The Transformation of Hg2+ during Anaerobic S0 Reduction by an AMD Environmental Enrichment Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of the AMD Enrichment
2.2. Experimental Setup
2.3. Analytical Methods
2.3.1. Physical and Chemical Characterization of the Culture Liquor
2.3.2. Surface Morphology and Chemical Composition of Solid Particulate and Microbial Cells
2.3.3. C, S and Hg Speciation on the Surface of Solid Particulate
2.3.4. Analysis of Microbial Diversity
3. Results and Discussion
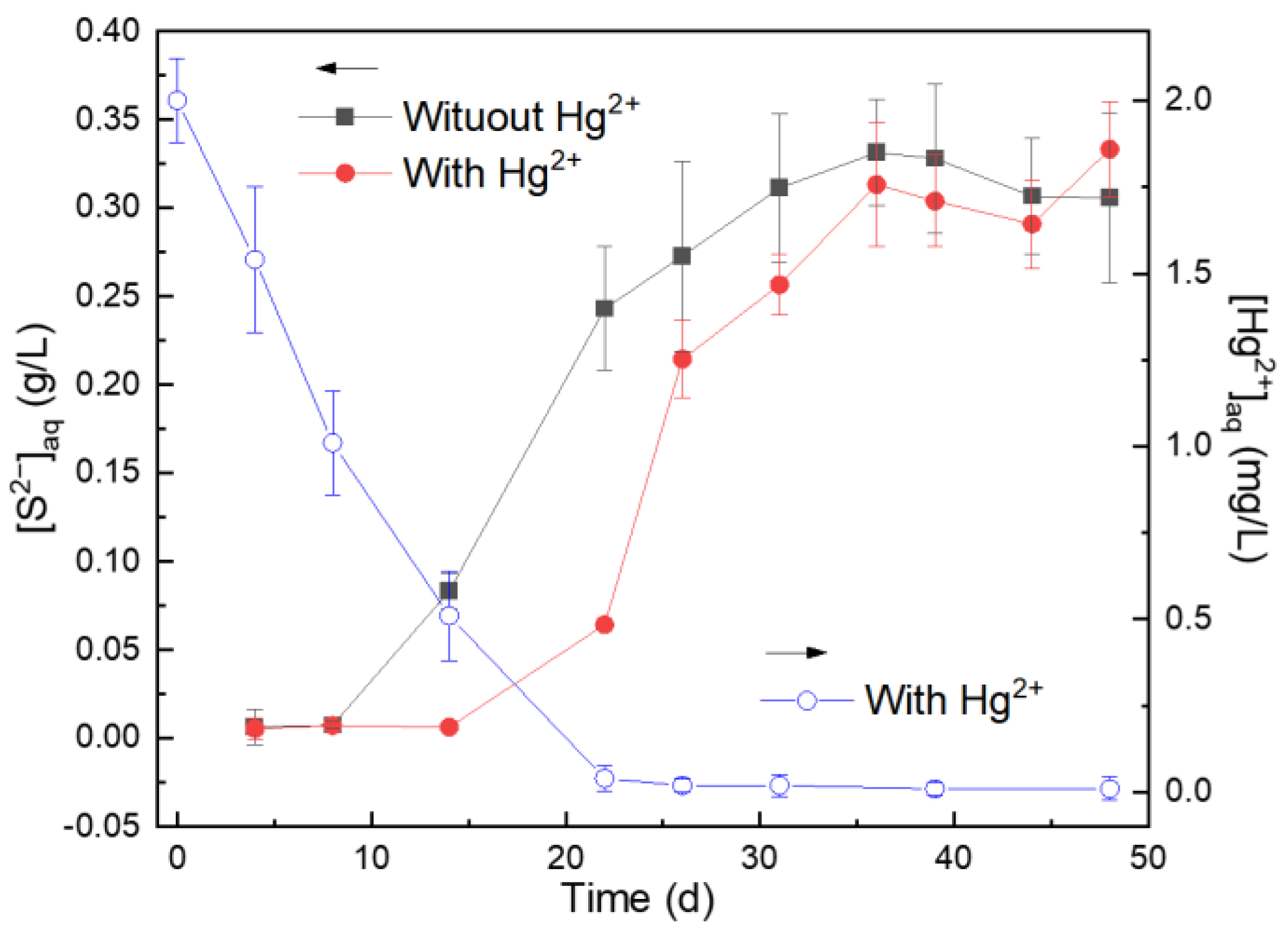
3.1. Effects of Hg2+ on Microbial S0 Reduction and Hg2+ Fixation
