Physiological and Genomic Characterization of a Novel Obligately Chemolithoautotrophic, Sulfur-Oxidizing Bacterium of Genus Thiomicrorhabdus Isolated from a Coastal Sediment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrichment and Isolation
2.2. DNA Extraction and Genomic Analyses
2.3. 16S rRNA Gene Phylogeny
2.4. Morphology, Physiology, and Chemotaxonomic Analysis
2.5. The Determination of Extracellular Sulfur via Scanning Electron Microscopy and Raman Spectromicroscopy
3. Results and Discussion
3.1. Phylogenetic Analysis Based on 16S rRNA Gene
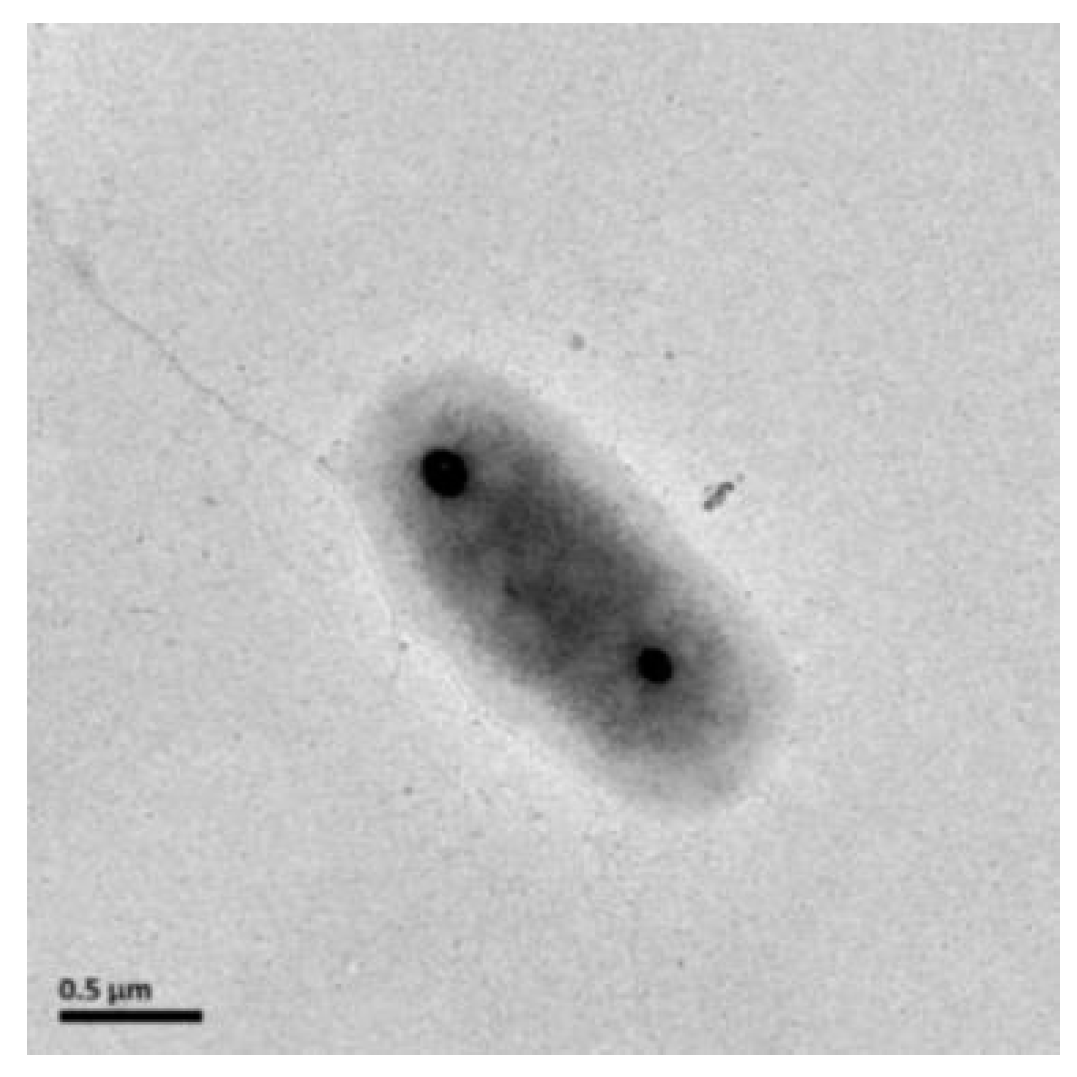
3.2. Morphology, Physiology, and Chemotaxonomic Analysis
3.3. Genome Features and Central Metabolism among Genus Thiomicrorhabdus
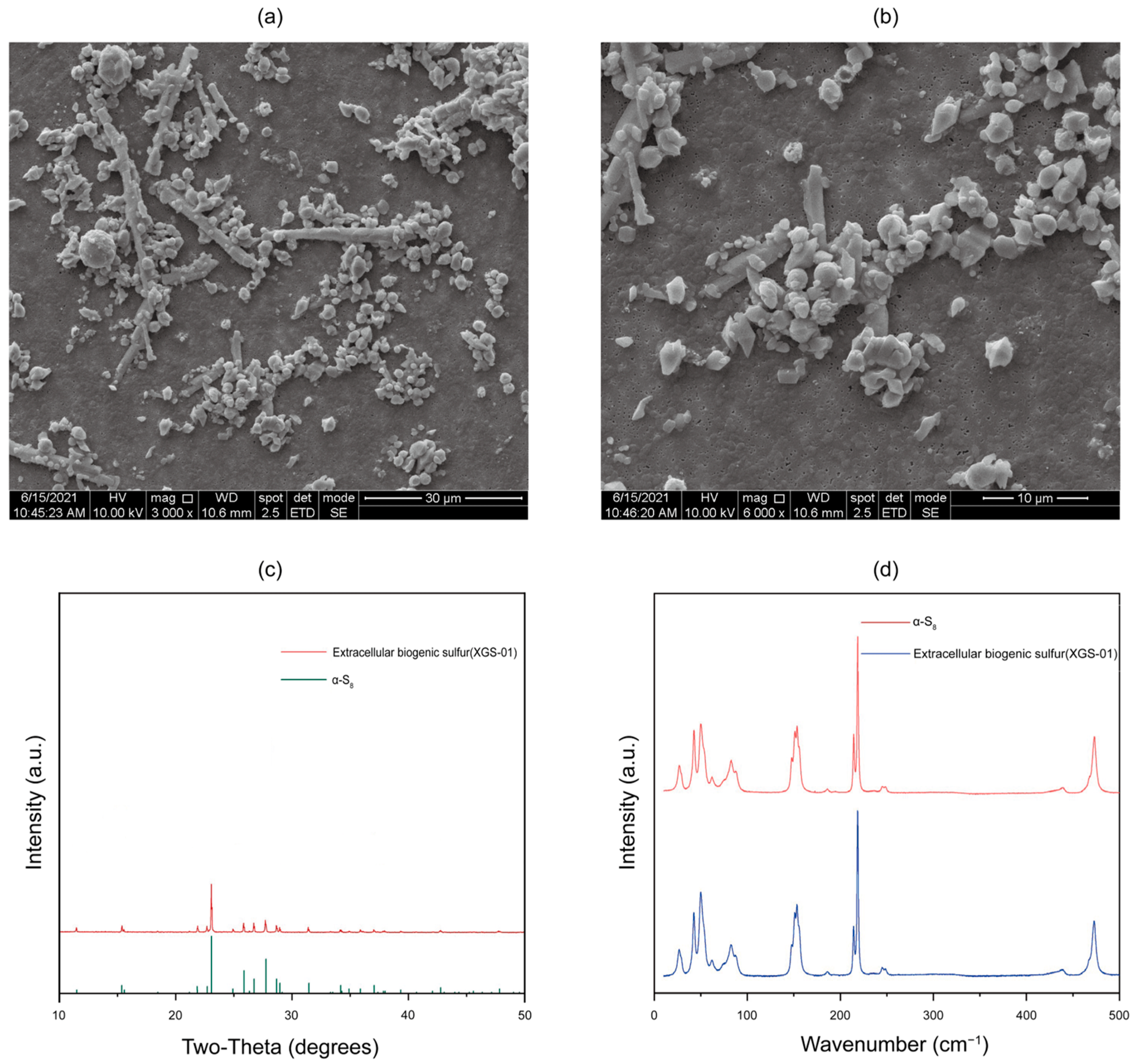
3.4. Characterization of Extracellular Sulfur Produced by Thiomicrorhabdus Species with SEM, XRD, and Raman Spectroscopy
4. Conclusions
Description of Thiomicrorhabdus lithotrophica sp. nov
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boden, R.; Scott, K.M.; Williams, J.; Russel, S.; Antonen, K.; Rae, A.W.; Hutt, L.P. An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: Reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov. and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira. Int. J. Syst. Evol. Microbiol. 2017, 67, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Brinkhoff, T.; Muyzer, G.; Wirsen, C.O.; Kuever, J. Thiomicrospira chilensis sp. nov., a mesophilic obligately chemolithoautotrophic sulfuroxidizing bacterium isolated from a Thioploca mat. Int. J. Syst. Bacteriol. 1999, 49 Pt 2, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Knittel, K.; Kuever, J.; Meyerdierks, A.; Meinke, R.; Amann, R.; Brinkhoff, T. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int. J. Syst. Evol. Microbiol. 2005, 55, 781–786. [Google Scholar] [CrossRef]
- Scott, K.M.; Williams, J.; Porter, C.M.B.; Russel, S.; Harmer, T.L.; Paul, J.H.; Antonen, K.M.; Bridges, M.K.; Camper, G.J.; Campla, C.K.; et al. Genomes of ubiquitous marine and hypersaline Hydrogenovibrio, Thiomicrorhabdus and Thiomicrospira spp. encode a diversity of mechanisms to sustain chemolithoautotrophy in heterogeneous environments. Environ. Microbiol. 2018, 20, 2686–2708. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Muyzer, G.; Wirsen, C.O.; Kuever, J. Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol. 1999, 49 Pt 2, 385–392. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, L.; Hu, Q.; Lyu, J.; Shao, Z. Thiomicrorhabdus indica sp. nov., an obligately chemolithoautotrophic, sulfur-oxidizing bacterium isolated from a deep-sea hydrothermal vent environment. Int. J. Syst. Evol. Microbiol. 2020, 70, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Xuewen, L.; Baoping, C.; Qiliang, L.; Zongze, S.; Lijing, J. Thiomicrorhabdus sediminis sp. nov. and Thiomicrorhabdus xiamenensis sp. nov., novel sulfur-oxidizing bacteria isolated from coastal sediments and an emended description of the genus Thiomicrorhabdus. Int. J. Syst. Evol. Microbiol. 2021, 71, 004660. [Google Scholar] [CrossRef]
- Watsuji, T.O.; Hada, E.; Miyazaki, M.; Ichimura, M.; Takai, K. Thiomicrospira hydrogeniphila sp. nov., an aerobic, hydrogen- and sulfur-oxidizing chemolithoautotroph isolated from a seawater tank containing a block of beef tallow. Int. J. Syst. Evol. Microbiol. 2016, 66, 3688–3693. [Google Scholar] [CrossRef]
- Kojima, H.; Fukui, M. Thiomicrorhabdus aquaedulcis sp. nov., a sulfur-oxidizing bacterium isolated from lake water. Int. J. Syst. Evol. Microbiol. 2019, 69, 2849–2853. [Google Scholar] [CrossRef]
- Tatum, U.; Grayson, S.C.; Hunter, G.; Sheila, P.; Rebecca, P.; Robert, W.; Clare, D.; Madison, D.; James, B.; Rich, B.; et al. Thiomicrorhabdus heinhorstiae sp. nov. and Thiomicrorhabdus cannonii sp. nov.: Novel sulphur-oxidizing chemolithoautotrophs isolated from the chemocline of Hospital Hole, an anchialine sinkhole in Spring Hill, Florida, USA. Int. J. Syst. Evol. Microbiol. 2022, 72, 005233. [Google Scholar] [CrossRef]
- Kojima, H.; Mochizuki, J.; Kanda, M.; Watanabe, T.; Fukui, M. Thiomicrorhabdus immobilis sp. nov., a mesophilic sulfur-oxidizing bacterium isolated from sediment of a brackish lake in northern Japan. Arch. Microbiol. 2022, 204, 605. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Inagaki, F.; Nakagawa, S.; Hirayama, H.; Nunoura, T.; Sako, Y.; Nelson, K.H.; Horikoshi, K. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 2003, 218, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R. Methanogens: Reevaluation of a unique biological group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar] [CrossRef]
