Bioactivity and Metabolomic Profile of Extracts Derived from Mycelial Solid Cultures of Hypsizygus marmoreus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mushrooms
2.2. Molecular Identification
2.3. Phylogeny
2.4. Samples’ Preparation
2.5. Total Phenolic Content
2.6. In Vitro Antioxidant Assays
2.7. Untargeted Ultra-Performance Liquid Chromatography–Mass Spectrometry (UHPLC)-Quadrupole Time of Flight (QTOF)-Based Metabolomics and Statistical Analysis
2.8. Antimicrobial Effects
3. Results
3.1. Molecular Identification of Hypsizygus marmoreus (PeruMyc2422)
3.2. Untargeted LC–MS/MS-Based Metabolomics
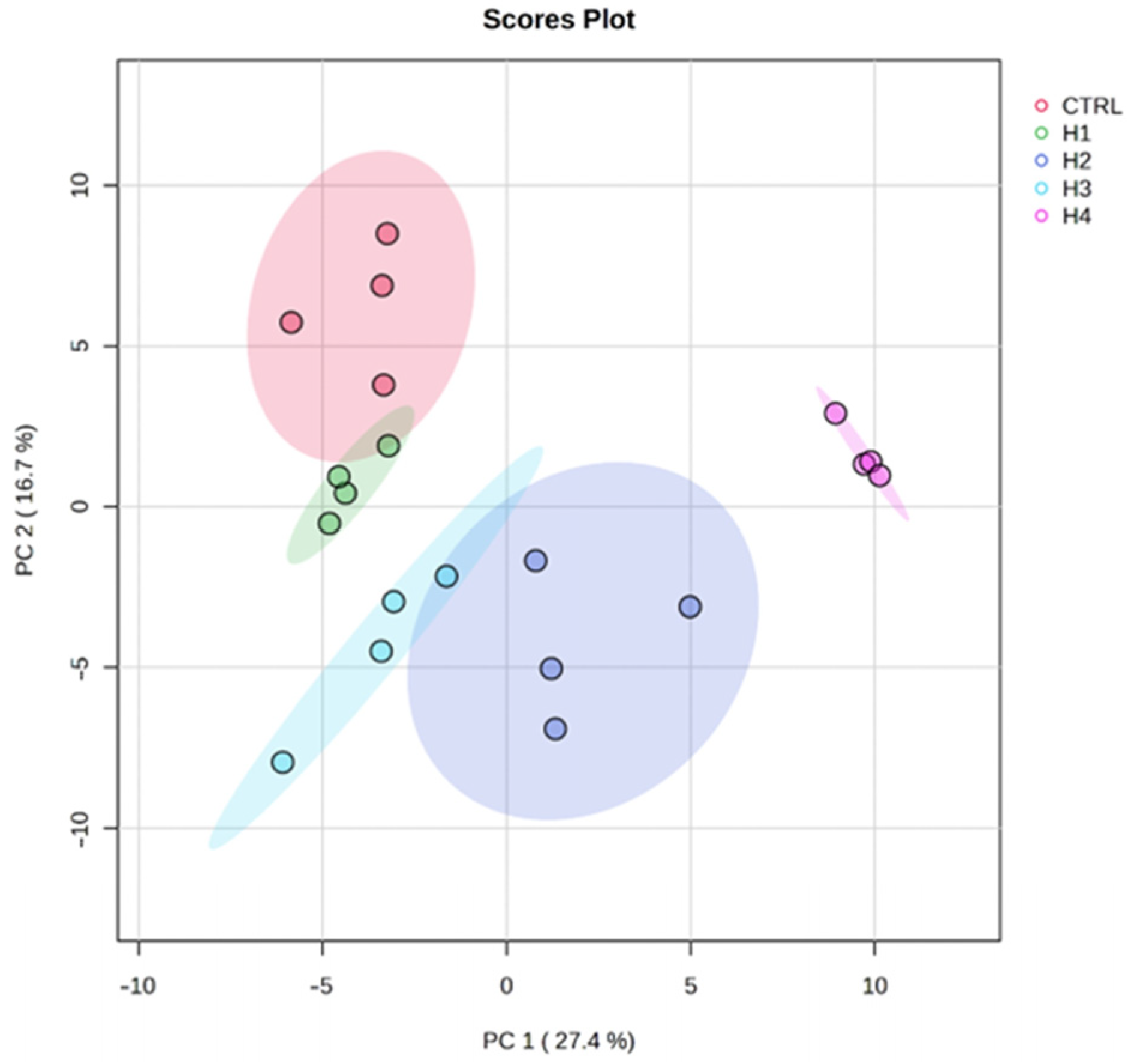
3.3. Statistical Data Analysis
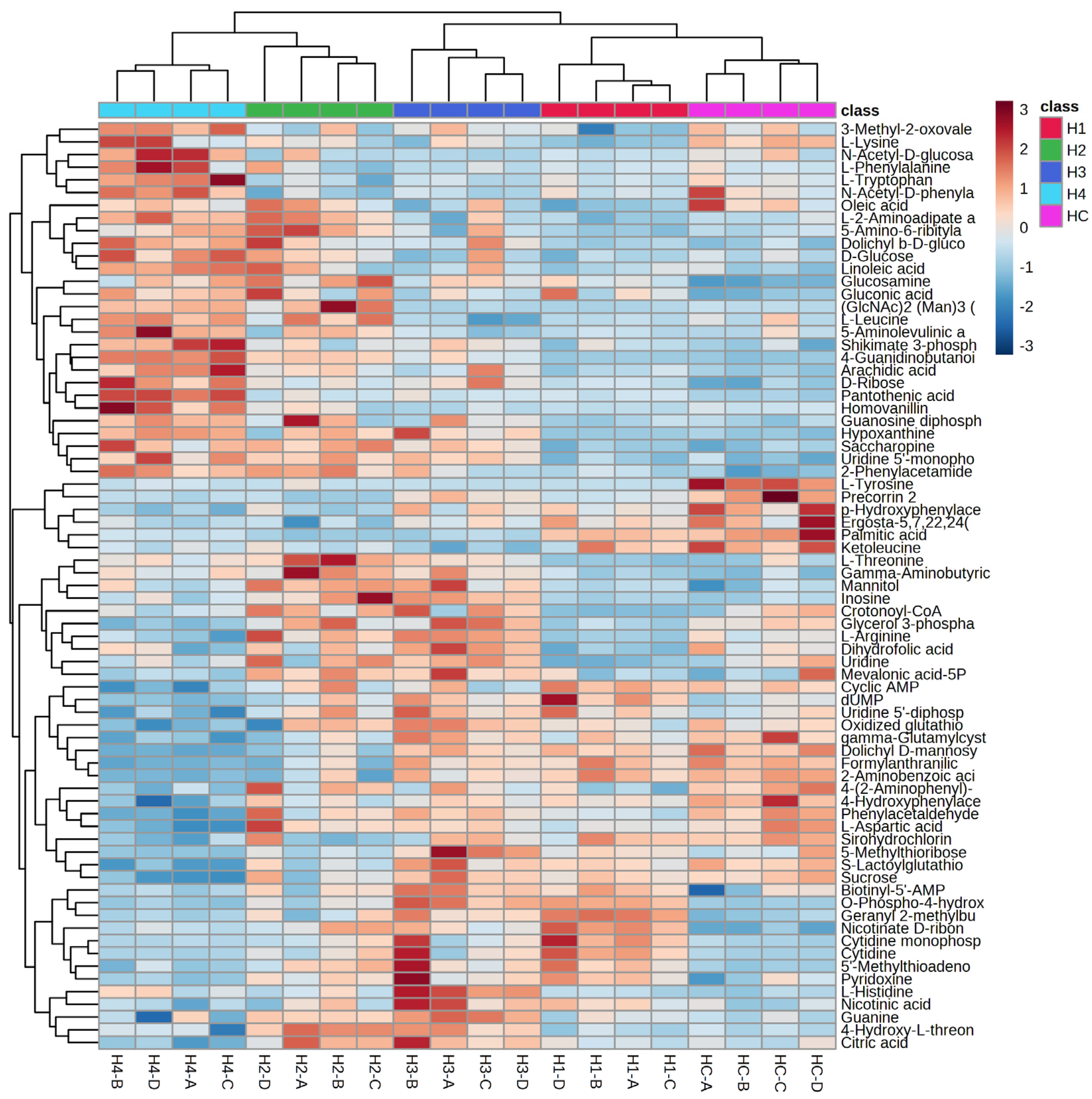
3.4. Cluster Analysis
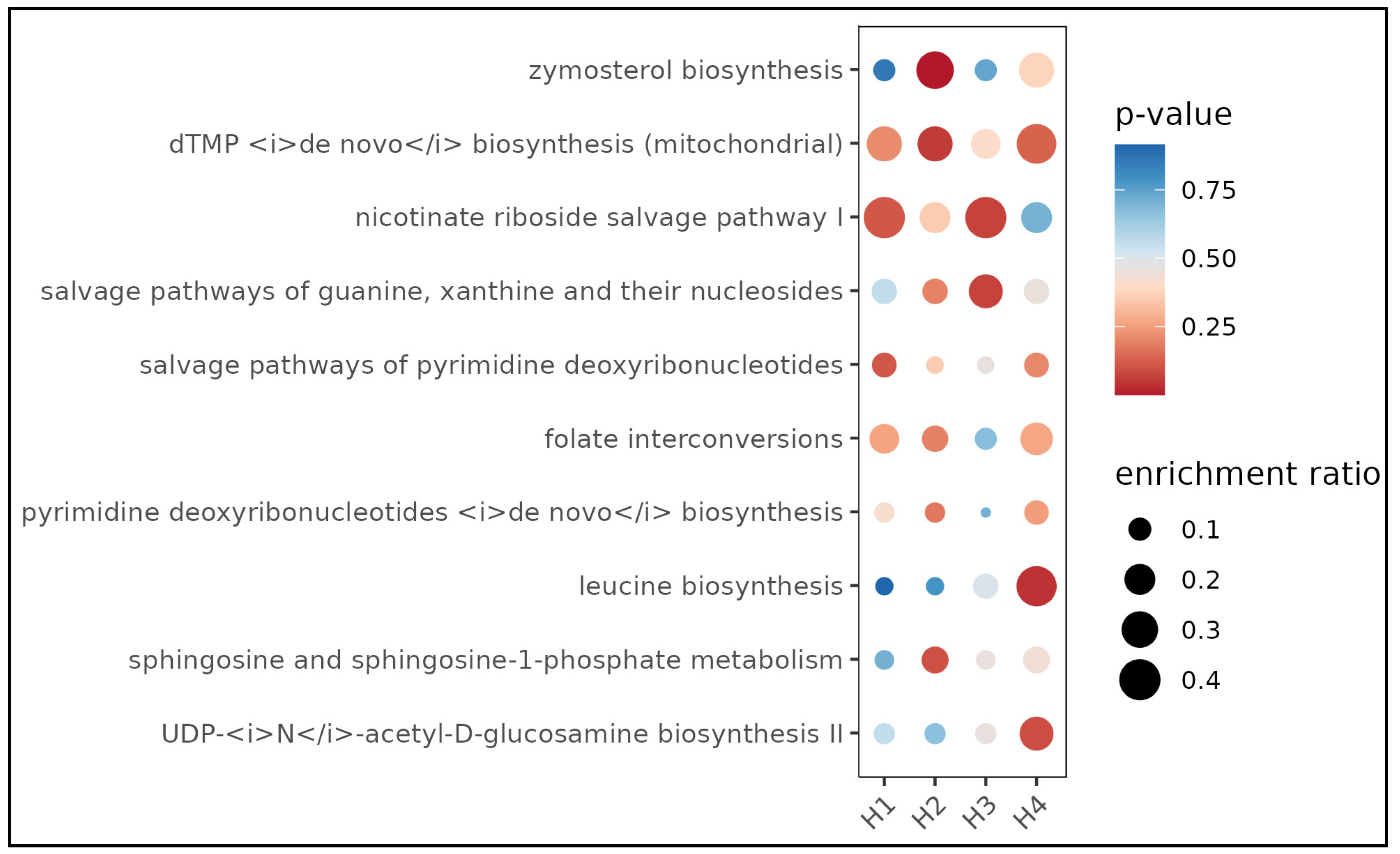
3.5. Functional Analysis
3.6. Total Phenol Content and Antioxidant Effects
3.7. Antimicrobial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, N.; Cohen, J.; Asatiani, M.D.; Varshney, V.K.; Yu, H.-T.; Yang, Y.-C.; Li, Y.-H.; Mau, J.-L.; Wasser, S.P. Chemical composition and nutritional and medicinal value of fruit bodies and submerged cultured mycelia of culinary-medicinal higher Basidiomycetes mushrooms. Int. J. Med. Mushrooms 2014, 16, 273–291. [Google Scholar] [CrossRef]
