Genetic- and Fiber-Diet-Mediated Changes in Antibiotic Resistance Genes in Pig Colon Contents and Feces and Their Driving Factors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animal Trial and Sample Collection
2.3. DNA Extraction, Library Construction, and Metagenomic Sequencing
2.4. Sequence Quality Control and Genome Assembly
2.5. Gene Prediction, Taxonomy, and Functional Annotation
2.6. SCFA Determination
2.7. qPCR
2.8. In Vitro Fermentation Assay
2.9. Statistics
3. Results
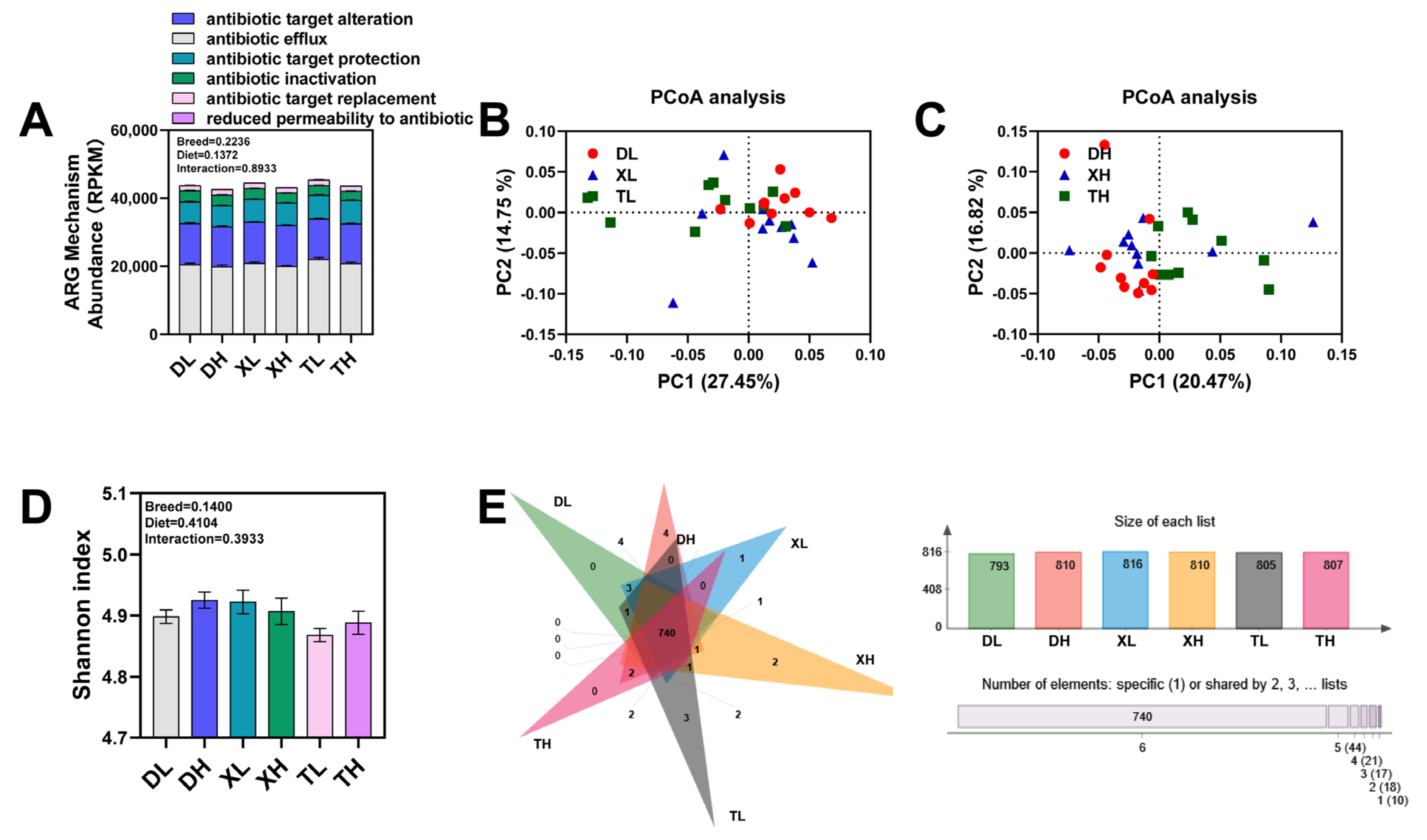
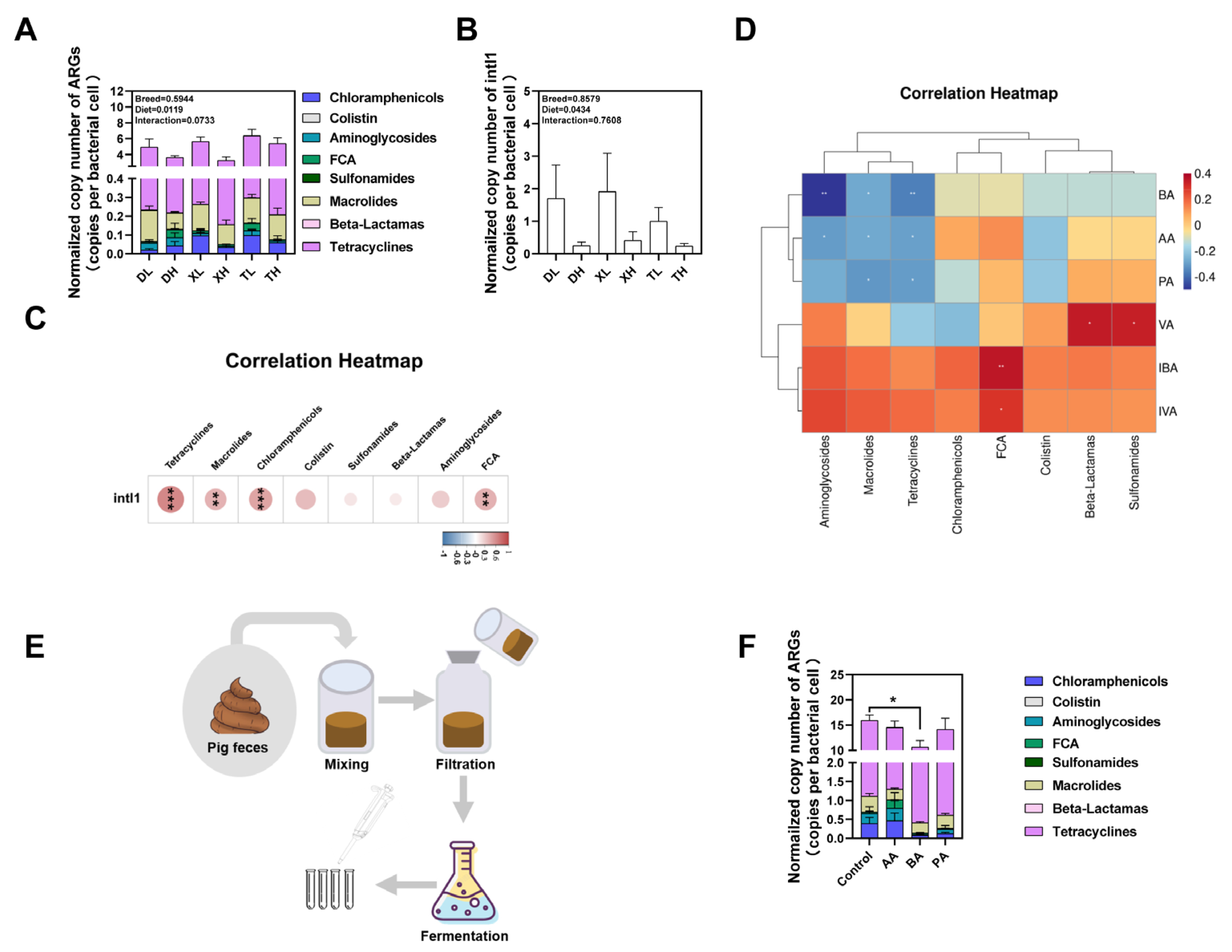
3.1. Expression Profiling of ARGs in Colonic Contents of Different Pig Breeds
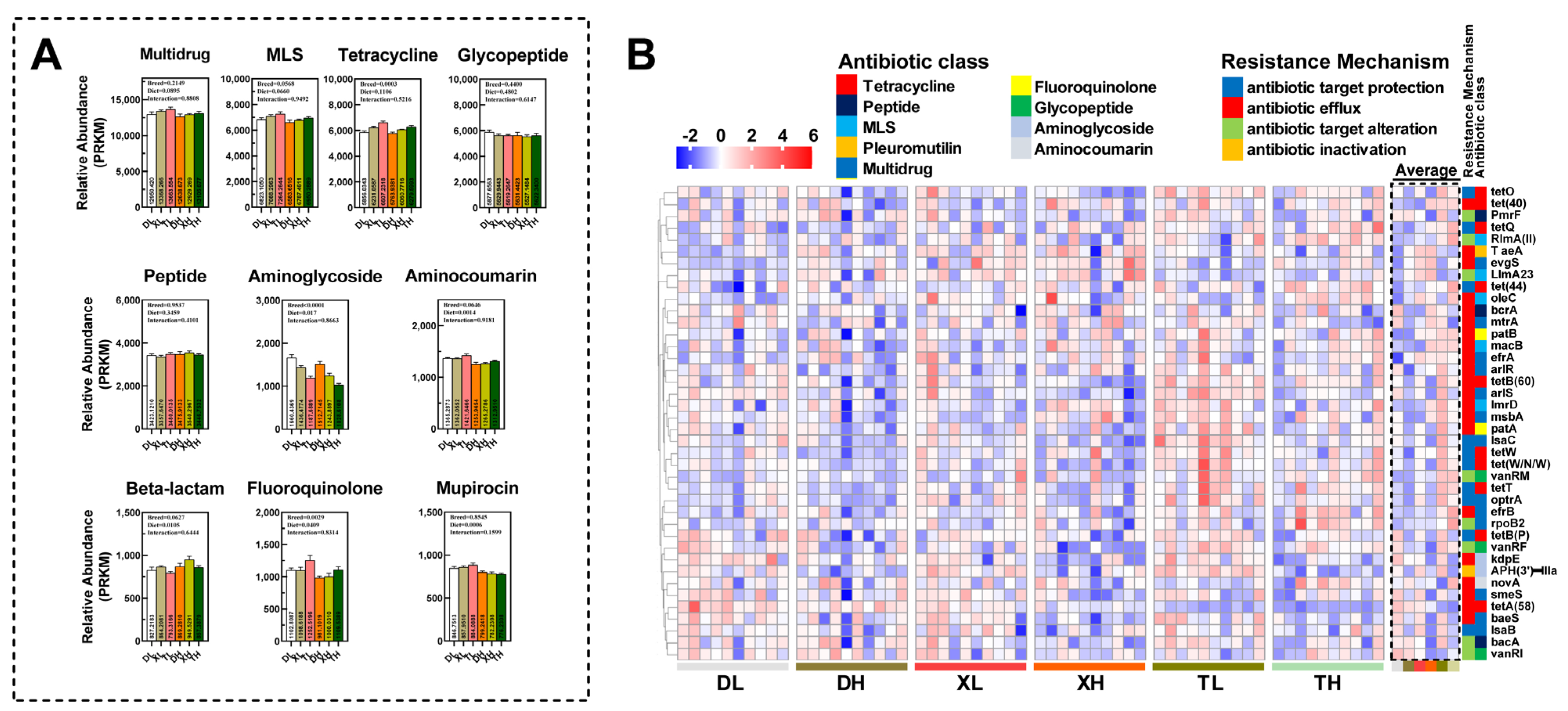
3.2. Fiber Diet Mediated Variation in the Profiling of Intestinal ARGs
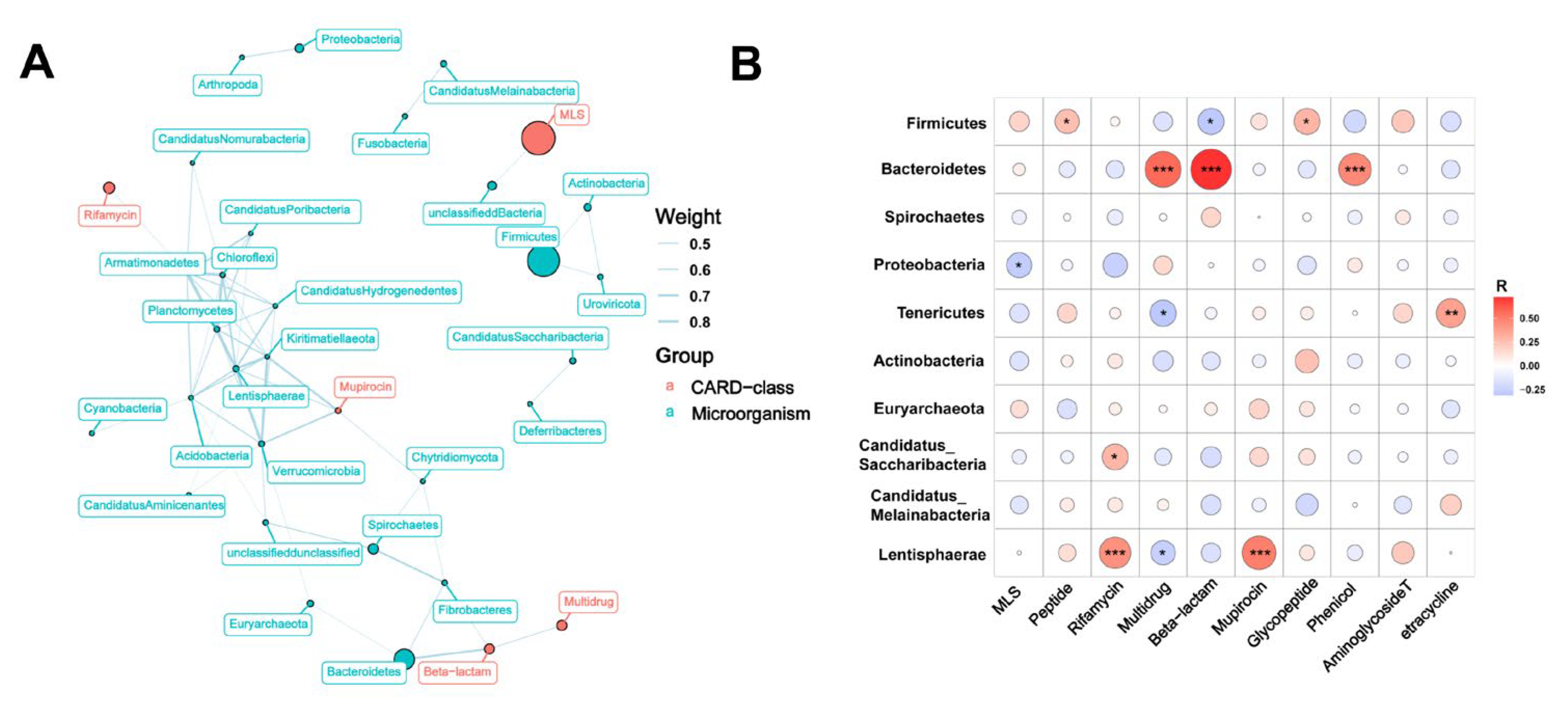
3.3. Relationship between Bacterial Taxonomic Dynamics and Antibiotic Resistance
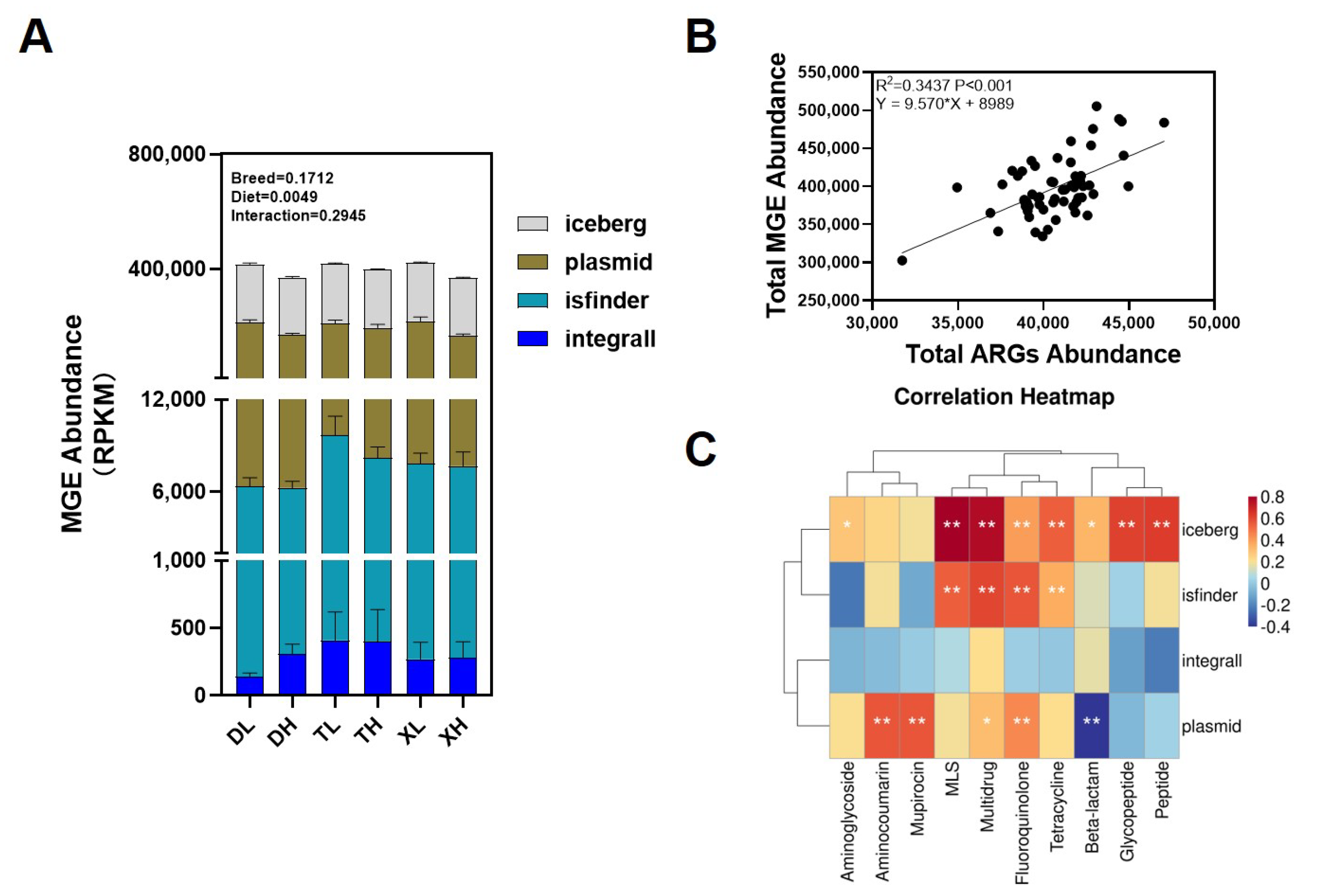
3.4. Effects of Breed and Fiber Diet on the Distribution of ARGs in Swine Feces and Their Potential Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallinga, D.; Smit, L.A.M.; Davis, M.F.; Casey, J.A.; Nachman, K.E. A Review of the Effectiveness of Current US Policies on Antimicrobial Use in Meat and Poultry Production. Curr. Environ. Health Rep. 2022, 9, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, S.; Li, F. Damage and elimination of soil and water antibiotic and heavy metal pollution caused by livestock husbandry. Environ. Res. 2022, 215 Pt 2, 114188. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, VMBF-0016-2015. [Google Scholar] [CrossRef] [PubMed]
