Characterizing the Gut Microbial Communities of Native and Invasive Freshwater Bivalves after Long-Term Sample Preservation
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Extraction, Amplification, and Sequencing
2.2. Statistical Analysis
3. Results
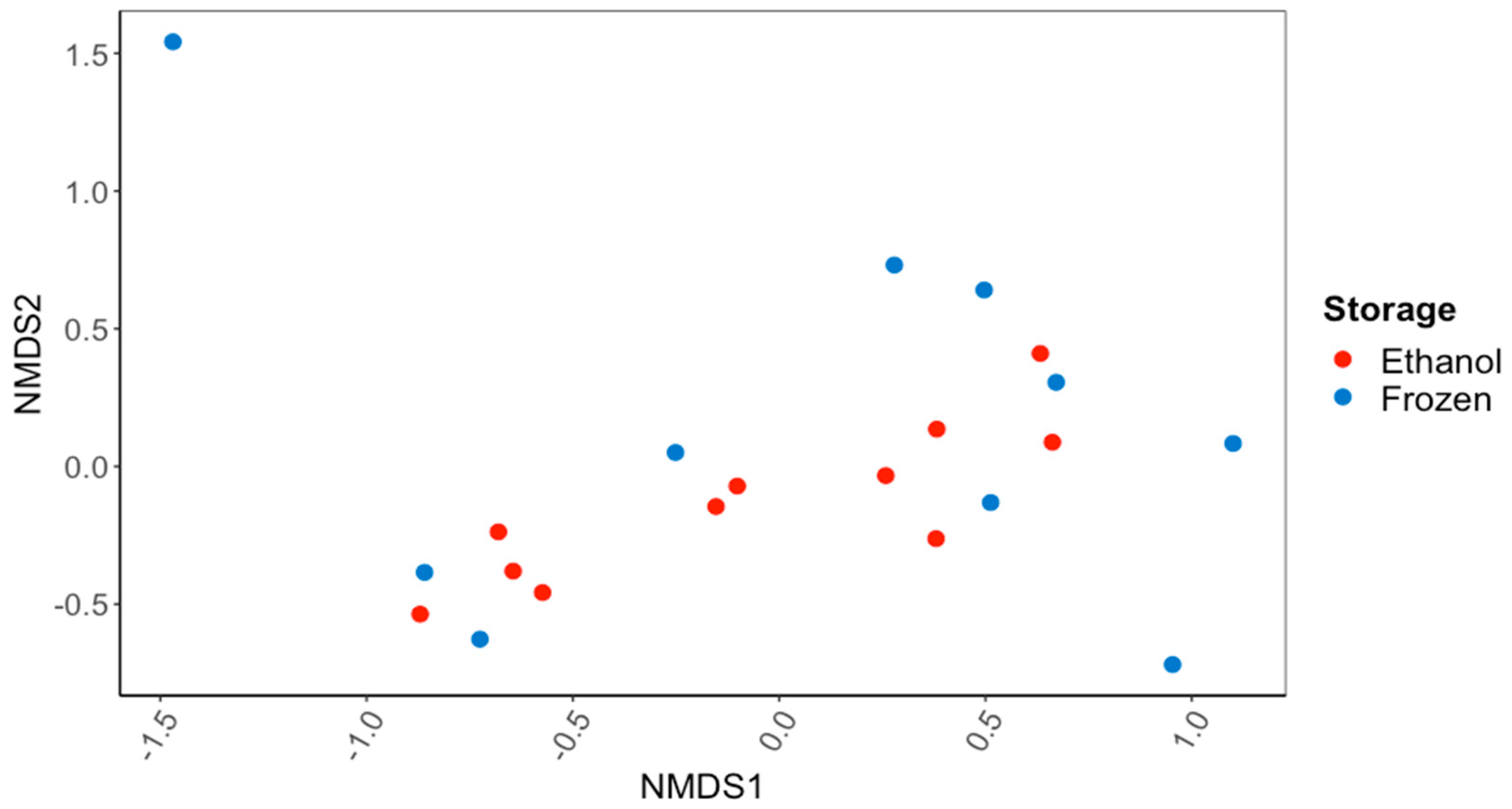
3.1. Characterization of the Gut Microbiomes of Ethanol-Preserved and Frozen C. fluminea
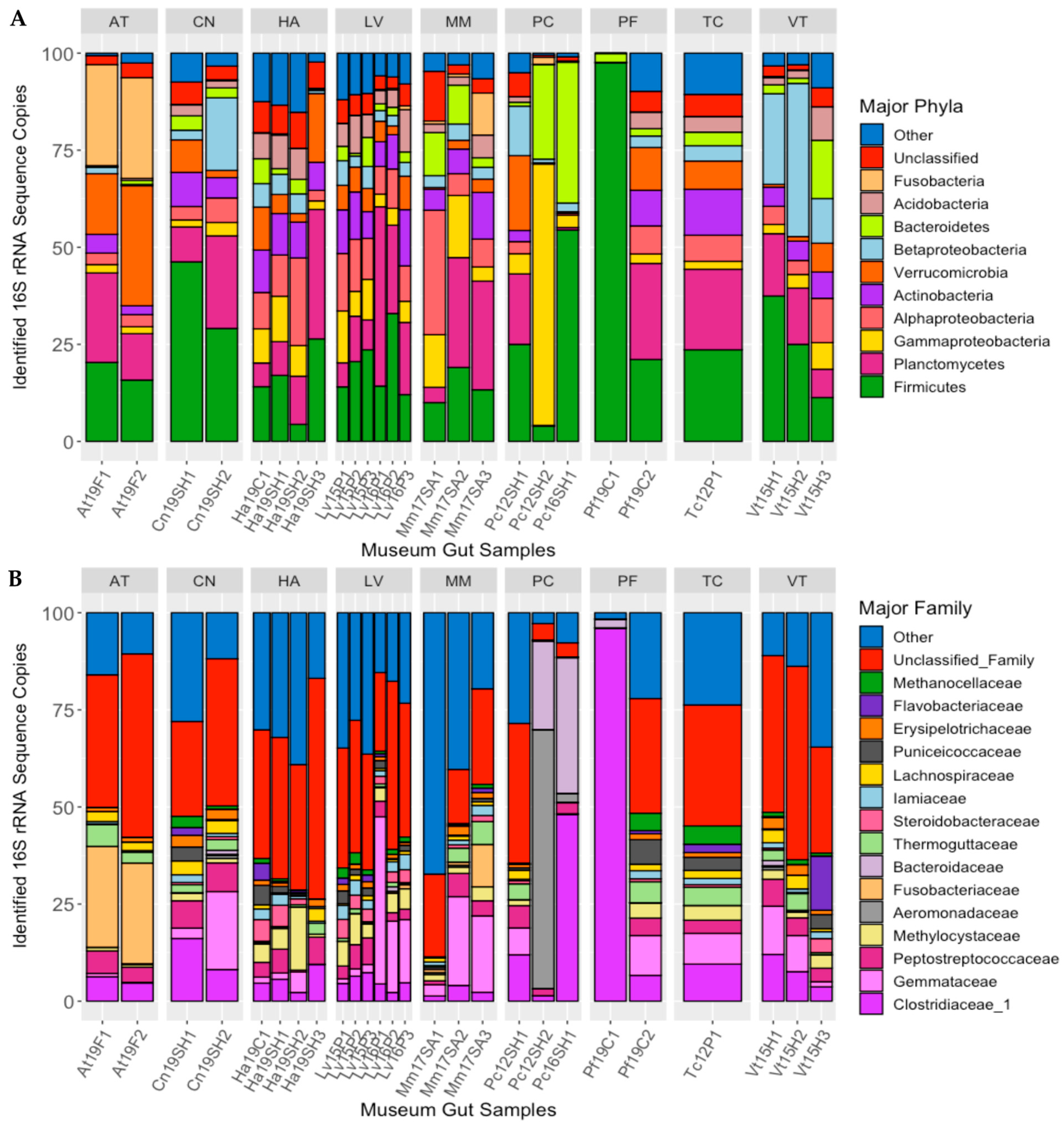
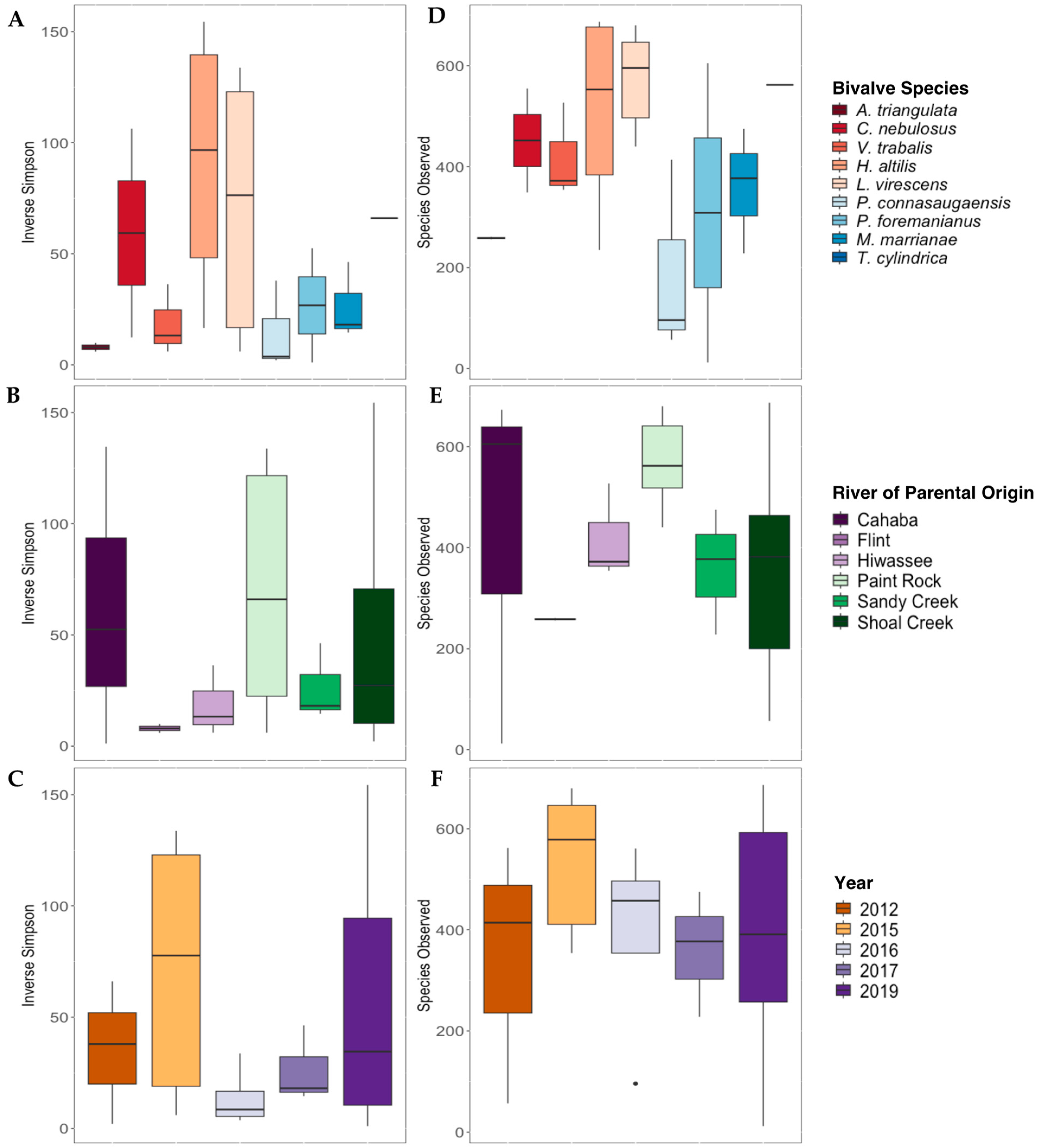
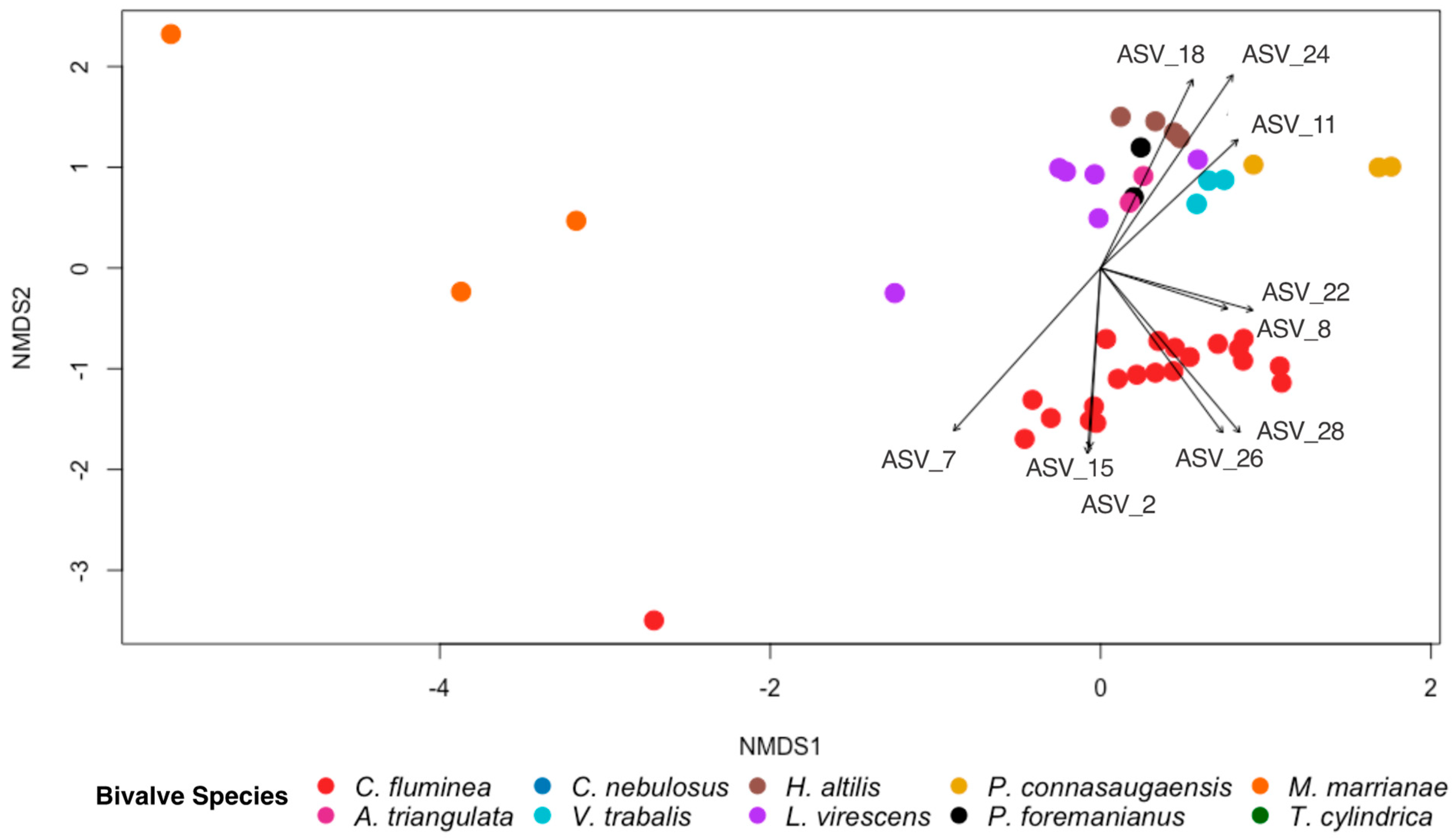
3.2. Characterization of the Gut Microbiomes of Long-Term-Stored Museum Mussels
3.3. Amplicon Sequence Variants
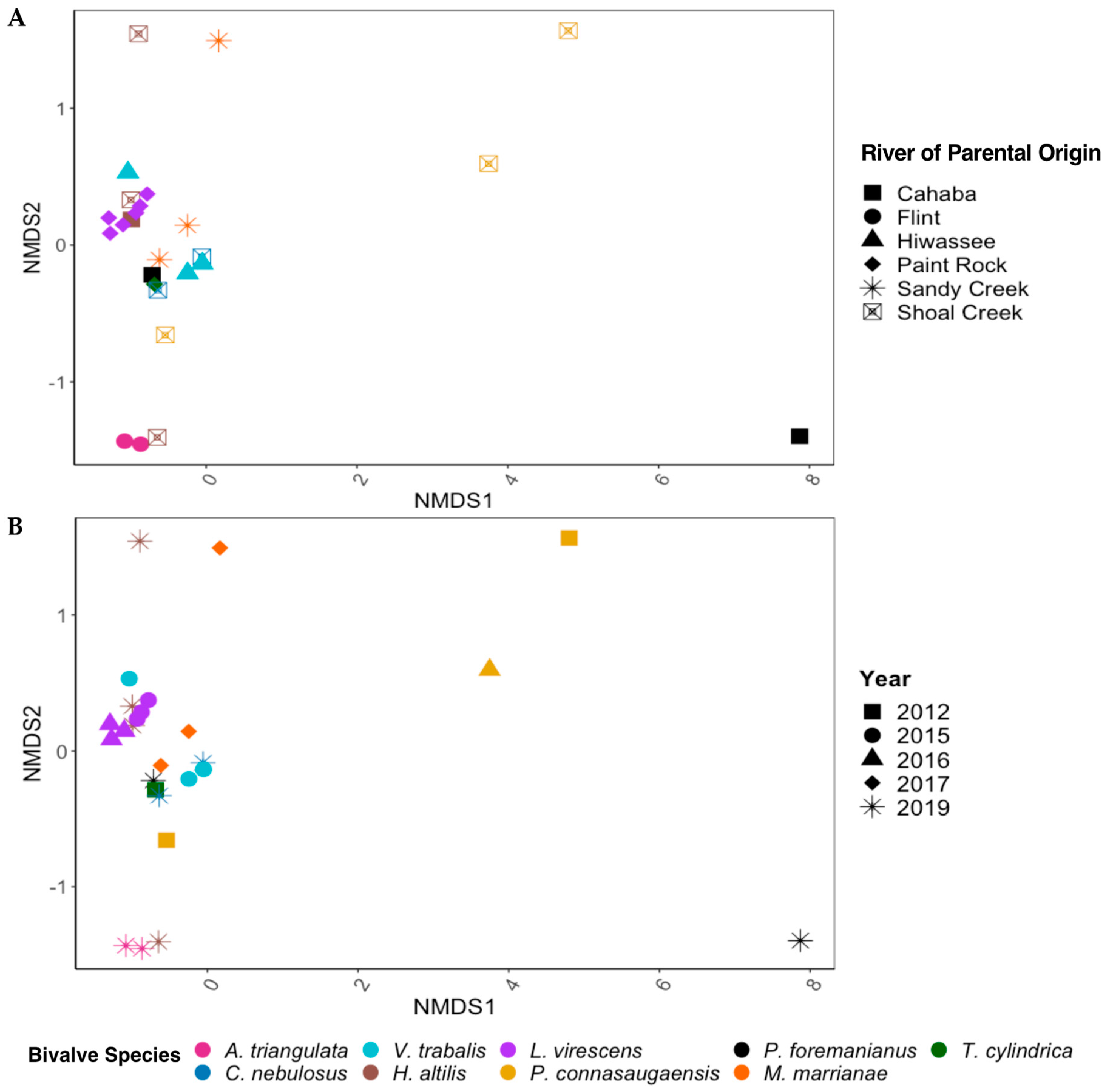
3.4. Comparison of the C. fluminea and Native Mussel Gut Microbiomes
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prather, C.M.; Pelini, S.L.; Laws, A.; Rivest, E.; Woltz, M.; Bloch, C.P.; Del Toro, I.; Ho, C.-K.; Kominoski, J.; Newbold, T.A.S.; et al. Invertebrates, ecosystem services and climate change. Biol. Rev. 2013, 88, 327–348. [Google Scholar]
- Vaughn, C.C. Ecosystem services provided by freshwater mussels. Hydrobiologia 2018, 810, 15–27. [Google Scholar]
- Weingarten, E.A.; Atkinson, C.L.; Jackson, C.R. The gut microbiome of freshwater Unionidae mussels is determined by host species and is selectively retained from filtered seston. PLoS ONE 2019, 14, e0224796. [Google Scholar] [CrossRef]