- Flores, G.A.; Girometta, C.E.; Cusumano, G.; Pellegrino, R.M.; Silviani, S.; Bistocchi, G.; Arcangeli, A.; Ianni, F.; Blasi, F.; Cossignani, L. Diversity of Pleurotus spp. (Agaricomycetes) and Their Metabolites of Nutraceutical and Therapeutic Importance. Int. J. Med. Mushrooms 2023, 25, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.A.; Girometta, C.E.; Cusumano, G.; Angelini, P.; Tirillini, B.; Ianni, F.; Blasi, F.; Cossignani, L.; Pellegrino, R.M.; Emiliani, C. Untargeted Metabolomics Used to Describe the Chemical Composition, Antioxidant and Antimicrobial Effects of Extracts from Pleurotus spp. Mycelium Grown in Different Culture Media. Antibiotics 2022, 11, 1468. [Google Scholar] [CrossRef] [PubMed]
- Gruen, H.E.; Wong, W.M. Distribution of cellular amino acids, protein, and total organic nitrogen during fruitbody development in Flammulina velutipes. I. Growth on sawdust medium. Can. J. Bot. 1982, 60, 1330–1341. [Google Scholar] [CrossRef]
- Pellegrino, R.M.; Blasi, F.; Angelini, P.; Ianni, F.; Alabed, H.B.; Emiliani, C.; Venanzoni, R.; Cossignani, L. LC/MS Q-TOF metabolomic investigation of amino acids and dipeptides in Pleurotus ostreatus grown on different substrates. J. Agric. Food Chem. 2022, 70, 10371–10382. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, R.M.; Ianni, F.; Blasi, F.; Angelini, P.; Emiliani, C.; Venanzoni, R.; Cossignani, L. Lipidomic profiling of Pleurotus ostreatus by LC/MS Q-TOF analysis. Food Res. Int. 2022, 156, 111335. [Google Scholar] [CrossRef]
- Bach, F.; Zielinski, A.A.F.; Helm, C.V.; Maciel, G.M.; Pedro, A.C.; Stafussa, A.P.; Ávila, S.; Haminiuk, C.W.I. Bio compounds of edible mushrooms: In vitro antioxidant and antimicrobial activities. Lwt 2019, 107, 214–220. [Google Scholar] [CrossRef]
- Liu, M.; Li, S.; Wang, X.; Zhu, Y.; Zhang, J.; Liu, H.; Jia, L. Characterization, anti-oxidation and anti-inflammation of polysaccharides by Hypsizygus marmoreus against LPS-induced toxicity on lung. Int. J. Biol. Macromol. 2018, 111, 121–128. [Google Scholar] [CrossRef]
- Rathore, H.; Prasad, S.; Kapri, M.; Tiwari, A.; Sharma, S. Medicinal importance of mushroom mycelium: Mechanisms and applications. J. Funct. Foods 2019, 56, 182–193. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, H.; Li, T.; Chen, L.; Zheng, B.; Liu, R.H. Comparison of phenolics, antioxidant, and antiproliferative activities of two Hypsizygus marmoreus varieties. J. Food Sci. 2020, 85, 2227–2235. [Google Scholar] [CrossRef]
- Available online: https://www.indexfungorum.org/names/Names.asp-16 (accessed on 9 October 2023).