- Mąka, Ł.; Maćkiw, E.; Ścieżyńska, H.; Modzelewska, M.; Popowska, M. Resistance to Sulfonamides and Dissemination of sul Genes Among Salmonella spIsolated from Food in Poland. Foodborne Pathog. Dis. 2015, 12, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Nové, M.; Kincses, A.; Molnár, J.; Amaral, L.; Spengler, G. The Role of Efflux Pumps and Environmental pH in Bacterial Multidrug Resistance. In Vivo 2020, 34, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Abdelrazik, E.; El-Hadidi, M. Tracking Antibiotic Resistance from the Environment to Human Health. Methods Mol. Biol. 2023, 2649, 289–301. [Google Scholar] [PubMed]
- Zhou, Y.; Fu, H.; Yang, H.; Wu, J.; Chen, Z.; Jiang, H.; Liu, M.; Liu, Q.; Huang, L.; Gao, J.; et al. Extensive metagenomic analysis of the porcine gut resistome to identify indicators reflecting antimicrobial resistance. Microbiome 2022, 10, 39. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, N.; Sun, L.; Zhang, Y.; Wu, Y.; Wang, Y.; Liao, X.; Mi, J. Short-term cold stress can reduce the abundance of antibiotic resistance genes in the cecum and feces in a pig model. J. Hazard. Mater. 2021, 416, 125868. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Czatzkowska, M.; Wolak, I.; Harnisz, M.; Korzeniewska, E. Impact of Anthropogenic Activities on the Dissemination of ARGs in the Environment—A Review. Int. J. Environ. Res. Public Health 2022, 19, 12853. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Hsu, C.W. Differentially transcribed genes in skeletal muscle of Duroc and Taoyuan pigs. J. Anim. Sci. 2005, 83, 2075–2086. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Peng, Y.; He, J.; Xiao, D.; Chen, C.; Li, F.; Huang, R.; Yin, Y. Dietary mulberry leaf powder affects growth performance, carcass traits and meat quality in finishing pigs. J. Anim. Physiol. Anim. Nutr. 2019, 103, 1934–1945. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, L.; Li, H.; Smidt, H.; Wright, A.G.; Zhang, K.; Ding, X.; Zeng, Q.; Bai, S.; Wang, J.; et al. Different Types of Dietary Fibers Trigger Specific Alterations in Composition and Predicted Functions of Colonic Bacterial Communities in BALB/c Mice. Front. Microbiol. 2017, 8, 966. [Google Scholar] [CrossRef]
- Franklin, M.A.; Mathew, A.G.; Vickers, J.R.; Clift, R.A. Characterization of microbial populations and volatile fatty acid concentrations in the jejunum, ileum, and cecum of pigs weaned at 17 vs. 24 days of age. J. Anim. Sci. 2002, 80, 2904–2910. [Google Scholar] [CrossRef]
- Perez-Burillo, S.; Molino, S.; Navajas-Porras, B.; Valverde-Moya, A.J.; Hinojosa-Nogueira, D.; Lopez-Maldonado, A.; Pastoriza, S.; Rufian-Henares, J.A. An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality. Nat. Protoc. 2021, 16, 3186–3209. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Hallowell, H.A.; Koutouvalis, K.; Suez, J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes 2022, 14, 2055944. [Google Scholar] [CrossRef]