- Jiang, L.; Lyu, J.; Shao, Z. Sulfur Metabolism of Hydrogenovibrio thermophilus Strain S5 and Its Adaptations to Deep-Sea Hydrothermal Vent Environment. Front. Microbiol. 2017, 8, 2513. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zheng, Y.; Peng, X.; Zhou, H.; Zhang, C.; Xiao, X.; Wang, F. Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 2009, 70, 93–106. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Joen, Y.S.; Lee, J.-H.; Yi, H. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 3, 716–721. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Naruya, S.; Masatoshi, N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Boden, R.; Scott, K.M.; Rae, A.W.; Hutt, L.P. Reclassification of Thiomicrospira hydrogeniphila (Watsuji et al. 2016) to Thiomicrorhabdus hydrogeniphila comb. nov., with emended description of Thiomicrorhabdus (Boden et al., 2017). Int. J. Syst. Evol. Microbiol. 2017, 67, 4205–4209. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G+ C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Marnocha, C.L.; Sabanayagam, C.R.; Modla, S.; Powell, D.H.; Henri, P.A.; Steele, A.S.; Hanson, T.E.; Webb, S.M.; Chan, C.S. Insights into the Mineralogy and Surface Chemistry of Extracellular Biogenic S(0) Globules Produced by Chlorobaculum tepidum. Front. Microbiol. 2019, 10, 271. [Google Scholar] [CrossRef]
- Pickering, I.J.; George, G.N.; Yu, E.Y.; Brune, D.C.; Tuschak, C.; Overmann, J.; Beatty, T.; Prince, R.C. Analysis of sulfur biochemistry of sulfur bacteria using X-ray absorption spectroscopy. Biochemistry 2001, 40, 8138–8145. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, L.; Hu, Q.; Cui, L.; Zhu, B.; Fu, X.; Lai, Q.; Shao, Z.; Yang, S. Characterization of Sulfurimonas hydrogeniphila sp. nov., a Novel Bacterium Predominant in Deep-Sea Hydrothermal Vents and Comparative Genomic Analyses of the Genus Sulfurimonas. Front. Microbiol. 2021, 12, 626705. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Bardischewsky, F.; Rother, D.; Quentmeier, A.; Fischer, J. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 2005, 8, 253–259. [Google Scholar] [CrossRef]