- Vlasenko, A.; Budsuren, D.; Nazyn, C. The First Records of a Rare Species of Reticularia olivacea (Myxomycetes) and Hypsizygus marmoreus (Fungi) in Inner Asia. In BIO Web of Conferences; EDP Sciences: Les Ulis, France, 2021; p. 00135. [Google Scholar]
- Qiu, C.; Yan, W.; Deng, W.; Song, B.; Li, T. Genetic diversity analysis of Hypsizygus marmoreus with target region amplification polymorphism. Sci. World J. 2014, 2014, 619746. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Ishihara, A.; Sakaguchi, N.; Nishino, S.; Parada, R.Y.; Nakagiri, A.; Otani, H. Antifungal Activity of Volatile Compounds Produced by an Edible Mushroom Hypsizygus marmoreus against Phytopathogenic Fungi. J. Phytopathol. 2015, 163, 987–996. [Google Scholar] [CrossRef]
- Shiono, Y.; Haga, M.; Koyama, H.; Murayama, T.; Koseki, T. Antifungal activity of a polyacetylene against the fungal pathogen of Japanese oak from the liquid culture of the edible mushroom, Hypsizygus marmoreus. Z. Für Naturforschung B 2013, 68, 293–295. [Google Scholar] [CrossRef]
- Wong, J.; Ng, T.; Hui, M.; Cheung, R.; Chan, Y.; Dan, X.; Wang, H. Screening of aqueous and organic extracts from a variety of fungi for their ability to antagonise the pathogenic yeast Candida albicans. Hong Kong Med. J. 2016, 22 (Suppl. S7), 26–29. [Google Scholar] [PubMed]
- Chang, J.-S.; Son, J.-K.; Li, G.; Oh, E.-J.; Kim, J.-Y.; Park, S.-H.; Bae, J.-T.; Kim, H.-J.; Lee, I.-S.; Kim, O.-M. Inhibition of cell cycle progression on HepG2 cells by hypsiziprenol A9, isolated from Hypsizigus marmoreus. Cancer Lett. 2004, 212, 7–14. [Google Scholar] [CrossRef]
- Wang, G.; Lin, J.; Shi, Y.; Chang, X.; Wang, Y.; Guo, L.; Wang, W.; Dou, M.; Deng, Y.; Ming, R. Mitochondrial genome in Hypsizygus marmoreus and its evolution in Dikarya. BMC Genom. 2019, 20, 765. [Google Scholar] [CrossRef]
- Zervakis, G.I.; Koutrotsios, G.; Katsaris, P. Composted versus raw olive mill waste as substrates for the production of medicinal mushrooms: An assessment of selected cultivation and quality parameters. BioMed Res. Int. 2013, 2013, 546830. [Google Scholar] [CrossRef]
- Sánchez, C. Cultivation of Pleurotus ostreatus and other edible mushrooms. Appl. Microbiol. Biotechnol. 2010, 85, 1321–1337. [Google Scholar] [CrossRef]
- Flores, G.A.; Cusumano, G.; Ianni, F.; Blasi, F.; Angelini, P.; Cossignani, L.; Pellegrino, R.M.; Emiliani, C.; Venanzoni, R.; Zengin, G. Fomitopsis officinalis: Spatial (Pileus and Hymenophore) Metabolomic Variations Affect Functional Components and Biological Activities. Antibiotics 2023, 12, 766. [Google Scholar] [CrossRef]
- Shih, L.; Tsai, K.-L.; Hsieh, C. Effects of culture conditions on the mycelial growth and bioactive metabolite production in submerged culture of Cordyceps militaris. Biochem. Eng. J. 2007, 33, 193–201. [Google Scholar] [CrossRef]