- Hooper, D.U.; Adair, E.C.; Cardinale, B.J.; Byrnes, J.E.K.; Hungate, B.A.; Matulich, K.L.; Gonzalez, A.; Duffy, J.E.; Gamfeldt, L.; O’Connor, M.I. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 2012, 486, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Isbel, F.; Gonzalez, A.; Loreau, M.; Cowles, J.; Diaz, S.; Hector, A.; Mace, G.M.; Wardle, D.A.; O’Connor, M.I.; Duffy, J.E.; et al. Linking the influence and dependence of people on biodiversity across scales. Nature 2017, 546, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Haag, W.R. Reassessing enigmatic mussel declines in the United States. FMBC 2019, 22, 43–60. [Google Scholar]
- Haag, W.R. The decline of the North American mussel fauna: Chronology and causes. In North American Freshwater Mussels: Natural History, Ecology, and Conservation; Cambridge University Press: Cambridge, UK, 2012; pp. 316–389. [Google Scholar]
- Osterling, M.E.; Arvidsson, B.L.; Greenberg, L.A. Habitat degradation and the decline of the threatened mussel Margaritifera margaritifera: Influence of turbidity and sedimentation on the mussel and its host. J. Appl. Ecol. 2010, 47, 759–768. [Google Scholar] [CrossRef]
- Downing, J.A.; Meter, P.V.; Woolnough, D.A. Suspects and evidence: A review of the causes of extirpation and decline in freshwater mussels. Anim. Biodivers. Conserv. 2010, 33, 151–185. [Google Scholar] [CrossRef]
- Wang, N.; Ingersoll, C.G.; Hardesty, D.K.; Kunz, J.L.; May, T.W.; Dwyer, F.J.; Roberts, A.D.; Augspurger, T.; Kane, C.M.; Neves, R.J.; et al. Acute toxicity of copper, ammonia, and chlorine to glochidia and juveniles of freshwater mussels (Unionidae). Environ. Toxicol. Chem. 2007, 26, 2036–2047. [Google Scholar] [CrossRef]