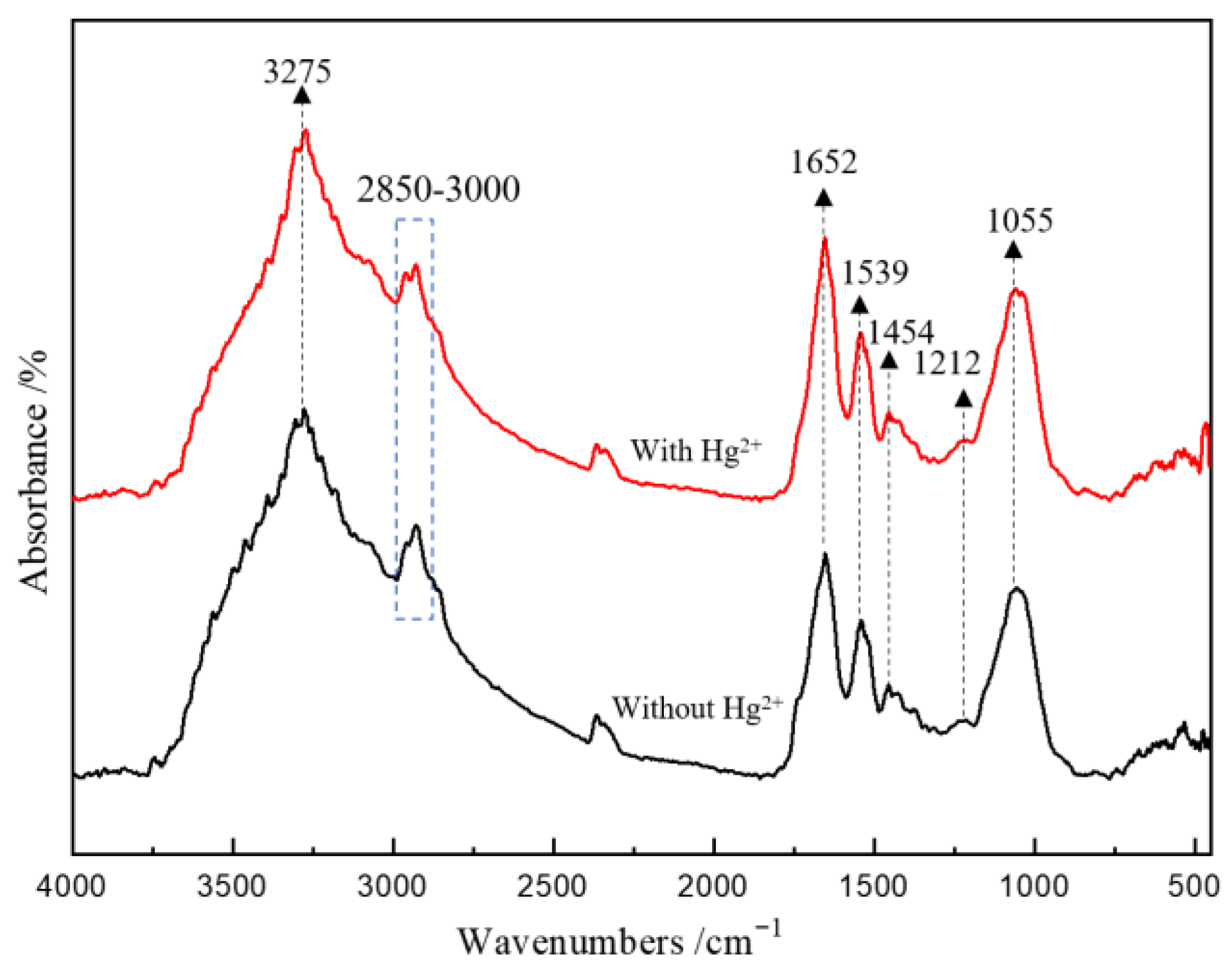
3.2. Microbial Cell Morphology and Surface Composition
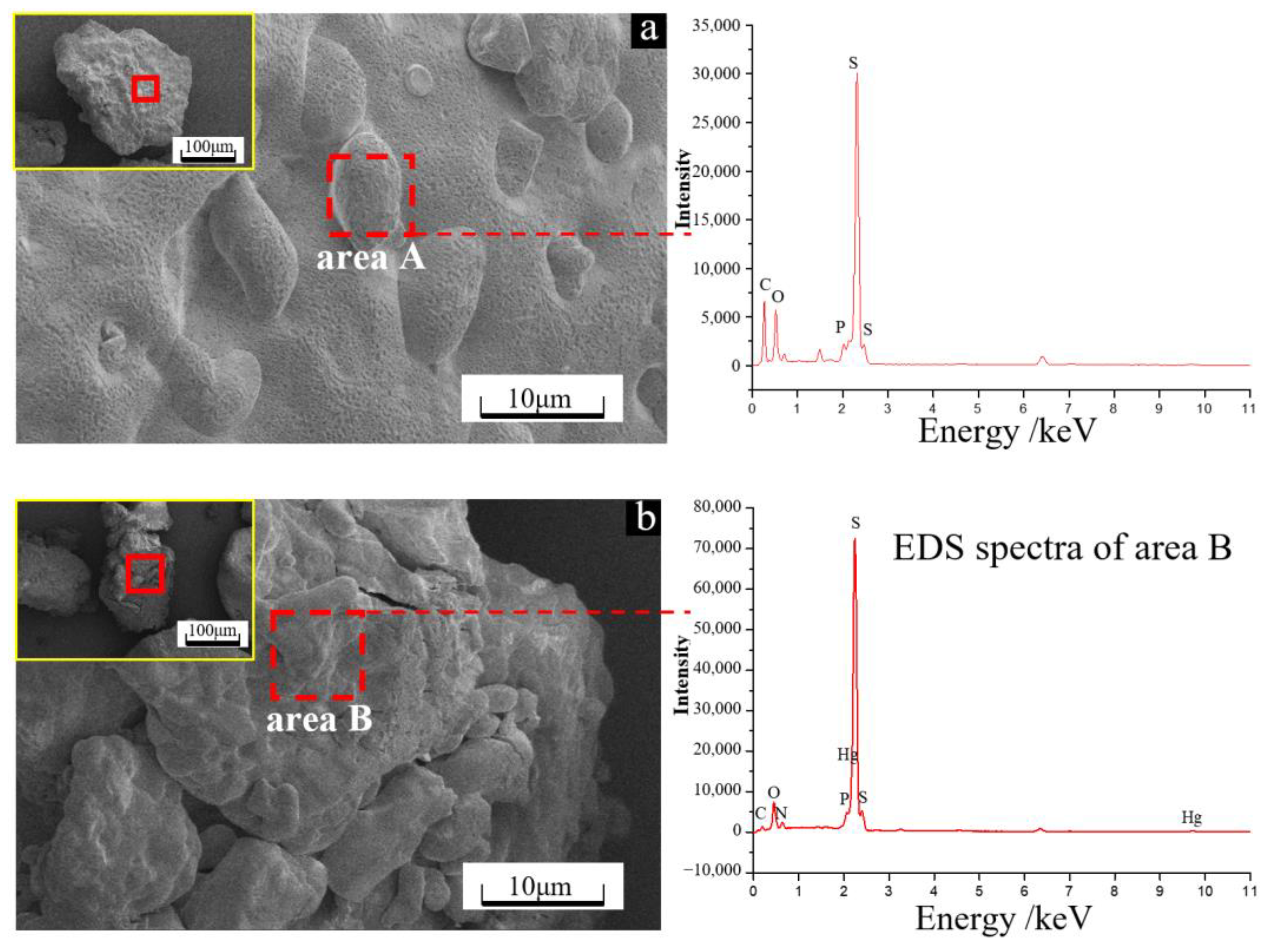
3.3. Surface Morphology and Composition of Solid Particulate
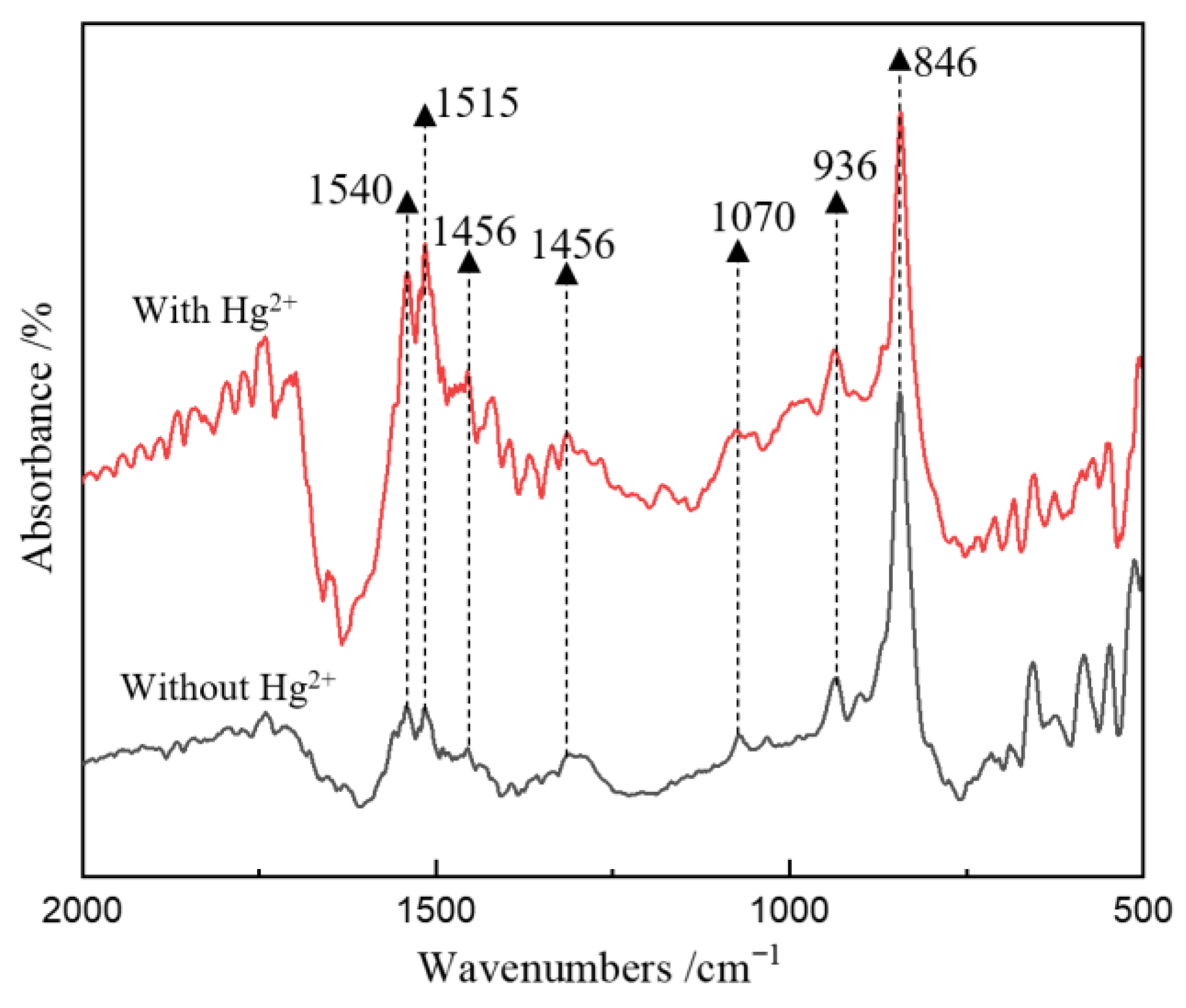
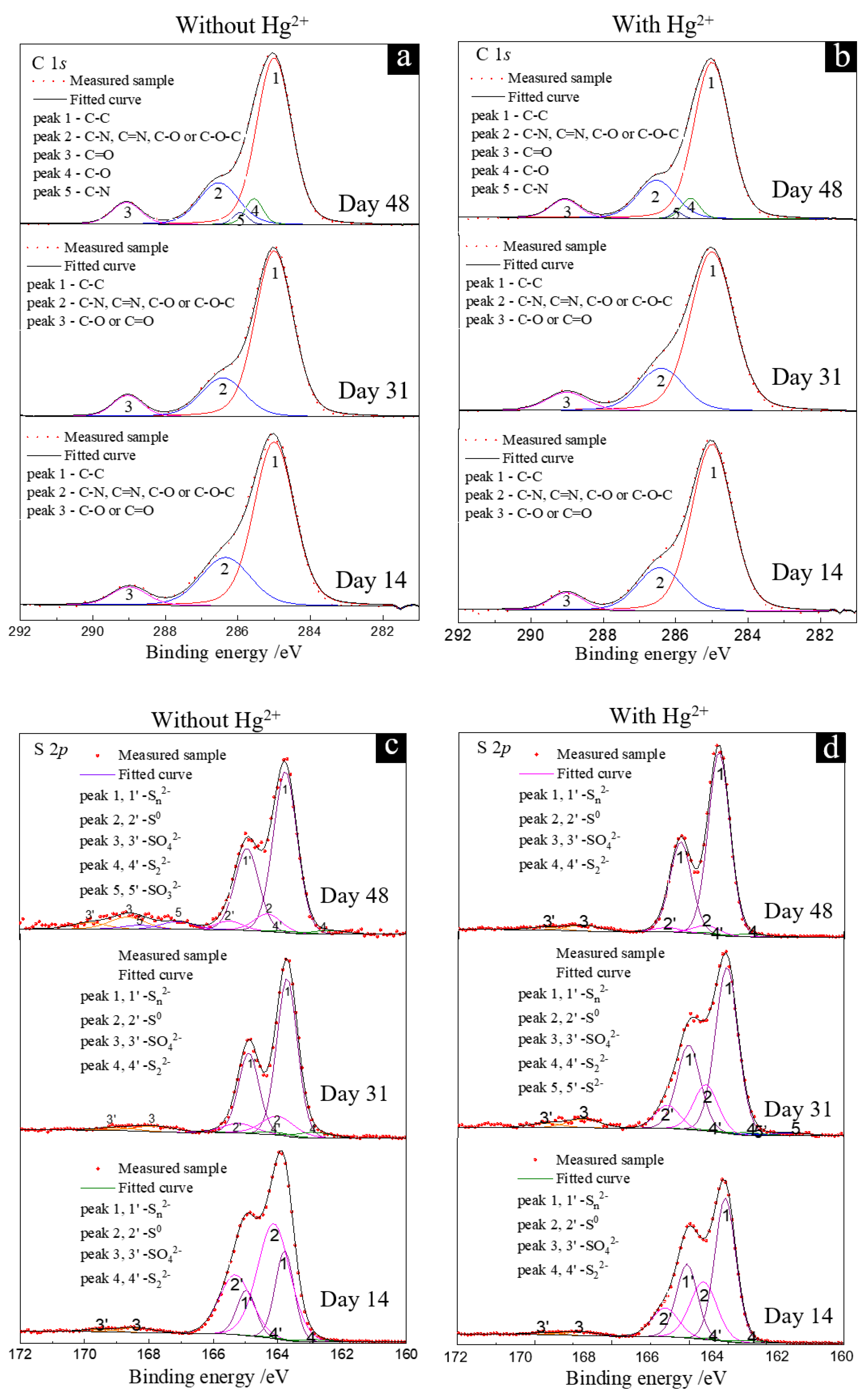
3.4. Dynamics of C, S and Hg Speciation on Solid Particulate Surface
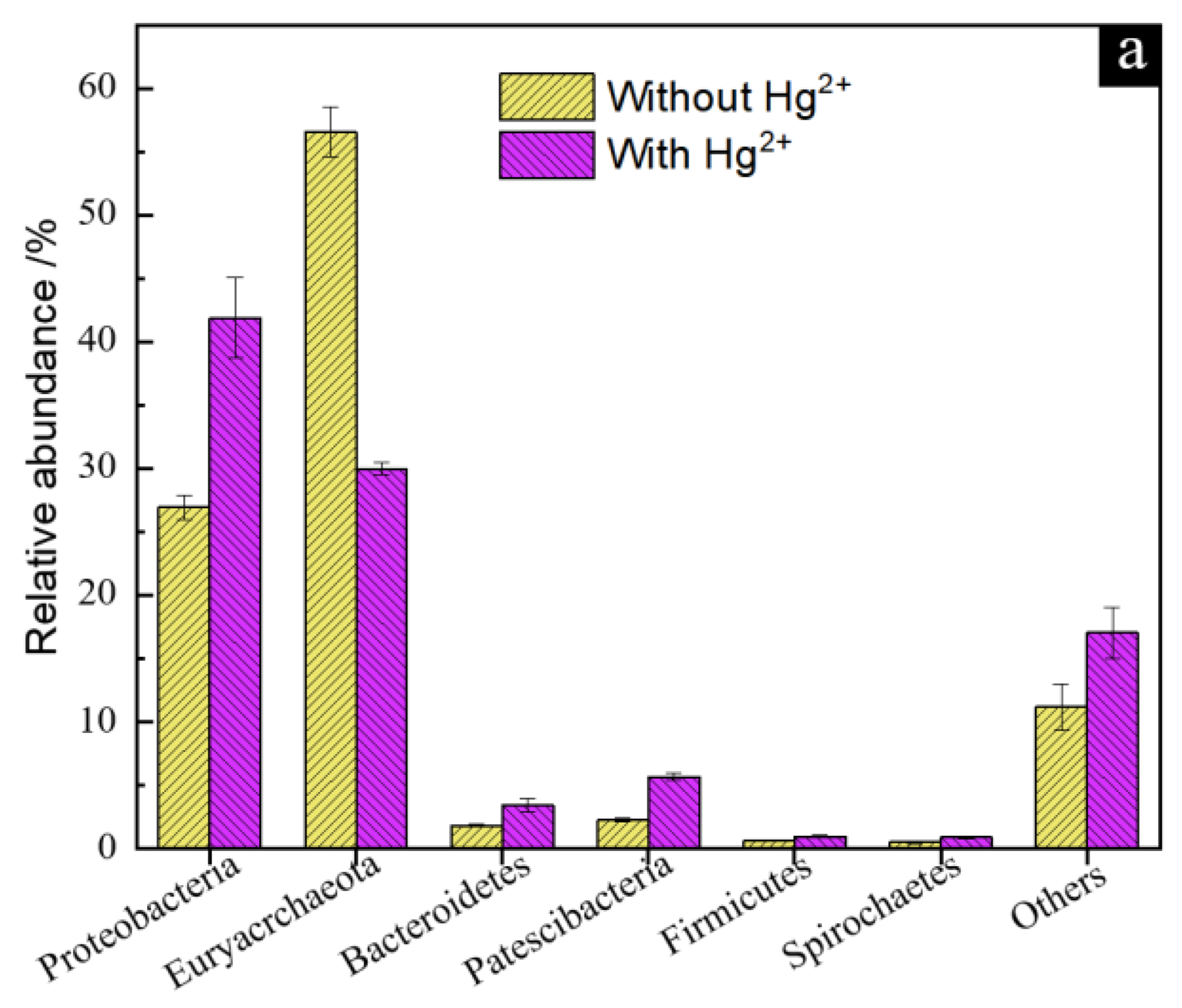
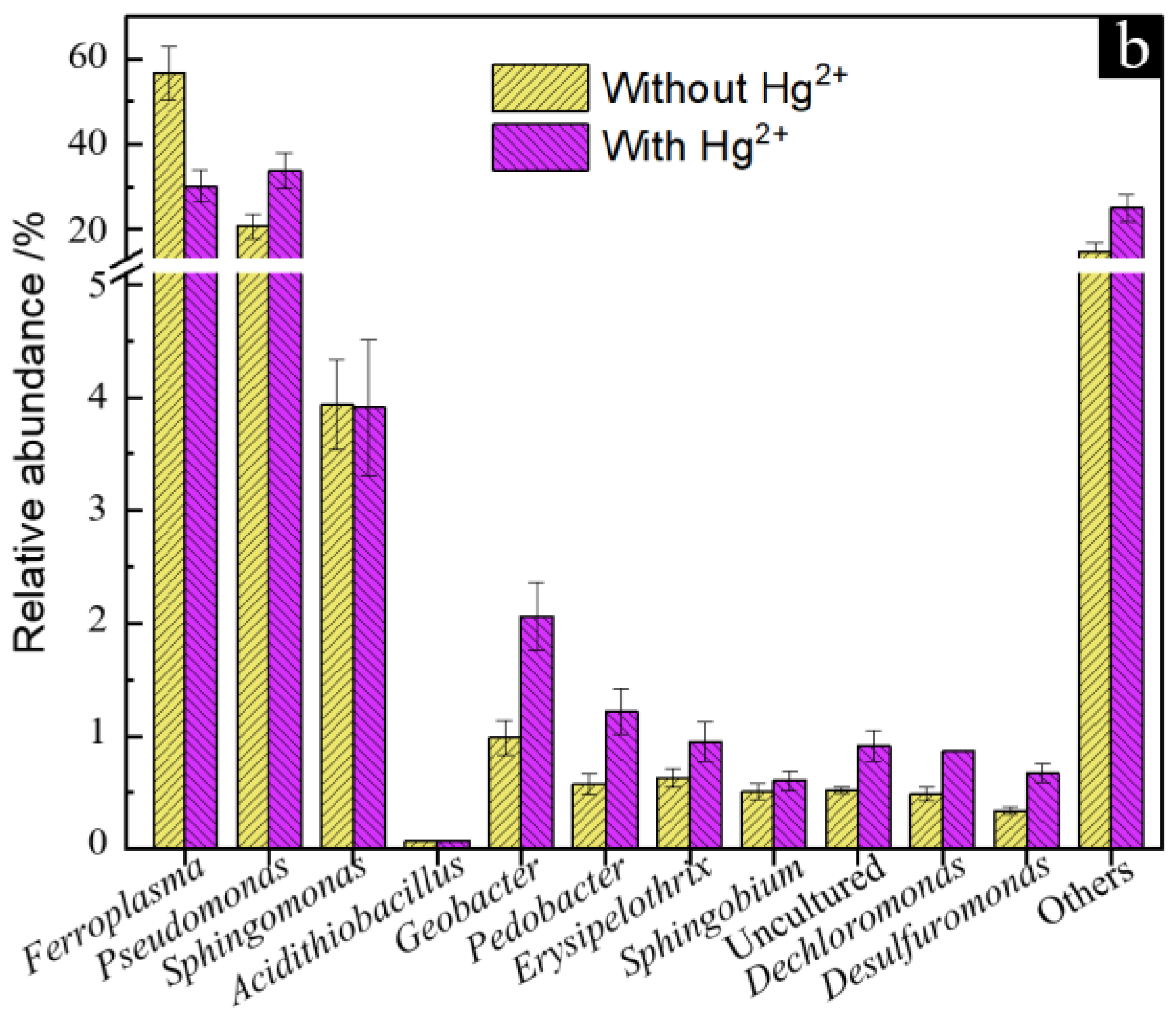
3.5. Microbial Community Structure
4. Environmental Significance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez-Galán, M.; Baena-Moreno, F.M.; Vázquez, S.; Arroyo-Torralvo, F.; Vilches, L.F.; Zhang, Z.E. Remediation