Pseudomonassin, a New Bioactive Ribosomally Synthesised and Post-Translationally Modified Peptide from Pseudomonas sp. SST3
Abstract
1. Introduction
2. Materials and Methods
2.1. Organism and Fermentation
2.2. Extraction and Isolation
2.3. HCD-MS/MS Analysis
2.4. Metabolomics Analysis
2.5. Antibacterial Assay
2.6. Planktonic Assay Solution for Biofim Inhibition
3. Results
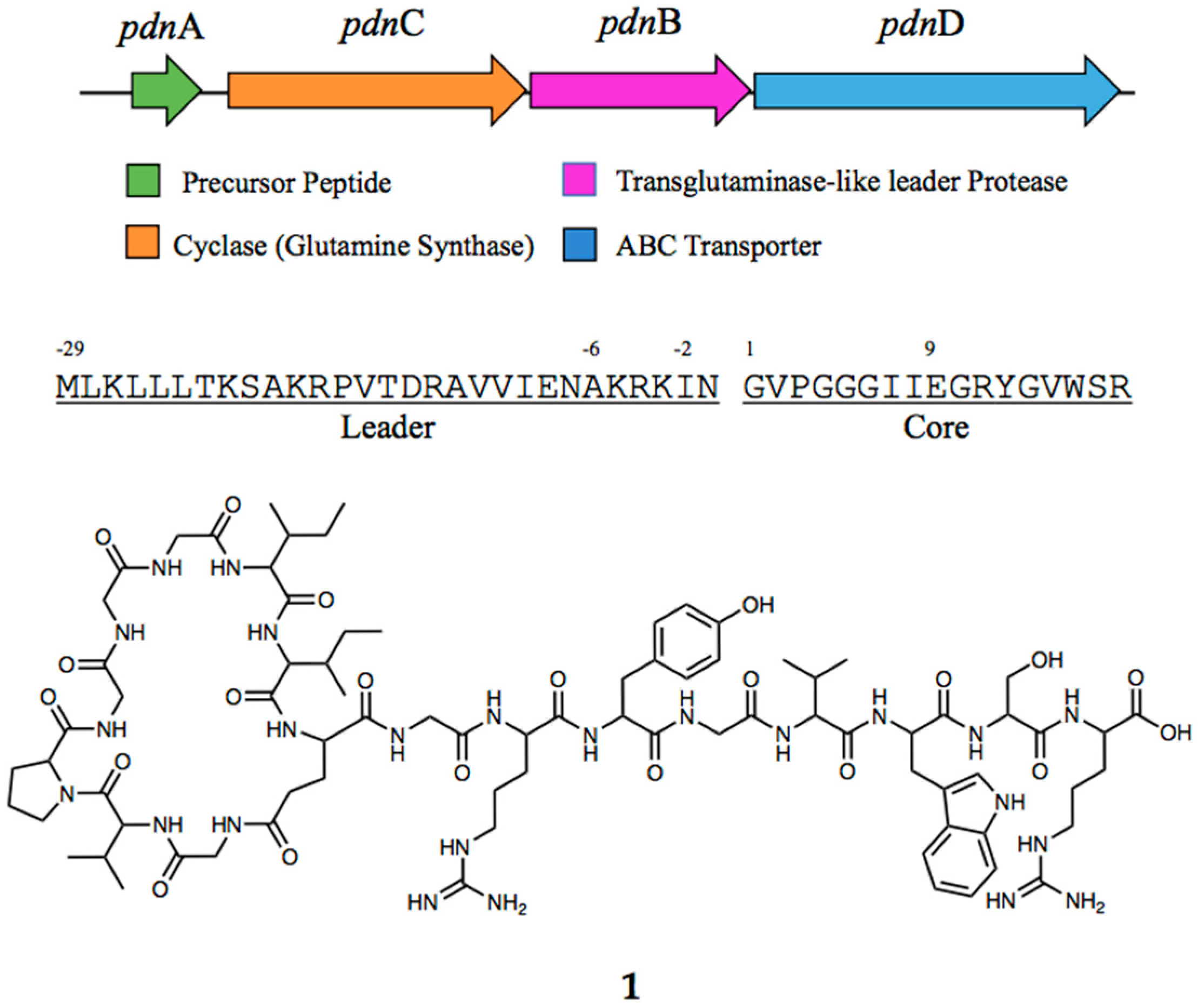
3.1. Genome Mining and Prediction of the Structure of Pseusomonassim
3.2. Elicitation of the Target RiPP
3.3. Pseudomonassin Characterisation Using High-Energy Collision Dissociation Tandem Mass Spectrometry (HDC-MS/MS)
3.4. Metabolomics Analysis Reveals Potential New Metabolites from the Co-Cultivation of Two Pseudomonas Species
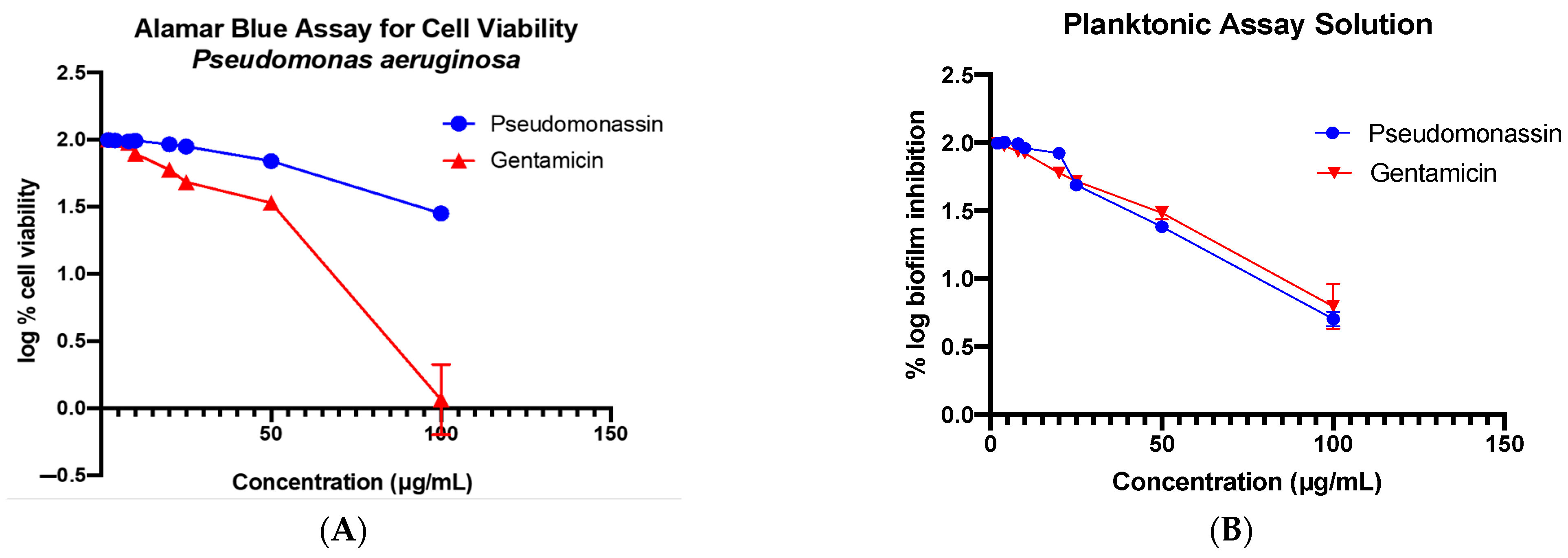
3.5. Antibacterial and Biofilm Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes-a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Trivella, D.B.B.; de Felicio, R. The Tripod for Bacterial Natural Product Discovery: Genome Mining, Silent Pathway Induction, and Mass Spectrometry-Based Molecular Networking. mSystems 2018, 3, e00160-17. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 47, D94–D99. [Google Scholar] [CrossRef]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef]
- Jones, J.A.; Wang, X. Use of bacterial co-cultures for the efficient production of chemicals. Curr. Opin. Biotechnol. 2018, 53, 33–38. [Google Scholar] [CrossRef]
- Wakefield, J.; Hassan, H.M.; Jaspars, M.; Ebel, R.; Rateb, M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 2017, 8, 1284. [Google Scholar] [CrossRef] [PubMed]
- Tawfike, A.F.; Viegelmann, C.; Edrada-Ebel, R. Metabolomics and Dereplication Strategies in Natural Products. Methods Mol. Biol. 2013, 1055, 227–244. [Google Scholar]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.J.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.R.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R.A. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef]
- Stuart, K.A.; Welsh, K.; Walker, M.C.; Edrada-Ebel, R.A. Metabolomic tools used in marine natural product drug discovery. Expert Opin. Drug Discov. 2020, 15, 499–522. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gómez, H.; Tulla-Puche, J. Lasso peptides: Chemical approaches and structural elucidation. Org. Biomol. Chem. 2018, 16, 5065–5080. [Google Scholar] [CrossRef]