- Karatayev, A.Y.; Miller, T.D.; Burlakova, L.E. Long-term changes in unionid assemblages in the Rio Grande, one of the World’s top 10 rivers at risk. Aquat. Conserv. 2012, 22, 206–219. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Makhrov, A.A.; Gofarov, M.Y.; Aksenova, O.V.; Aspholm, P.E.; Bespalaya, Y.V.; Kabakov, M.B.; Kolosova, Y.S.; Kondakov, A.V.; Ofenbock, T.; et al. Climate warming as a possible trigger of keystone mussel population decline in oligotrophic rivers at the continental scale. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Gallardo, B.; Bogan, A.E.; Harun, S.; Jainih, L.; Lopes-Lima, M.; Pizzaro, M.; Rahim, K.A.; Sousa, R.; Virdis, S.G.P.; Zieritz, A. M Current and future effects of global change on a hotspot’s freshwater diversity. Sci. Total Environ. 2018, 635, 750–760. [Google Scholar] [CrossRef]
- Atkinson, C.L.; Julian, J.P.; Vaughn, C.C. Species and function lost: Role of drought in structuring stream communities. Biol. Conserv. 2014, 176, 30–38. [Google Scholar] [CrossRef]
- Aldridge, D.C.; Ollard, I.S.; Bespalaya, Y.V.; Bolotov, I.N.; Douda, K.; Geist, J.; Haag, W.R.; Klunzinger, M.W.; Lopes-Lima, M.; Mlambo, M.C.; et al. Freshwater mussel conservation: A global horizon scan of emerging threats and opportunities. Glob. Chang. Biol. 2022, 29, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, N.B.; Heisterkamp, I.M.; Sigby-Clausen, M.; Larsen, L.H.; Nielsen, L.P.; Stief, P.; Schramm, A. Shell biofilm nitrification and gut denitrification contribute to emission of nitrous oxide by the invasive freshwater mussel Dreissena polymorpha (Zebra Mussel). Appl. Environ. Microbiol. 2012, 78, 4505–4509. [Google Scholar] [CrossRef]