of acid mine drainage. Environ. Chem. Lett. 2019, 17, 1529–1538. [Google Scholar] [CrossRef]
- O′Connor, D.; Hou, D.Y.; Ok, Y.K.; Mulder, J.; Duan, L.; Wu, Q.R.; Wang, S.; Tack, F.M.G.; Rinklebe, J. Mercury speciation, transformation, and transportation in soils, atmospheric flux, and implications for risk management: A critical review. Environ. Int. 2019, 126, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Fisher, N.S. Bioaccumulation of methylmercury in a marine copepod. Environ. Toxicol. Chem. 2016, 36, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, C.T.; Mason, R.P.; Chan, H.M.; Jacob, D.J.; Pirrone, N. Mercury as a global pollutant: Sources, pathways, and effects. Environ. Sci. Technol. 2013, 47, 4967–4983. [Google Scholar] [CrossRef]
- Klein, R.; Tischler, J.S.; Mühling, M.; Schlömann, M. Bioremediation of mine water. Adv. Biochem. Eng. Biotechnol. 2014, 141, 109–172. [Google Scholar]
- Ndu, U.; Barkay, T.; Schartup, A.T.; Mason, R.P.; Reinfelder, J.R. The effect of aqueous speciation and cellular ligand binding on the biotransformation and bioavailability of methylmercury