- Das, A.R.; Borthakur, M.; Saha, A.; Joshi, S.; Das, P. Growth of mycelial biomass and fruit body cultivation of Lentinus squarrosulus collected from home garden of Tripura in Northeast India. J. Appl. Biol. Biotechnol. 2015, 3, 017–019. [Google Scholar]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Angelini, P.; Girometta, C.; Tirillini, B.; Moretti, S.; Covino, S.; Cipriani, M.; D’Ellena, E.; Angeles, G.; Federici, E.; Savino, E. A comparative study of the antimicrobial and antioxidant activities of Inonotus hispidus fruit and their mycelia extracts. Int. J. Food Prop. 2019, 22, 768–783. [Google Scholar] [CrossRef]
- Acquaviva, A.; Nilofar, N.; Bouyahya, A.; Zengin, G.; Di Simone, S.C.; Recinella, L.; Leone, S.; Brunetti, L.; Uba, A.I.; Cakilcioğlu, U. Chemical characterization of different extracts from Artemisia annua and their antioxidant, enzyme inhibitory and anti-inflammatory properties. Chem. Biodivers. 2023, 20, e202300547. [Google Scholar] [CrossRef]
- Uysal, S.; Zengin, G.; Locatelli, M.; Bahadori, M.B.; Mocan, A.; Bellagamba, G.; De Luca, E.; Mollica, A.; Aktumsek, A. Cytotoxic and enzyme inhibitory potential of two Potentilla species (P. speciosa L. and P. reptans Willd.) and their chemical composition. Front. Pharmacol. 2017, 8, 290. [Google Scholar] [CrossRef]
- Angelini, P.; Pellegrino, R.M.; Tirillini, B.; Flores, G.A.; Alabed, H.B.; Ianni, F.; Blasi, F.; Cossignani, L.; Venanzoni, R.; Orlando, G. Metabolomic profiling and biological activities of Pleurotus columbinus Quél. Cultivated on different agri-food byproducts. Antibiotics 2021, 10, 1245. [Google Scholar] [CrossRef]
- Angelini, P.; Venanzoni, R.; Angeles Flores, G.; Tirillini, B.; Orlando, G.; Recinella, L.; Chiavaroli, A.; Brunetti, L.; Leone, S.; Di Simone, S.C. Evaluation of antioxidant, antimicrobial and tyrosinase inhibitory activities of extracts from Tricholosporum goniospermum, an edible wild mushroom. Antibiotics 2020, 9, 513. [Google Scholar] [CrossRef]
- Menghini, L.; Leporini, L.; Vecchiotti, G.; Locatelli, M.; Carradori, S.; Ferrante, C.; Zengin, G.; Recinella, L.; Chiavaroli, A.; Leone, S. Crocus sativus L. stigmas and byproducts: Qualitative fingerprint, antioxidant potentials and enzyme inhibitory activities. Food Res. Int. 2018, 109, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Mingyi, Y.; Belwal, T.; Devkota, H.P.; Li, L.; Luo, Z. Trends of Utilizing Mushroom Polysaccharides (MPs) as Potent Nutraceutical Components in Food and Medicine: A Comprehensive Review. Trends Food Sci. Technol. 2019, 92, 94–110. [Google Scholar] [CrossRef]
- Kała, K.; Pająk, W.; Sułkowska-Ziaja, K.; Krakowska, A.; Lazur, J.; Fidurski, M.; Marzec, K.; Zięba, P.; Fijałkowska, A.; Szewczyk, A. Hypsizygus marmoreus as a source of indole compounds and other bioactive substances with health-promoting activities. Molecules 2022, 27, 8917. [Google Scholar] [CrossRef] [PubMed]
- Maffei, M.E. 5-Hydroxytryptophan (5-HTP): Natural occurrence, analysis, biosynthesis, biotechnology, physiology and toxicology. Int. J. Mol. Sci. 2020, 22, 181. [Google Scholar] [CrossRef]
- Muszyńska, B.; Komendacki, P.; Kała, K.; Opoka, W.; Rojowski, J. L-Tryptophan and its derivatives in edible mushrooms species. Med. Int. Rev. 2014, 103, 82–88. [Google Scholar]