- Jerez, C.A.; Albar, J.P.; Paradela, A.; Beard, S. Growth of Acidithiobacillus ferrooxidans ATCC 23270 in Thiosulfate under Oxygen-Limiting Conditions Generates Extracellular Sulfur Globules by Means of a Secreted Tetrathionate Hydrolase. Front. Microbiol. 2011, 2, 79. [Google Scholar] [CrossRef]
- Javor, B.J.; Wilmot, D.B.; Vetter, R.D. pH-Dependent metabolism of thiosulfate and sulfur globules in the chemolithotrophic marine bacterium Thiomicrospira crunogena. Arch. Microbiol. 1990, 154, 231–238. [Google Scholar] [CrossRef]
- Houghton, J.L.; Foustoukos, D.I.; Flynn, T.M.; Vetriani, C.; Bradley, A.S.; Fike, D.A. Thiosulfate oxidation by Thiomicrospira thermophila: Metabolic flexibility in response to ambient geochemistry. Environ. Microbiol. 2016, 18, 3057–3072. [Google Scholar] [CrossRef]

| Characteristic | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Shape | Rod | Rod | Rod | Rod | Rod |
| G+C content (mol%) | 39.1 | 39.6 | 39.6 | 42.5 | 42.2 |
| Maximum growth rate (h−1) | 0.15 | 0.40 | 0.45 | 0.20 | 0.14 |
| Temperature range (°C) | 4.0–45.0 | 2.0–40.0 | 3.5–39.0 | −2.0–20.8 | −2.0–20.8 |
| Optimal temperature (°C) | 30.0 | 30.0 | 25.0–32.0 | 14.6–15.4 | 11.5–13.2 |
| Optimal Na+ concentration (mM) | 510 | 270 | 470 | 250 | 250 |
| pH range | 5.0–9.0 | 5.0–8.0 | 4.2–8.5 | 6.5–9.0 | 6.5–9.0 |
| Optimal pH | 7.0 | 6.0 | 6.5 | 7.5–8.5 | 7.3–8.0 |
| Electron donor: | |||||
| Thiosulfate | + | + | + | + | + |
| Sulfide | + | + | + | + | + |
| Elemental sulfur | + | + | + | + | + |
| Sulfite | − | − | − | − | − |
| Tetrathionate | + | + | + | + | + |
| Hydrogen | − | + | − | − | − |
| Fatty Acid | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Saturated: | ||||
| C9:0 | 0.23 | ND | 1.11 | 2.85 |
| C11:0 | 0.06 | ND | ND | 1.01 |
| C12:0 | 0.43 | 1.00 | 1.63 | 1.08 |
| C14:0 | 1.38 | 0.80 | 3.61 | 2.90 |
| C16:0 | 15.97 | 16.90 | 27.03 | 25.65 |
| C17:0 | ND | ND | 1.34 | 1.18 |
| C18:0 | 5.91 | 1.90 | 33.79 | 29.42 |
| C19:0 | 0.17 | ND | ND | 2.24 |
| Unsaturated: | ||||
| C16:1 | 6.90 | 44.40 | 3.60 | 3.17 |
| C17:1 | 0.81 | ND | 4.87 | 3.17 |
| C18:1 | 50.14 | 30.40 | 9.21 | 7.47 |
| C18:2 | 0.56 | ND | 3.60 | 2.59 |
| C18:3 | 0.42 | ND | 1.32 | 1.27 |
| Hydroxy: | ||||
| C8:0 3OH | 0.25 | ND | 0.68 | 0.57 |
| C12:0 3OH | 5.01 | ND | 1.76 | 3.03 |
| C18:1 2OH | 0.49 | ND | 4.25 | ND |