- Black, E.M.; Chimenti, M.S.; Just, C.L. Effect of freshwater mussels on vertical distribution of anaerobic ammonia oxidizers and other nitrogen-transforming microorganisms in upper Mississippi river sediment. PeerJ 2016, 5, e3536. [Google Scholar] [CrossRef]
- Hoellein, T.J.; Zarnoch, C.B.; Bruesewitz, D.A.; DeMartini, J. Contributions of freshwater mussels (Unionidae) to nutrient cycling in an urban river: Filtration, recycling, storage, and removal. Biogeochemistry 2017, 135, 307–324. [Google Scholar] [CrossRef]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 10, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Minich, J.J.; Morris, M.M.; Brown, M.; Doane, M.; Edwards, M.S.; Michael, T.P.; Dinsdale, E.A. Elevated temperature drives kelp microbiome dysbiosis, while elevated carbon dioxide induces water microbiome disruption. PLoS ONE 2018, 13, e0192772. [Google Scholar] [CrossRef]
- Lauber, C.L.; Zhou, N.; Gordon, J.I.; Knight, R.; Fierer, N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 2010, 307, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Moreau, C.S.; Wray, B.D.; Czekanski-Moir, J.E.; Rubin, B.E.R. DNA preservation: A test of commonly used preservatives for insects. Invertebr. Syst. 2013, 27, 81–86. [Google Scholar] [CrossRef]
- Hale, V.L.; Tan, C.L.; Knight, R.; Amato, K.R. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J. Microbiol. Methods 2015, 113, 16–26. [Google Scholar] [CrossRef]
- Hammer, T.J.; Dickerson, J.C.; Fierer, N. Evidence-based recommendations on storing and handling specimens for analyses of insect microbiota. PeerJ 2015, 3, e1190. [Google Scholar] [CrossRef]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 2016, 1, e00021-16. [Google Scholar] [CrossRef]