- de Bary, A. Morphologie und Physiologie der Pilze, Flechten und Myxomyceten; Wilhelm Engelmann: Leipzig, Germany, 1866. [Google Scholar]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Lasso Peptides: An Intriguing Class of Bacterial Natural Products. Acc. Chem. Res. 2015, 48, 1909–1919. [Google Scholar] [CrossRef]
- Tan, S.; Moore, G.; Nodwell, J. Put a bow on it: Knotted antibiotics take center stage. Antibiotics 2019, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Naimi, S.; Zirah, S.; Hammami, R.; Fernandez, B.; Rebuffat, S.; Fliss, I. Fate and biological activity of the antimicrobial lasso peptide microcin J25 under gastrointestinal tract conditions. Front. Microbiol. 2018, 9, 1764. [Google Scholar] [CrossRef]
- Li, Y.; Rebuffat, S. The manifold roles of microbial ribosomal peptide-based natural products in physiology and ecology. J. Biol. Chem. 2020, 295, 34–54. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Parry, M.E.; Cartagena, A.J.; Darst, S.A.; James Link, A. Discovery and structure of the antimicrobial lasso peptide citrocin. J. Biol. Chem. 2019, 294, 6822–6830. [Google Scholar] [CrossRef]
- Zimmermann, M.; Hegemann, J.D.; Xie, X.; Marahiel, M.A. The astexin-1 lasso peptides: Biosynthesis, stability, and structural studies. Chem. Biol. 2013, 20, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Knappe, T.A.; Linne, U.; Robbel, L.; Marahiel, M.A. Insights into the Biosynthesis and Stability of the Lasso Peptide Capistruin. Chem. Biol. 2009, 16, 1290–1298. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Lehri, B.; Jarmusch, S.A.; Miranda, K.J.; Al-Wahaibi, L.H.; Stewart, H.A.; Jamieson, A.J.; Jaspars, M.; Karlyshev, A.V. Whole genome sequencing of four bacterial strains from South Shetland Trench revealing biosynthetic and environmental adaptation gene clusters. Mar. Genomics 2020, 54, 100782. [Google Scholar] [CrossRef] [PubMed]
- Zuo, M.Q.; Sun, R.X.; Fang, R.Q.; He, S.M.; Dong, M.Q. Characterization of collision-induced dissociation of deprotonated peptides of 4–16 amino acids using high-resolution mass spectrometry. Int. J. Mass Spectrom. 2019, 445, 116186. [Google Scholar] [CrossRef]