- Covino, S.; D’Ellena, E.; Tirillini, B.; Angeles, G.; Arcangeli, A.; Bistocchi, G.; Venanzoni, R.; Angelini, P. Characterization of biological activities of methanol extract of Fuscoporia torulosa (Basidiomycetes) from Italy. Int. J. Med. Mushrooms 2019, 21, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Franzblau, S.G.; Tokuda, H.; Tagata, M.; Ukiya, M.; Matsuzawa, T.; Metori, K.; Kimura, Y.; Suzuki, T.; Yasukawa, K. Antitubercular Activity and Inhibitory Effect on Epstein-Barr Virus Activation of Sterols and Polyisoprenepolyols from an Edible Mushroom, Hypsizigus marmoreus. Biol. Pharm. Bull. 2005, 28, 1117–1119. [Google Scholar] [CrossRef] [PubMed]


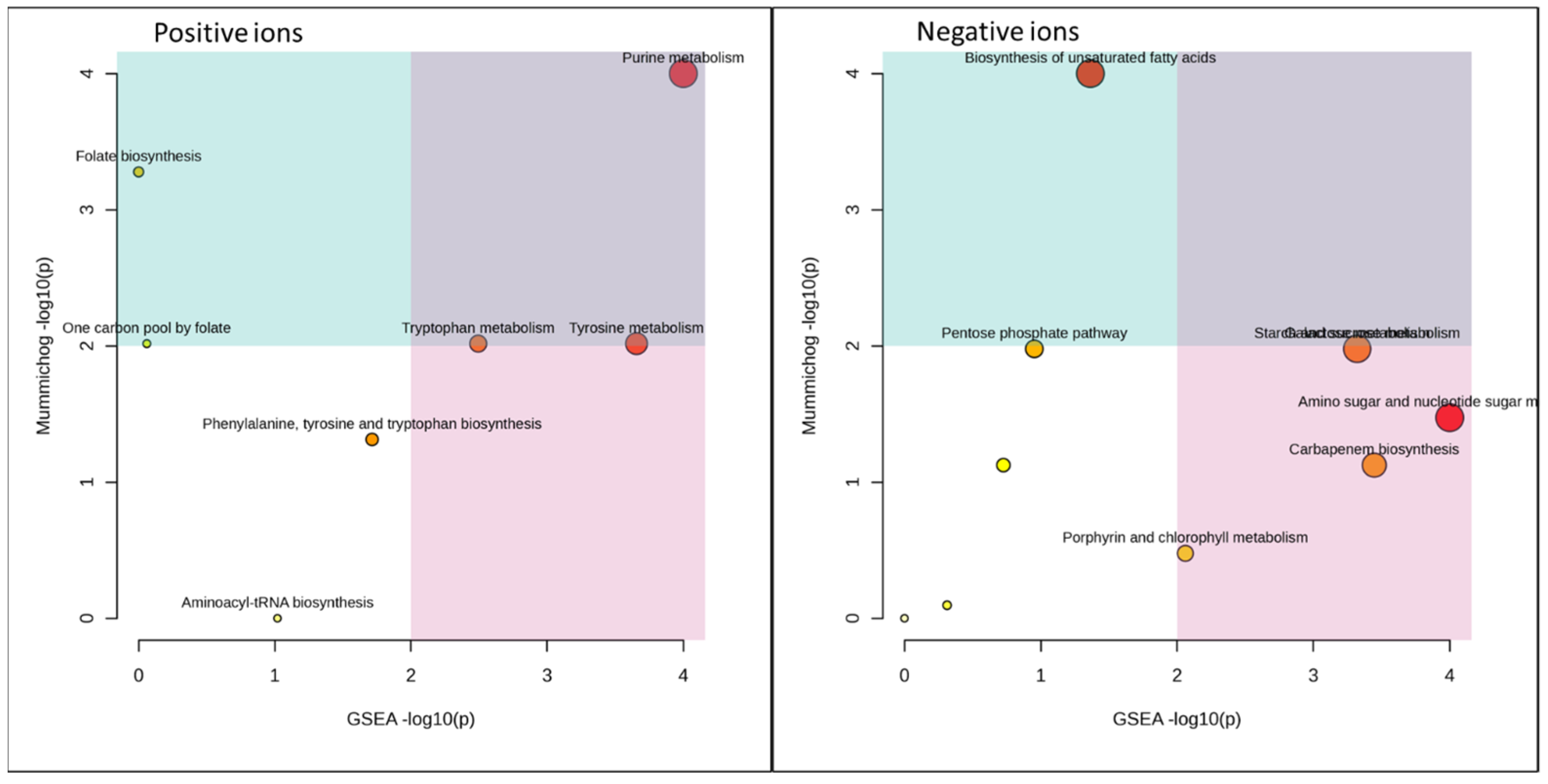
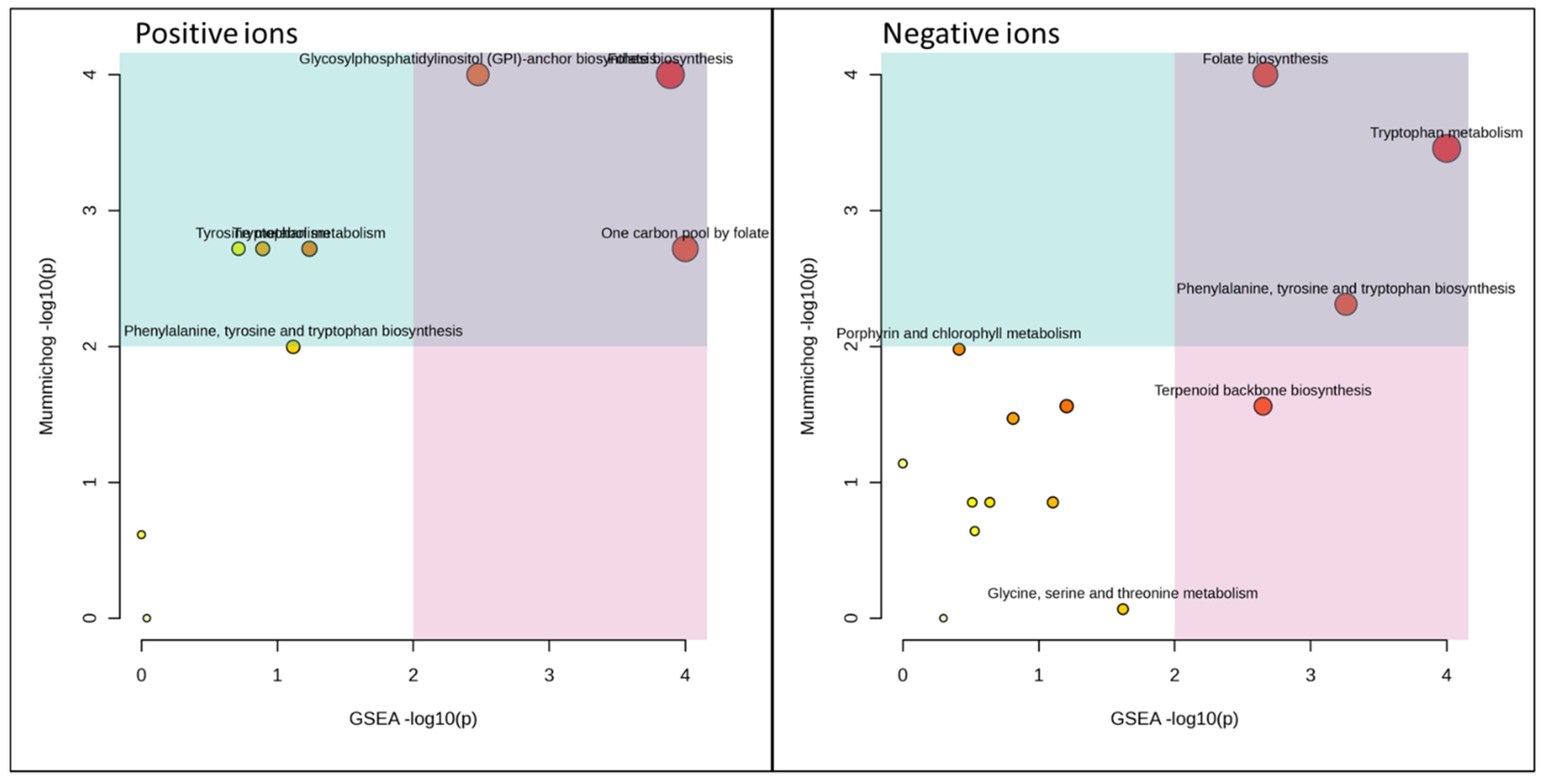
| Barley Malt vs. Control | Total Size | Hits | Sig_Hits | Mummichog p-Values | GSEA p-Values | Combined p-Values |
|---|---|---|---|---|---|---|
| Purine metabolism | 62 | 12 | 4 | 0.06893 | 0.01538 | 0.00832 |
| Tyrosine metabolism | 15 | 2 | 1 | 0.1915 | 0.02128 | 0.0265 |
| Amino sugar and nucleotide sugar metabolism | 22 | 6 | 1 | 0.1549 | 0.03509 | 0.03378 |
| Biosynthesis of unsaturated fatty acids | 23 | 3 | 2 | 0.0228 | 0.2419 | 0.0342 |
| Galactose metabolism | 17 | 7 | 2 | 0.1056 | 0.05769 | 0.03718 |
| Starch and sucrose metabolism | 12 | 5 | 3 | 0.1056 | 0.05769 | 0.03718 |
| Carbapenem biosynthesis | 3 | 2 | 1 | 0.2018 | 0.05263 | 0.0589 |
| Tryptophan metabolism | 30 | 2 | 1 | 0.1915 | 0.06383 | 0.06607 |
| Pentose phosphate pathway | 18 | 2 | 1 | 0.1056 | 0.3269 | 0.1508 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 21 | 3 | 1 | 0.2752 | 0.1333 | 0.1579 |
| Porphyrin and chlorophyll metabolism | 20 | 5 | 1 | 0.3298 | 0.1452 | 0.1934 |
| Folate biosynthesis | 23 | 1 | 1 | 0.1 | 0.6735 | 0.249 |
| Pentose and glucuronate interconversions | 12 | 4 | 2 | 0.2018 | 0.386 | 0.2767 |
| One carbon pool by folate | 8 | 2 | 1 | 0.1915 | 0.6364 | 0.3784 |
| Aminoacyl-tRNA biosynthesis | 22 | 7 | 1 | 0.5414 | 0.2571 | 0.4137 |
| Glycine, serine and threonine metabolism | 28 | 9 | 1 | 0.4403 | 0.5224 | 0.568 |
| Arginine and proline metabolism | 25 | 9 | 2 | 0.4735 | 0.6567 | 0.6742 |
| Grape Pomace vs. Control | Total Size | Hits | Sig_Hits | Mummichog p-Values | GSEA p-Values | Combined p-Values |
|---|---|---|---|---|---|---|
| Amino sugar and nucleotide sugar metabolism | 22 | 6 | 1 | 0.2487 | 0.9375 | 0.5726 |
| Aminoacyl-tRNA biosynthesis | 22 | 7 | 1 | 0.4746 | 0.8529 | 0.7708 |
| Arginine and proline metabolism | 25 | 9 | 2 | 0.6641 | 0.6818 | 0.8116 |
| Carbapenem biosynthesis | 3 | 2 | 1 | 0.3183 | 0.2889 | 0.3114 |
| Folate biosynthesis | 23 | 2 | 1 | 0.02103 | 0.05455 | 0.00891 |
| Galactose metabolism | 17 | 7 | 2 | 0.1728 | 0.2593 | 0.184 |
| Glycine, serine and threonine metabolism | 28 | 9 | 1 | 0.6272 | 0.1667 | 0.3407 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 11 | 1 | 1 | 0.08333 | 0.07547 | 0.03817 |
| N-Glycan biosynthesis | 28 | 3 | 2 | 0.161 | 0.4237 | 0.2514 |
| One carbon pool by folate | 8 | 2 | 1 | 0.161 | 0.01695 | 0.01884 |