- Heindler, F.M.; Christiansen, H.; Frédérich, B.; Dettaï, A.; Lepoint, G.; Maes, G.E.; Van de Putte, A.P.; Morgan, K. Historical DNA metabarcoding of the prey and microbiome of trematomid fishes using museum samples. Front. Ecol. Evol. 2018, 6, 51. [Google Scholar] [CrossRef]
- Horng, K.R.; Ganz, H.H.; Eisen, J.A.; Marks, S.L. Effects of preservation method on canine (Canis lupus familiaris) fecal microbiota. PeerJ 2018, 6, e4827. [Google Scholar] [CrossRef]
- Neu, A.T.; Hughes, I.V.; Allen, E.E.; Roy, K. Decade-scale stability and change in marine bivalve microbiome. Mol. Ecol. 2021, 30, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Chalifour, B.N.; Elder, L.E.; Li, J. Gut microbiome of century-old snail specimens stable across time in preservation. Microbiome 2022, 10, 99. [Google Scholar] [CrossRef]
- Vaughn, S.N.; Jackson, C.R. Evaluating methods of preserving aquatic invertebrates for microbiome analysis. Microorganisms 2022, 10, 811. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Jackson, C.R.; Stone, B.W.G.; Tyler, H.L. Emerging perspectives on the natural microbiome of fresh produce vegetables. Agriculture 2015, 5, 170–187. [Google Scholar] [CrossRef]
- Stone, B.W.G.; Jackson, C.R. Biogeographic patterns between bacterial phyllosphere communities of the Southern Magnolia (Magnolia grandiflora) in a small forest. Microb. Ecol. 2016, 71, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 21 August 2022).
