA | 138 | 61 |
| RdRp-HRSV-R | ATGCATACACCCAATCCAAT | ||||
| HRV-B | 5′-UTR | pp-HRV-F | GTGTTCGATCAGGTGAGTTT | 143 | 61 |
| pp-HRV-R | CGAGTCTTCACACCATGTC | ||||
| Targets | |||||
|---|---|---|---|---|---|
| RdRp-SARS-CoV-2 | N-SARS-CoV-2 | M2-FLU-A | L-HRSV | 5′-UTR-HRV-B | |
| Positive Control | + | + | + | + | + |
| SARS-CoV-2 LMM 38135 | + | + | − | − | − |
| SARS-CoV-2 MAM 87209 | + | + | − | − | − |
| SARS-CoV-2 HM11-20 | + | + | − | − | − |
| SARS-CoV-2 LFL11-20 | + | + | − | − | − |
| SARS-CoV-2 AGBCS11-20 | + | + | − | − | − |
| SARS-CoV-2 PQB12-20 | + | + | − | − | − |
| SARS-CoV-2 AOC12-20 | + | + | − | − | − |
| SARS-CoV-2 CNPB11-20 | + | + | − | − | − |
| SARS-CoV-2 MPC12-20 | + | + | − | − | − |
| SARS-CoV-2 MKA01-21 | + | + | − | − | − |
| SARS-CoV-2 BWS02-21 | + | + | − | − | − |
| SARS-CoV-2 VCS02-21 | + | + | − | − | − |
| SARS-CoV-2 EOC02-21 | + | + | − | − | − |
| SARS-CoV-2 SMGC02-21 | + | + | − | − | − |
| SARS-CoV-2 VRS02-21 | + | + | − | − | − |
| SARS-CoV-2 SS09-21 | + | + | − | − | − |
| SARS-CoV-2 ARM09-21 | + | + | − | − | − |
| SARS-CoV-2 CHB09-21 | + | + | − | − | − |
| SARS-CoV-2 ALMO12-21 | + | + | − | − | − |
| SARS-CoV-2 MPV10-21 | + | + | − | − | − |
| SARS-CoV-2 DIBM12-21 | + | + | − | − | − |
| SARS-CoV-2 PLCSC01-22 | + | + | − | − | − |
| Human Rhinovirus HRV 3760 | − | − | − | − | + |
| Human Rhinovirus HRV HRV01 | − | − | − | − | + |
| Human Rhinovirus HRV HRV02 | − | − | − | − | + |
| Human Respiratory Syncytial Virus HRSV 3760 | − | − | − | + | − |
| Human Respiratory Syncytial Virus HRSV 4226 | − | − | − | + | − |
| Human Respiratory Syncytial Virus HRSV 4122 | − | − | − | + | − |
| Human Respiratory Syncytial Virus HRSV IVC1 | − | − | − | + | − |
| Human Respiratory Syncytial Virus HRSV IVC2 | − | − | − | + | − |
| Human Respiratory Syncytial Virus HRSV IVC3 | − | − | − | + | − |
| Influenza Virus A FLU-A/H1N1 FLU | − | − | + | − | − |
| Influenza Virus A FLU-A/H3N2 | − | − | + | − | − |
| Influenza Virus A FLU-A CS1 | − | − | + | − | − |
| Influenza Virus A FLU-A CS2 | − | − | + | − | − |
| Influenza Virus A FLU-A CS3 | − | − | + | − | − |
| Influenza Virus A FLU-A CS4 | − | − | + | − | − |
| Influenza Virus A FLU-A CS5 | − | − | + | − | − |
| Influenza Virus B | − | − | − | − | − |
| Human Enterovirus (HEV) | − | − | − | − | − |
| Seasonal Coronavirus (HCoVs) | − | − | − | − | − |
| Parainfluenza Virus 1 (HPIV1) | − | − | − | − | − |
| Parainfluenza Virus 2 (HPIV2) | − | − | − | − | − |
| Parainfluenza Virus 3 (HPIV3) | − | − | − | − | − |
| Parainfluenza Virus 4 (HPIV4) | − | − | − | − | − |
| Human Adenovirus ADV 3226 | − | − | − | − | − |
| Human Adenovirus ADH 4122 | − | − | − | − | − |
| Staphylococcus aureus ATCC 25923 | − | − | − | − | − |
| Staphylococcus epidermidis ATCC 35984 | − | − | − | − | − |
| Staphylococcus haemolyticus ATCC 29968 | − | − | − | − | − |
| Staphylococcus saprophyticus ATCC 15305 | − | − | − | − | − |
| Staphylococcus pseudintermedius SIG34 | − | − | − | − | − |
| Staphylococcus schleiferi | − | − | − | − | − |
| Pseudomonas aeruginosa ATCC 27858 | − | − | − | − | − |
| Klebsiella pneumonia ATCC 10031 | − | − | − | − | − |
| Klebsiella pneumoniae kp39 | − | − | − | − | − |
| Enterococcus faecalis ATCC 51299 | − | − | − | − | − |
| Enterococcus faecium ATCC 6569 | − | − | − | − | − |
| Escherichia coli ATCC 25922 | − | − | − | − | − |
| Candida albicans ATCC 25923 | − | − | − | − | − |