- Kowalska-krochmal, B.; Dudek-wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility 30th ed CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Oliveira, A.; Cunha, M.D.L.R.S. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res. Notes 2010, 3, 260. [Google Scholar] [CrossRef]
- Macià, M.D.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Pascal Andreu, V.; De Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Yu, H.; Li, N.; Zeng, X.; Liu, L.; Wang, Y.; Wang, G.; Cai, S.; Huang, S.; Ding, X.; Song, Q.; et al. A comprehensive antimicrobial activity evaluation of the recombinant microcin j25 against the foodborne pathogens salmonella and e. Coli O157:H7 by using a matrix of conditions. Front. Microbiol. 2019, 10, 1954. [Google Scholar] [CrossRef]
- Rintoul, M.R.; De Arcuri, B.F.; Salomón, R.A.; Farías, R.N.; Morero, R.D. The antibacterial action of microcin J25: Evidence for disruption of cytoplasmic membrane energization in Salmonella newport. FEMS Microbiol. Lett. 2001, 204, 265–270. [Google Scholar] [CrossRef][Green Version]
- Diedrich, J.K.; Pinto, A.F.M.; Yates, J.R. Energy dependence of HCD on peptide fragmentation: Stepped collisional energy finds the sweet spot. J. Am. Soc. Mass Spectrom. 2013, 24, 1690–1699. [Google Scholar] [CrossRef]
- Frese, C.K.; Altelaar, A.F.M.; Hennrich, M.L.; Nolting, D.; Zeller, M.; Griep-Raming, J.; Heck, A.J.R.; Mohammed, S. Improved Peptide Identification by Targeted Fragmentation Using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 2011, 10, 2377–2388. [Google Scholar] [CrossRef]
- Tu, C.; Li, J.; Shen, S.; Sheng, Q.; Shyr, Y.; Qu, J. Performance Investigation of Proteomic Identification by HCD/CID Fragmentations in Combination with High/Low-Resolution Detectors on a Tribrid, High-Field Orbitrap Instrument. PLoS ONE 2016, 11, e0160160. [Google Scholar] [CrossRef] [PubMed]
- Stieger, C.E.; Doppler, P.; Mechtler, K. Optimized Fragmentation Improves the Identification of Peptides Cross-Linked by MS-Cleavable Reagents. J. Proteome Res. 2019, 18, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Clauser, K.; Baker, P. Protein Prospector. 1999. Available online: Http://Prospector.Ucsf.Edu/ (accessed on 15 November 2019).