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Mikryukov, V. metagMisc: Miscellaneous functions for metagenomic analysis. Zenodo 2017. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Lahti, L.; Sudarshan, S.; Blake, T.; Salojarvi, J. Tools for microbiome analysis in R. Version 2017, 1, 28. [Google Scholar]
- Aceves, A. Studies on the Digestive Gland Microbiome of Freshwater Mussels. Ph.D. Thesis, University of Auburn, Auburn, AL, USA, 2019. [Google Scholar]
- Pierce, M.L.; Ward, J.E. Gut microbiomes of the Eastern Oyster (Crassostrea virginica) and the Blue Mussel (Mytilus edulis): Temporal variation and the influence of marine aggregate-associated microbial communities. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Griffin, T.W.; Baer, J.G.; Ward, J.E. Direct comparisons of fecal and gut microbiota in the Blue Mussel (Mytilus edulis) discourages fecal sampling as a proxy for resident gut community. Invertebr. Biol. 2021, 81, 180–192. [Google Scholar] [CrossRef] [PubMed]
- McCauley, M.; Chiarello, M.; Atkinson, C.L.; Jackson, C.R. Gut microbiomes of freshwater mussels (Unionidae) are taxonomically and phylogenetically variable across years but remain functionally stable. Microorganisms 2021, 9, 411. [Google Scholar] [CrossRef] [PubMed]
- Lawson, L.A.; Atkinson, C.L.; Jackson, C.R. The gut bacterial microbiome of the Threeridge mussel, Amblema plicata, varies between rivers but shows a consistent core community. Freshw. Biol. 2022, 67, 1125–1136. [Google Scholar] [CrossRef]
- Chiarello, M.; Bucholz, J.R.; McCauley, M.; Vaughn, S.N.; Hopper, G.W.; Sánchez González, I.; Atkinson, C.L.; Lozier, J.D.; Jackson, C.R. Environment and co-occurring native mussel species, but not host genetics, impact the microbiome of a freshwater invasive species (Corbicula fluminea). Front. Microbiol. 2022, 13, 800061. [Google Scholar] [CrossRef]
- Millar, E.N.; Kiss, K.A.; Surette, M.G.; Bennett, C.J.; Salerno, J.; Gillis, P.L. Effects of municipal wastewater effluents on the digestive gland microbiome of wild freshwater mussels (Lasmigona costata). Ecotoxicoll. Environ. Saf. 2022, 241, 113774. [Google Scholar] [CrossRef]
- Santibáñez, P.; Romalde, J.; Maldonado, J.; Fuentes, D.; Figueroa, J. First characterization of the gut microbiome associated with Mytilus chilensis collected at a mussel farm and from a natural environment in Chile. Aquaculture 2022, 548, 737644. [Google Scholar] [CrossRef]
- Mioduchowska, M.; Zajac, K.; Bartoszek, K.; Madanecki, P.; Kur, J.; Zajac, T. 16S rRNAgene-based metagenomic analysis of the gut microbial community associated with the DUI species Unio crassus (Bivalvia: Unionidae). J. Zoolog. Syst. Evol. Res. 2020, 58, 615–623. [Google Scholar] [CrossRef]
- Higgins, E.; Parr, T.B.; Vaughn, C.C. Mussels and local conditions interact to influence. Microbial communities in mussel beds. Front. Microbiol. 2022, 12, 790554. [Google Scholar] [CrossRef]
- Bahrndorff, S.; Alemu, T.; Alemneh, T.; Lund Nielsen, J. The microbiome of animals: Implications for conservation biology. Int. J. Genom. 2016, 2016, 5304028. [Google Scholar] [CrossRef]
- Goddard-Dwyer, M.; López-Legentil, S.; Erwin, P.M. Microbiome variability across the native and invasive ranges of the ascidian Clavelina oblonga. Appl. Environ. Microbiol. 2021, 87, e2233-20. [Google Scholar] [CrossRef]
- Ferreira-Rodríguez, N.; Gangloff, M.; Shafer, G.; Atkinson, C.L. Drivers of ecosystem vulnerability to Corbicula invasions in southeastern North America. Biol. Invasions. 2022, 24, 1677–1688. [Google Scholar] [CrossRef]
- Kelley, T.E.; Hopper, G.W.; Sánchez González, I.; Bucholz, J.R.; Atkinson, C.L. Identifying potential drivers of distribution patterns of invasive Corbicula fluminea relative to native freshwater mussels (Unionidae) across spatial scales. Ecol. Evol. 2022, 12, e8737. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.L.; Opsahl, S.P.; Covich, A.P.; Golladay, S.W.; Conner, L.M. Stable isotopic signatures, tissue stoichiometry, and nutrient cycling (C and N) of native and invasive freshwater bivalves. J. N. Am. Benthol. 2010, 29, 496–505. [Google Scholar] [CrossRef]