- Fraser, C.; Lythgoe, K.; Leventhal, G.E.; Shirreff, G.; Hollingsworth, T.D.; Alizon, S.; Bonhoeffer, S. Virulence and pathogenesis of HIV-1 infection: An evolutionary perspective. Science 2014, 343, 1243727. [Google Scholar] [CrossRef] [PubMed]
- Mahnert, A.; Moissl-Eichinger, C.; Zojer, M.; Bogumil, D.; Mizrahi, I.; Rattei, T.; Martinez, J.L.; Berg, G. Man-made microbial resistances in built environments. Nat. Commun. 2019, 10, 968. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Chen, Z.; Tan, L.; Wang, X.; Xue, Y.; Wang, S.; Wang, Q.; Das, R.; Lin, H.; Hou, J.; et al. Gut resistomes, microbiota and antibiotic residues in Chinese patients undergoing antibiotic administration and healthy individuals. Sci. Total Environ. 2020, 705, 135674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ren, Q.; Ma, S.; Wu, J.; Yang, X.; Yu, Y. Intergenerational transfer of Dechlorane Plus and the associated long-term effects on the structure and function of gut microbiota in offspring. Environ. Int. 2020, 141, 105770. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pirolo, M.; Subramani, P.; Gehring, R.; Damborg, P.; Franzyk, H.; Guardabassi, L. Macrolide Resistance and In Vitro Potentiation by Peptidomimetics in Porcine Clinical Escherichia coli. mSphere 2022, 7, e0040222. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Letellier, A. Resistance to colistin: What is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 2016, 48, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Momoh, A.H.; Kwaga, J.K.P.; Bello, M.; Sackey, A.K.B.; Larsen, A.R. Antibiotic resistance and molecular characteristics of Staphylococcus aureus isolated from backyard-raised pigs and pig workers. Trop. Anim. Health Prod. 2018, 50, 1565–1571. [Google Scholar] [CrossRef]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef]