- Mazlan, N.W.; Tate, R.; Yusoff, Y.M.; Clements, C.; Edrada-Ebel, R. Metabolomics-Guided Isolation of Anti-Trypanosomal Compounds from Endophytic Fungi of the Mangrove plant Avicennia Lanata. Curr. Med. Chem. 2019, 27, 1815–1835. [Google Scholar] [CrossRef] [PubMed]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R.A. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1006–1007, 71–83. [Google Scholar] [CrossRef] [PubMed]
- do Nascimento, T.G.; dos Santos Arruda, R.E.; da Cruz Almeida, E.T.; dos Santos Oliveira, J.M.; Basílio-Júnior, I.D.; Celerino de Moraes Porto, I.C.; Rodrigues Sabino, A.; Tonholo, J.; Gray, A.; Ebel, R.A.E.; et al. Comprehensive multivariate correlations between climatic effect, metabolite-profile, antioxidant capacity and antibacterial activity of Brazilian red propolis metabolites during seasonal study. Sci. Rep. 2019, 9, 18293. [Google Scholar] [CrossRef]
- Sebak, M.; Saafan, A.E.; AbdelGhani, S.; Bakeer, W.; El-Gendy, A.O.; Espriu, L.C.; Duncan, K.; Edrada-Ebel, R. Bioassay- And metabolomics-guided screening of bioactive soil actinomycetes from the ancient city of Ihnasia, Egypt. PLoS ONE 2019, 14, e0226959. [Google Scholar] [CrossRef]
- El-Sayed, A.K.; Hothersall, J.; Thomas, C.M. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antiobiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 2001, 147, 2127–2139. [Google Scholar] [CrossRef]
- Gao, S.S.; Hothersall, J.; Wu, J.; Murphy, A.C.; Song, Z.; Stephens, E.R.; Thomas, C.M.; Crump, M.P.; Cox, R.J.; Simpson, T.J.; et al. Biosynthesis of mupirocin by pseudomonas fluorescens NCIMB 10586 involves parallel pathways. J. Am. Chem. Soc. 2014, 136, 5501–5507. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [PubMed]
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.P.; Agrawal, R.K. Validation of QSAR Models—Strategies and Importance. Int. J. Drug Des. Disocovery 2011, 2, 511–519. [Google Scholar]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Lawson, A.J.; Swienty-Busch, J.; Géoui, T.; Evans, D. The making of reaxys—Towards unobstructed access to relevant chemistry information. In The Future of the History of Chemical Information; ACS Symposium Series; ACS Publications: Washington, DC, USA, 2014. [Google Scholar]

| Amino Acid | b-ions | y-ions | ||||
|---|---|---|---|---|---|---|
| Theoretical MW Fragment | Observed MW Fragment | Error | Theoretical MW Fragment | Observed MW Fragment | Error | |
| G | - | - | - | |||
| V | 157.0972 | 157.0971 | 0.64 ppm | 1684.8918 | - | - |