- Strayer, D.L. Effects of alien species on freshwater mollusks in North America. J. N. Am. Benthol. 1999, 18, 74–98. [Google Scholar] [CrossRef]
- Cherry, D.S.; Scheller, J.L.; Cooper, N.L.; Bidwell, J.R. Potential effects of Asian clam (Corbicula fluminea) dieoffs on native freshwater mussels (Unionidae): Watercolumn ammonia levels and ammonia toxicity. J. North Am. Benthol. 2005, 24, 369–380. [Google Scholar] [CrossRef]
- Ferreira-Rodríguez, N.; Sousa, N.; Pardo, I. Negative effects of Corbicula fluminea over native freshwater mussels. Hydrobiologia 2018, 810, 85–95. [Google Scholar] [CrossRef]
- Zhao, Z.; Mao, Z.; Xing, P.; Tao, X.; Wu, Q. Intrahabitat differences in bacterial communities associated with Corbicula fluminea in the large shallow eutrophic Lake Taihu. Appl. Environ. Microbiol. 2021, 88, e02328-21. [Google Scholar]
- Gerritsen, J.; Hotnung, B.; Ritari, J.; Paulin, L.; Rijkers, G.T.; Schaap, P.J.; de Vos, W.M.; Smidt, H. A comparative and functional genomics analysis of the genus Romboutsia provides insight into adaptation to an intestinal lifestyle. bioRxiv 2019, 845511. [Google Scholar] [CrossRef]
- Das, T.; Jayasudha, R.; Chakravarthy, S.; Sai Prashanthi, G.; Bhargava, A.; Tyagi, M.; Kumari Rani, P.; Reddy Pappuru, R.; Sharma, S.; Shivaji, S. Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Sci. Rep. 2021, 11, 2738. [Google Scholar] [CrossRef]
| Mussel Tribe | Mussel Species | Preservation Year | River of Origin | Sample ID |
|---|---|---|---|---|
| Alasmidontini | Alasmidonta triangulata | 2019 | Flint (AL) | At19F1 |
| Alasmidonta triangulata | 2019 | Flint (AL) | At19F2 | |
| Cambarunio nebulosus | 2019 | Shoal Creek (AL) | Cn19SH1 | |
| Cambarunio nebulosus | 2019 | Shoal Creek (AL) | Cn19SH2 | |
| Venustaconcha trabalis | 2015 | Hiwassee (TN) | Vt15H1 | |
| Venustaconcha trabalis | 2015 | Hiwassee (TN) | Vt15H2 | |
| Venustaconcha trabalis | 2015 | Hiwassee (TN) | Vt15H3 | |
| Lampsilini | Hamiota altilis | 2019 | Cahaba (AL) | Ha19C1 |
| Hamiota altilis | 2019 | Cahaba (AL) | Ha19C2 1 | |
| Hamiota altilis | 2019 | Shoal Creek (AL) | Ha19SH1 | |
| Hamiota altilis | 2019 | Shoal Creek (AL) | Ha19SH2 | |
| Hamiota altilis | 2019 | Shoal Creek (AL) | Ha19SH3 | |
| Lampsilis virescens | 2015 | Paint Rock (AL) | Lv15P1 | |
| Lampsilis virescens | 2015 | Paint Rock (AL) | Lv15P2 | |
| Lampsilis virescens | 2015 | Paint Rock (AL) | Lv15P3 | |
| Lampsilis virescens | 2016 | Paint Rock (AL) | Lv16P1 | |
| Lampsilis virescens | 2016 | Paint Rock (AL) | Lv16P2 | |
| Lampsilis virescens | 2016 | Paint Rock (AL) | Lv16P3 | |
| Pseudodontoideus connasaugaensis | 2012 | Shoal Creek (AL) | Pc12SH1 | |
| Pseudodontoideus connasaugaensis | 2012 | Shoal Creek (AL) | Pc12SH2 | |
| Pseudodontoideus connasaugaensis | 2016 | Shoal Creek (AL) | Pc16SH1 | |
| Ptychobranchus foremanianus | 2019 | Cahaba (AL) | Pf19C1 | |
| Ptychobranchus foremanianus | 2019 | Cahaba (AL) | Pf19C2 | |
| Margaritifera marrianae | 2017 | Sandy Creek (AL) | Mm17SA1 | |
| Margaritiferini | Margaritifera marrianae | 2017 | Sandy Creek (AL) | Mm17SA2 |
| Margaritifera marrianae | 2017 | Sandy Creek (AL) | Mm17SA3 | |
| Pleurobemini | Theliderma cylindrica | 2012 | Paint Rock (AL) | Tc12P1 |
| Theliderma cylindrica | 2012 | Paint Rock (AL) | Tc12P2 1 | |
| Theliderma cylindrica | 2012 | Paint Rock (AL) | Tc12P3 1 |
| Specimen | ASV | Identification | Frequency a |
|---|---|---|---|
| Ethanol C. fluminea | ASV 8 | Romboutsia sedimentorum (Firmicutes) | 10/11 |
| ASV 15 | Peptostreptococcaceae (Firmicutes) | 9/11 | |
| ASV 2 | Paeniclostridium (Firmicutes) | 9/11 | |
| ASV 26 | Lacipirellula (Planctomycetes) | 8/11 | |