| Characteristic | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Genome size (Mb) | 2.5 | 2.7 | 2.6 | 2.4 | 2.6 |
| GC content (%) | 39.1 | 39.6 | 42.2 | 49.9 | 38.0 |
| ANI | − | 71.1 | 70.1 | 70.4 | 71.5 |
| Gene count | 2292 | 2526 | 2337 | 2285 | 2382 |
| rRNA operons | 9 | 3 | 3 | 2 | 3 |
| tRNA count | 47 | 45 | 45 | 43 | 46 |
| Sox | ABCDXYZ | ABCDXYZ | ABCDXYZ | ABCDXYZ | ABCDXYZ |
| Fcc | + | + | − | + | + |
| Sqr | + | + | + | + | + |
| TCA Cycle | + | + | + | + | + |
| Pentose phosphate pathway | + | + | + | + | + |
| Glycolysis/Gluconeogenesis | + | + | + | + | + |
| Carboxysome | + | + | + | + | + |
| Carbonic anhydrase | 1α-class, 1γ-class | 1α-class, 1γ-class, 1β-class | 1α-class, 1γ-class, 1β-class | 1α-class, 2γ-class | 1α-class, 1γ-class, 1β-class |
| Nitrate reductase | − | + | − | − | − |
| Nitrite reductase | − | + | − | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Zhu, H.; Wang, J.; Shao, Z.; Wei, S.; Wang, R.; Cheng, R.; Jiang, L. Physiological and Genomic Characterization of a Novel Obligately Chemolithoautotrophic, Sulfur-Oxidizing Bacterium of Genus Thiomicrorhabdus Isolated from a Coastal Sediment. Microorganisms 2023, 11, 2569. https://doi.org/10.3390/microorganisms11102569
Gao Y, Zhu H, Wang J, Shao Z, Wei S, Wang R, Cheng R, Jiang L. Physiological and Genomic Characterization of a Novel Obligately Chemolithoautotrophic, Sulfur-Oxidizing Bacterium of Genus Thiomicrorhabdus Isolated from a Coastal Sediment. Microorganisms. 2023; 11(10):2569. https://doi.org/10.3390/microorganisms11102569
Chicago/Turabian StyleGao, Yu, Han Zhu, Jun Wang, Zongze Shao, Shiping Wei, Ruicheng Wang, Ruolin Cheng, and Lijing Jiang. 2023. "Physiological and Genomic Characterization of a Novel Obligately Chemolithoautotrophic, Sulfur-Oxidizing Bacterium of Genus Thiomicrorhabdus Isolated from a Coastal Sediment" Microorganisms 11, no. 10: 2569. https://doi.org/10.3390/microorganisms11102569
APA StyleGao, Y., Zhu, H., Wang, J., Shao, Z., Wei, S., Wang, R., Cheng, R., & Jiang, L. (2023). Physiological and Genomic Characterization of a Novel Obligately Chemolithoautotrophic, Sulfur-Oxidizing Bacterium of Genus Thiomicrorhabdus Isolated from a Coastal Sediment. Microorganisms, 11(10), 2569. https://doi.org/10.3390/microorganisms11102569