| P | 254.1499 | 254.1495 | 1.57 ppm | 1585.8234 | - | - |
| G | 311.1714 | 311.1710 | 1.29 ppm | 1488.7706 | 1488.7701 | −0.34 ppm |
| G | 368.1928 | 368.1923 | 1.36 ppm | 1431.7492 | 1431.7497 | 0.34 ppm |
| G | 425.2143 | 425.2137 | 1.41 ppm | 1374.7277 | 1374.7292 | 1.09 ppm |
| I | 538.2984 | 538.2979 | 0.93 ppm | 1317.7062 | - | |
| I | 651.3824 | 651.3818 | 0.92 ppm | 1204.6222 | 1204.6205 | −1.41 ppm |
| E | 762.4145 | 762.4138 | 0.92 ppm | 1091.5381 | 1091.5361 | −1.83 ppm |
| G | 819.4359 | 819.4351 | 0.98 ppm | 980.5061 | 980.5026 | −3.57 ppm |
| R | 975.5370 | 975.5354 | 1.64 ppm | 923.4846 | 923.4833 | −1.41 ppm |
| Y | 1138.6004 | 1138.5991 | 1.14 ppm | 767.3835 | 767.3825 | −1.30 ppm |
| G | 1195.6218 | 1195.6205 | 1.09 ppm | 604.3202 | 604.3201 | −0.17 ppm |
| V | 1294.6908 | 1294.6897 | 0.85 ppm | 547.2987 | 547.2984 | −0.55 ppm |
| W | 1480.7696 | 1480.7712 | −1.08 ppm | 448.2303 | 448.23 | −0.67 ppm |
| S | 1567.8016 | 1567.8020 | −0.26 ppm | 262.151 | 262.1507 | −1.14 ppm |
| R | - | - | - | 175.1190 | 175.1188 | −1.14 ppm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda, K.J.; Jaber, S.; Atoum, D.; Arjunan, S.; Ebel, R.; Jaspars, M.; Edrada-Ebel, R. Pseudomonassin, a New Bioactive Ribosomally Synthesised and Post-Translationally Modified Peptide from Pseudomonas sp. SST3. Microorganisms 2023, 11, 2563. https://doi.org/10.3390/microorganisms11102563
Miranda KJ, Jaber S, Atoum D, Arjunan S, Ebel R, Jaspars M, Edrada-Ebel R. Pseudomonassin, a New Bioactive Ribosomally Synthesised and Post-Translationally Modified Peptide from Pseudomonas sp. SST3. Microorganisms. 2023; 11(10):2563. https://doi.org/10.3390/microorganisms11102563
Chicago/Turabian StyleMiranda, Kevin Jace, Saif Jaber, Dana Atoum, Subha Arjunan, Rainer Ebel, Marcel Jaspars, and RuAngelie Edrada-Ebel. 2023. "Pseudomonassin, a New Bioactive Ribosomally Synthesised and Post-Translationally Modified Peptide from Pseudomonas sp. SST3" Microorganisms 11, no. 10: 2563. https://doi.org/10.3390/microorganisms11102563
APA StyleMiranda, K. J., Jaber, S., Atoum, D., Arjunan, S., Ebel, R., Jaspars, M., & Edrada-Ebel, R. (2023). Pseudomonassin, a New Bioactive Ribosomally Synthesised and Post-Translationally Modified Peptide from Pseudomonas sp. SST3. Microorganisms, 11(10), 2563. https://doi.org/10.3390/microorganisms11102563