| Pantothenate and CoA biosynthesis | 18 | 4 | 1 | 0.3183 | 0.4737 | 0.436 |
| Pentose and glucuronate interconversions | 12 | 4 | 1 | 0.3183 | 0.5439 | 0.4767 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 21 | 4 | 1 | 0.09046 | 0.02899 | 0.01821 |
| Porphyrin and chlorophyll metabolism | 20 | 5 | 2 | 0.1204 | 0.6029 | 0.263 |
| Purine metabolism | 62 | 12 | 1 | 0.6508 | 0.8205 | 0.869 |
| Starch and sucrose metabolism | 12 | 5 | 3 | 0.1728 | 0.2593 | 0.184 |
| Terpenoid backbone biosynthesis | 13 | 2 | 1 | 0.1728 | 0.05556 | 0.05421 |
| Tryptophan metabolism | 30 | 7 | 2 | 0.0336 | 0.01316 | 0.00386 |
| Tyrosine metabolism | 15 | 2 | 1 | 0.161 | 0.3559 | 0.2212 |
| Valine, leucine and isoleucine biosynthesis | 20 | 7 | 2 | 0.1868 | 0.3947 | 0.2659 |
| Valine, leucine and isoleucine degradation | 16 | 5 | 1 | 0.382 | 0.5333 | 0.5278 |
| Sample | mg GAE (Gallic Acid Equivalents)/gdm | SD |
|---|---|---|
| H1 | 3.063 | 0.073 |
| H2 | 3.418 | 0.400 |
| H3 | 3.250 | 1.634 |
| H4 | 2.720 | 0.070 |
| HC | 3.545 | 0.924 |
| Sample | Decolorization (%) (2 mg/mL) | SD | mg TE (Trolox Equivalents/gdm) | SD |
|---|---|---|---|---|
| H1 | 21.25 | 1.64 | 14.323 | 1.339 |
| H2 | 18.92 | 1.06 | 12.428 | 0.862 |
| H3 | 18.25 | 1.957 | 11.878 | 1.594 |
| H4 | 18.19 | 3.356 | 11.831 | 2.734 |
| HC | 21.11 | 1.968 | 14.206 | 1.603 |
| Decolorization (%) (2 mg/mL) | SD | mg TE (Trolox Equivalents/gdm) | SD |
|---|---|---|---|
| 7.81 | 0.72 | 0.587 | 0.254 |
| 6.92 | 0.74 | 0.295 | 0.261 |
| 9.25 | 0.089 | 1.118 | 0.021 |
| 9.90 | 0.095 | 1.331 | 0.034 |
| 7.75 | 0.333 | 0.566 | 0.118 |
| MIC (µg mL−1) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Escherichia coli | Escherichia coli | Escherichia coli | Bacillus cereus | Pseudomonas aeruginosa | Bacillus subtilis | Salmonella typhi | Staphylococcus aureus |
| (ATCC 10536) | (PeruMycA 2) | (PeruMycA 3) | (PeruMycA 4) | (ATCC 15442) | (PeruMycA 6) | (PeruMycA 7) | (ATCC 6538) | |
| Extracts | ||||||||
| HC | 39.68 (25–50) | 9.92 (6.25–12.5) | 62.99 (50–100) | >200 | >200 | 79.37 (50–100) | 62.99 (50–100) | 125.99 (100–200) |
| H1 | 8.29 (6.75–12.5) | 3.93 (3.12–6.25) | 39.68 (25–50) | >200 | >200 | 31.49 (25–50) | 31.49 (25–50) | 79.37 (50–100) |
| H2 | 15.75 (12.5–25) | 3.93 (3.12–6.25) | 31.49 (25–50) | >200 | >200 | 62.99 (50–100) | 62.99 (50–100) | 125.99 (100–200) |
| H3 | 15.75 (12.5–25) | 19.84 (12.5–25) | 9.92 (6.25–12.5) | >200 | >200 | 62.99 (50–100) | 31.49 (25–50) | 62.99 (50–100) |
| H4 | 9.92 (6.25–12.5) | 3.93 (3.12–6.25) | 2.48 (1.56–3.125) | >200 | >200 | 79.37 (50–100) | 31.49 (25–50) | 62.99 (50–100) |
| Ciprofloxacin (µg mL−1) | 31.49 (25–50) | 9.92 (6.25–12.5) | 79.37 (50–100) | 125.99 (100–200) | 125.99 (100–200) | 125.99 (100–200) | 79.37 (50–100) | 200–>200 |
| MIC (μg mL−1) | ||||
|---|---|---|---|---|
| Yeast Strain | Candida tropicalis | Candida albicans | Candida parapsilosis | Candida albicans |
| (YEPGA 6184) | (YEPGA 6379) | (YEPGA 6551) | (YEPGA 6183) | |
| Extracts | ||||