- Morrison, K.E.; Jašarević, E.; Howard, C.D.; Bale, T.L. It’s the fiber, not the fat: Significant effects of dietary challenge on the gut microbiome. Microbiome 2020, 8, 15. [Google Scholar] [CrossRef]
- Horne, T.; Orr, V.T.; Hall, J. How do interactions between mobile genetic elements affect horizontal gene transfer? Curr. Opin. Microbiol. 2023, 73, 102282. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W.; et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef]
- Goren, M.G.; Carmeli, Y.; Schwaber, M.J.; Chmelnitsky, I.; Schechner, V.; Navon-Venezia, S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 2010, 16, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Fan, X.; Zhu, L.; Yang, X.; Liu, Y.; Gao, S.; Jin, X.; Liu, D.; Ding, J.; Guo, Y.; et al. Phylogenetic and genomic analysis reveals high genomic openness and genetic diversity of Clostridium perfringens. Microb. Genom. 2020, 6, mgen000441. [Google Scholar] [CrossRef] [PubMed]
- Knetsch, C.W.; Kumar, N.; Forster, S.C.; Connor, T.R.; Browne, H.P.; Harmanus, C.; Sanders, I.M.; Harris, S.R.; Turner, L.; Morris, T.; et al. Zoonotic Transfer of Clostridium difficile Harboring Antimicrobial Resistance between Farm Animals and Humans. J. Clin. Microbiol. 2018, 56, e01384-17. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Tang, X.; Fan, C.; Wang, L.; Shen, W.; Ren, S.; Zhang, L.; Zhang, Y. Gastrointestinal Tract and Dietary Fiber Driven Alterations of Gut Microbiota and Metabolites in Durco × Bamei Crossbred Pigs. Front. Nutr. 2021, 8, 806646. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Liu, X.; Xu, J.; Li, F.; Wang, J.; Zhang, X.; Yang, X.; Wang, L.; Ma, S.; Li, D.; et al. Effects of Silage Diet on Meat Quality through Shaping Gut Microbiota in Finishing Pigs. Microbiol. Spectr. 2023, 11, e0241622. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Y.H.; Zhou, D.; Wu, Q.; Song, D.; Dicksved, J.; Wang, J.F. Oral Administration of a Select Mixture of Bacillus Probiotics Affects the Gut Microbiota and Goblet Cell Function following Escherichia coli Challenge in Newly Weaned Pigs of Genotype MUC4 That Are Supposed to Be Enterotoxigenic E. coli F4ab/ac Receptor Negative. Appl. Environ. Microbiol. 2017, 83, e02747-16. [Google Scholar]
- De Groot, P.; Scheithauer, T.; Bakker, G.J.; Prodan, A.; Levin, E.; Khan, M.T.; Herrema, H.; Ackermans, M.; Serlie, M.J.M.; de Brauw, M.; et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 2020, 69, 502–512. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Sadurní, M.; Barroeta, A.C.; Sol, C.; Puyalto, M.; Castillejos, L. Effects of dietary crude protein level and sodium butyrate protected by medium-chain fatty acid salts on performance and gut health in weaned piglets. J. Anim. Sci. 2023, 101, skad090. [Google Scholar] [CrossRef]