| ASV 23 | Romboutsia (Firmicutes) | 8/11 | |
| ASV 19 | Clostridiales (Firmicutes) | 8/11 | |
| ASV 31 | Peptostreptococcaceae (Firmicutes) | 7/11 | |
| ASV 28 | Romboutsia (Firmicutes) | 7/11 | |
| ASV 22 | Methylocystis (Alphaproteobacteria) | 7/11 | |
| ASV 7 | Clostridium chauvoei (Firmicutes) | 7/11 | |
| Frozen C. fluminea | ASV 8 | Romboutsia sedimentorum (Firmicutes) | 10/10 |
| ASV 15 | Peptostreptococcaceae (Firmicutes) | 9/10 | |
| ASV 2 | Paeniclostridium (Firmicutes) | 9/10 | |
| ASV 23 | Romboutsia (Firmicutes) | 8/10 | |
| ASV 19 | Clostridiales (Firmicutes) | 8/10 | |
| ASV 31 | Peptostreptococcaceae (Firmicutes) | 7/10 | |
| ASV 26 | Lacipirellula (Planctomycetes) | 7/10 | |
| ASV 22 | Methylocystis (Alphaproteobacteria) | 7/10 | |
| ASV 7 | Clostridium chauvoei (Firmicutes) | 6/10 | |
| ASV 39 | Methylocystis (Alphaproteobacteria) | 6/10 |
| Mussel Tribe | Mussel Species | ASV | Identification | Frequency a |
|---|---|---|---|---|
| Alasmidonta triangulata | ASV 41 | Isosphaeraceae (Planctomycetes) | 2/2 | |
| Alasmidontini | ASV 16 | Pirellulales (Planctomycetes) | 2/2 | |
| ASV 12 | Pirellulales (Planctomycetes) | 2/2 | ||
| Cambarunio nebulosus | ASV 25 | Clostridiaceae_1 (Firmicutes) | 2/2 | |
| ASV 24 | Turicibacter (Firmicutes) | 2/2 | ||
| ASV 21 | Clostridiaceae_1 (Firmicutes) | 2/2 | ||
| Venustaconcha trabalis | ASV 24 | Turicibacter (Firmicutes) | 3/3 | |
| ASV 18 | Methylocystis (Alphaproteobacteria) | 3/3 | ||
| ASV 8 | Romboutsia sedimentorum (Firmicutes) | 3/3 | ||
| Lampsilini | Hamiota altilis | ASV 8 | Romboutsia sedimentorum (Firmicutes) | 3/4 |
| ASV 24 | Turicibacter (Firmicutes) | 2/4 | ||
| ASV 720 | Planctomicrobium (Planctomycetes) | 1/4 | ||
| Lampsilis virescens | ASV 22 | Methylocystis (Alphaproteobacteria) | 6/6 | |
| ASV 18 | Methylocystis (Alphaproteobacteria) | 6/6 | ||
| ASV 8 | Romboutsia sedimentorum (Firmicutes) | 6/6 | ||
| Pseudodontoideus connasaugaensis | ASV 16 | Pirellulales (Planctomycetes) | 3/3 | |
| ASV 12 | Pirellulales (Planctomycetes) | 3/3 | ||
| ASV 8 | Romboutsia sedimentorum (Firmicutes) | 3/3 | ||
| Ptychobranchus foremanianus | ASV 14 | Bacteroides luti (Bacteroidetes) | 2/2 | |
| ASV 10 | Aeromonas (Gammaproteobacteria) | 2/2 | ||
| ASV 965 | Peptostreptococcaceae (Firmicutes) | 1/2 | ||
| Margaritifera marrianae | ASV 4 | Gemmataceae (Planctomycetes) | 3/3 | |
| Margaritiferini | ASV 131 | Chryseobacterium (Bacteroidetes) | 2/3 | |
| ASV 52 | Pseudomonas (Gammaproteobacteria) | 2/3 | ||
| Theliderma cylindrica | ASV 471 | Selenomonadaceae (Firmicutes) | 1/1 | |
| Pleurobemini | ASV 14 | Bacteroides luti (Bacteroidetes) | 1/1 | |
| ASV 9 | Anaerobacter (Firmicutes) | 1/1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaughn, S.N.; Atkinson, C.L.; Johnson, P.D.; Jackson, C.R. Characterizing the Gut Microbial Communities of Native and Invasive Freshwater Bivalves after Long-Term Sample Preservation. Microorganisms 2023, 11, 2489. https://doi.org/10.3390/microorganisms11102489
Vaughn SN, Atkinson CL, Johnson PD, Jackson CR. Characterizing the Gut Microbial Communities of Native and Invasive Freshwater Bivalves after Long-Term Sample Preservation. Microorganisms. 2023; 11(10):2489. https://doi.org/10.3390/microorganisms11102489
Chicago/Turabian StyleVaughn, Stephanie N., Carla L. Atkinson, Paul D. Johnson, and Colin R. Jackson. 2023. "Characterizing the Gut Microbial Communities of Native and Invasive Freshwater Bivalves after Long-Term Sample Preservation" Microorganisms 11, no. 10: 2489. https://doi.org/10.3390/microorganisms11102489
APA StyleVaughn, S. N., Atkinson, C. L., Johnson, P. D., & Jackson, C. R. (2023). Characterizing the Gut Microbial Communities of Native and Invasive Freshwater Bivalves after Long-Term Sample Preservation. Microorganisms, 11(10), 2489. https://doi.org/10.3390/microorganisms11102489