| HC | 79.37 (50–100) | 62.99 (50–100) | 158.74 (100–200) | 79.37 (50–100) |
| H1 | 39.68 (25–50) | 19.84 (12.5–25) | 62.99 (50–100) | 125.99 (100–200) |
| H2 | 39.68 (25–50) | 79.37 (50–100) | 39.68 (25–50) | 79.37 (50–100) |
| H3 | 39.68 (25–50) | 39.68 (25–50) | 62.99 (50–100) | 39.68 (25–50) |
| H4 | 79.37 (50–100) | 79.37 (50–100) | 19.84 (12.5–25) | 62.99 (50–100) |
| Fluconazole (μg mL−1) | 2 | 1 | 4 | 2 |
| MIC (µg mL−1) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dermatophyte | Trichophyton mentagrophytes | Trichophyton tonsurans | Trichophyton rubrum | Arthroderma quadrifidum | Trichophyton mentagrophytes | Arthroderma gypseum | Arthroderma curreyi | Arthroderma insingulare |
| (CCF 4823) | (CCF 4834) | (CCF 4933) | (CCF 5792) | (CCF 5930) | (CCF 6261) | (CCF 5207) | (CCF 5417) | |
| Extracts | ||||||||
| HC | 158.74 (100–200) | 125.99 (100–200) | 125.99 (100–200) | 79.37 (50–100) | >200 | >200 | 158.74 (100–200) | 79.37 (50–100) |
| H1 | 39.68 (25–50) | 31.49 (25–50) | 62.99 (50–100) | 79.37 (50–100) | 125.99 (100–200) | 125.99 (100–200) | 125.99 (100–200) | 62.99 (50–100) |
| H2 | 62.99 (50–100) | 39.68 (25–50) | 31.49 (25–50) | 62.99 (50–100) | 125.99 (100–200) | 100–>200 | 100–>200 | 125.99 (100–200) |
| H3 | 62.99 (50–100) | 31.49 (25–50) | 62.99 (50–100) | 79.37 (50–100) | 158.74 (100–200) | 158.74 (100–200) | 100–>200 | 62.99 (50–100) |
| H4 | 19.84 (12.5–25) | 9.92 (6.25–12.5) | 19.84 (12.5–25) | >200 | >200 | 62.99 (50–100) | 125.99 (100–200) | 79.37 (50–100) |
| Griseofulvin (µg mL−1) | 2.52 (2–4) | 0.198 (0.125–0.25) | 1.26 (1–2) | >8 | 3.174 (2–4) | 1.587 (1–2) | >8 | >8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelini, P.; Flores, G.A.; Cusumano, G.; Venanzoni, R.; Pellegrino, R.M.; Zengin, G.; Di Simone, S.C.; Menghini, L.; Ferrante, C. Bioactivity and Metabolomic Profile of Extracts Derived from Mycelial Solid Cultures of Hypsizygus marmoreus. Microorganisms 2023, 11, 2552. https://doi.org/10.3390/microorganisms11102552
Angelini P, Flores GA, Cusumano G, Venanzoni R, Pellegrino RM, Zengin G, Di Simone SC, Menghini L, Ferrante C. Bioactivity and Metabolomic Profile of Extracts Derived from Mycelial Solid Cultures of Hypsizygus marmoreus. Microorganisms. 2023; 11(10):2552. https://doi.org/10.3390/microorganisms11102552
Chicago/Turabian StyleAngelini, Paola, Giancarlo Angeles Flores, Gaia Cusumano, Roberto Venanzoni, Roberto Maria Pellegrino, Gokhan Zengin, Simonetta Cristina Di Simone, Luigi Menghini, and Claudio Ferrante. 2023. "Bioactivity and Metabolomic Profile of Extracts Derived from Mycelial Solid Cultures of Hypsizygus marmoreus" Microorganisms 11, no. 10: 2552. https://doi.org/10.3390/microorganisms11102552
APA StyleAngelini, P., Flores, G. A., Cusumano, G., Venanzoni, R., Pellegrino, R. M., Zengin, G., Di Simone, S. C., Menghini, L., & Ferrante, C. (2023). Bioactivity and Metabolomic Profile of Extracts Derived from Mycelial Solid Cultures of Hypsizygus marmoreus. Microorganisms, 11(10), 2552. https://doi.org/10.3390/microorganisms11102552