- Zhen, R.; Liu, C.; Wei, C.; Luo, Y.; Hu, X.; Liu, G.; Yi, H.; Huang, Y. Effect of different dosages of sodium butyrate and niacin on growth, faecal microbiota and Vitamin B metabolism in weaned piglets. J. Appl. Microbiol. 2022, 132, 4466–4475. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Luo, Y.; Kong, X.; Yu, B.; Zheng, P.; Huang, Z.; Mao, X.; Yu, J.; Luo, J.; Yan, H.; et al. Genetic- and Fiber-Diet-Mediated Changes in Antibiotic Resistance Genes in Pig Colon Contents and Feces and Their Driving Factors. Microorganisms 2023, 11, 2370. https://doi.org/10.3390/microorganisms11102370
Wang T, Luo Y, Kong X, Yu B, Zheng P, Huang Z, Mao X, Yu J, Luo J, Yan H, et al. Genetic- and Fiber-Diet-Mediated Changes in Antibiotic Resistance Genes in Pig Colon Contents and Feces and Their Driving Factors. Microorganisms. 2023; 11(10):2370. https://doi.org/10.3390/microorganisms11102370
Chicago/Turabian StyleWang, Tao, Yuheng Luo, Xiangfeng Kong, Bing Yu, Ping Zheng, Zhiqing Huang, Xiangbing Mao, Jie Yu, Junqiu Luo, Hui Yan, and et al. 2023. "Genetic- and Fiber-Diet-Mediated Changes in Antibiotic Resistance Genes in Pig Colon Contents and Feces and Their Driving Factors" Microorganisms 11, no. 10: 2370. https://doi.org/10.3390/microorganisms11102370
APA StyleWang, T., Luo, Y., Kong, X., Yu, B., Zheng, P., Huang, Z., Mao, X., Yu, J., Luo, J., Yan, H., & He, J. (2023). Genetic- and Fiber-Diet-Mediated Changes in Antibiotic Resistance Genes in Pig Colon Contents and Feces and Their Driving Factors. Microorganisms, 11(10), 2370. https://doi.org/10.3390/microorganisms11102370