| Candida glabrata ATCC 2001 | − | − | − | − | − |
| Candida krusei ATCC 34135 | − | − | − | − | − |
| Candida parapsilosis ATCC 22019 | − | − | − | − | − |
| Candida tropicalis ATCC 28707 | − | − | − | − | − |
| Candida auris 10913 | − | − | − | − | − |
| Cryptococcus neoformans ATCC 34872 | − | − | − | − | − |
| Cryptococcus gattii ATCC 32269 | − | − | − | − | − |
| Paracoccidioides brasiliensis Pb1 8 | − | − | − | − | − |
| Histoplasma capsulatum I | − | − | − | − | − |
| Date | Viral Strain | Pango Lineage |
|---|---|---|
| 2020-11 | SARS-CoV-2 LAVIR-HM-UEL | B.1.1.33 |
| 2020-11 | SARS-CoV-2 LAVIR-AB-UEL | B.1.1.143 |
| 2020-11 | SARS-CoV-2 LAVIR-PB-UEL | B.1.1.28 |
| 2020-11 | SARS-CoV-2 LAVIR-LL-UEL | B.1.1.33 |
| 2020-11 | SARS-CoV-2 LAVIR-CB-UEL | P.2 |
| 2020-12 | SARS-CoV-2 LAVIR-AC-UEL | B.1.1.28 |
| 2020-12 | SARS-CoV-2 LAVIR-MC-UEL | P.2 |
| 2021-01 | SARS-CoV-2 LAVIR-MA-UEL | P.2 |
| 2021-02 | SARS-CoV-2 LAVIR-BS-UEL | B.1.1.7 |
| 2021-02 | SARS-CoV-2 LAVIR-VCS-UEL | B.1.1.7 |
| 2021-02 | SARS-CoV-2 LAVIR-SC-UEL | B.1.1.7 |
| 2021-02 | SARS-CoV-2 LAVIR-VRS-UEL | B.1.1.7 |
| 2021-02 | SARS-CoV-2 LAVIR-EC-UEL | B.1.1.7 |
| 2021-09 | SS09-21 | Delta (B.1.617.2-like) |
| 2021-09 | ARM09-21 | Delta (B.1.617.2-like) |
| 2021-09 | CHB09-21 | Delta (B.1.617.2-like) |
| 2021-12 | ALMO12-21 | Omicron (BA.1-like) |
| 2021-10 | MPV10-21 | Delta (B.1.617.2-like) |
| 2021-12 | DIBM12-21 | Omicron (BA.1-like) |
| 2022-01 | PLCSC01-22 | Delta (B.1.617.2-like) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavares, E.R.; de Lima, T.F.; Bartolomeu-Gonçalves, G.; de Castro, I.M.; de Lima, D.G.; Borges, P.H.G.; Nakazato, G.; Kobayashi, R.K.T.; Venancio, E.J.; Tarley, C.R.T.; et al. Development of a Melting-Curve-Based Multiplex Real-Time PCR Assay for the Simultaneous Detection of Viruses Causing Respiratory Infection. Microorganisms 2023, 11, 2692. https://doi.org/10.3390/microorganisms11112692
Tavares ER, de Lima TF, Bartolomeu-Gonçalves G, de Castro IM, de Lima DG, Borges PHG, Nakazato G, Kobayashi RKT, Venancio EJ, Tarley CRT, et al. Development of a Melting-Curve-Based Multiplex Real-Time PCR Assay for the Simultaneous Detection of Viruses Causing Respiratory Infection. Microorganisms. 2023; 11(11):2692. https://doi.org/10.3390/microorganisms11112692
Chicago/Turabian StyleTavares, Eliandro Reis, Thiago Ferreira de Lima, Guilherme Bartolomeu-Gonçalves, Isabela Madeira de Castro, Daniel Gaiotto de Lima, Paulo Henrique Guilherme Borges, Gerson Nakazato, Renata Katsuko Takayama Kobayashi, Emerson José Venancio, César Ricardo Teixeira Tarley, and et al. 2023. "Development of a Melting-Curve-Based Multiplex Real-Time PCR Assay for the Simultaneous Detection of Viruses Causing Respiratory Infection" Microorganisms 11, no. 11: 2692. https://doi.org/10.3390/microorganisms11112692
APA StyleTavares, E. R., de Lima, T. F., Bartolomeu-Gonçalves, G., de Castro, I. M., de Lima, D. G., Borges, P. H. G., Nakazato, G., Kobayashi, R. K. T., Venancio, E. J., Tarley, C. R. T., de Almeida, E. R. D., Pelisson, M., Vespero, E. C., Simão, A. N. C., Perugini, M. R. E., Kerbauy, G., Fornazieri, M. A., Tognim, M. C. B., Góes, V. M., ... Yamada-Ogatta, S. F. (2023). Development of a Melting-Curve-Based Multiplex Real-Time PCR Assay for the Simultaneous Detection of Viruses Causing Respiratory Infection. Microorganisms, 11(11), 2692. https://doi.org/10.3390/microorganisms11112692