in mercury-resistant bacteria. Biodegradation 2016, 27, 29–36. [Google Scholar] [CrossRef]
- Lovley, D.R. Dissimilatory metal reduction. Annu. Rev. Microbiol. 1993, 47, 263–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Shaheen, S.M.; Jing, M.; Anderson, C.W.N.; Swertz, A.C.; Wang, S.L.; Feng, X.; Rinklebe, J. Mobilization, methylation, and demethylation of mercury in a paddy soil under systematic redox changes. Environ. Sci. Technol. 2021, 55, 10133–10141. [Google Scholar] [CrossRef]
- Pak, K.R.; Bartha, R. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl. Environ. Microbiol. 1998, 64, 1013–1017. [Google Scholar] [CrossRef]
- Bravo, G.; Vega-Celedón, P.; Gentina, J.C.; Seeger, M. Effects of mercury II on Cupriavidus metallidurans strain MSR33 during mercury bioremediation under aerobic and anaerobic conditions. Processes 2020, 8, 893. [Google Scholar] [CrossRef]
- Hamlett, N.V.; Landale, E.C.; Davis, B.H.; Summers, A.O. Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J. Bacteriol. 1992, 174, 6377–6385. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Mason, R.P.; Porter, E.T.; Soulen, H.L. The impact of resuspension on sediment mercury dynamics, and methylmercury production and fate: A mesocosm study. Mar. Chem. 2006, 102, 300–315. [Google Scholar] [CrossRef]
- Du, H.; Ma, M.; Igarashi, Y. Biotic and abiotic degradation of methylmercury in aquatic ecosystems: A review. Bull. Environ. Contam. Toxicol. 2019, 102, 605–611. [Google Scholar] [CrossRef]
- Sasaki, Y.; Minakawa, T.; Miyazaki, A.; Silver, S.; Kusano, T. Functional dissection of a mercuric ion transporter, MerC, from Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 2005, 69, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Niane, B.; Devarajan, N.; Poté, J.; Moritz, R. Quantification and characterization of mercury resistant bacteria in sediments contaminated by artisanal small-scale gold mining activities, Kedougou region, Senegal. J. Geochem. Explor. 2019, 205, 106353. [Google Scholar] [CrossRef]
- Möller, A.; Grahn, A.; Welander, U. Precipitation of heavy metals from landfill leachates by microbially-produced sulphide. Environ. Technol. 2004, 25, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.M.; Ning, D.L.; He, Z.L.; Zhang, P.; Spencer, S.J.; Gao, S.H.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef]
- Sun, R.R.; Li, Y.; Lin, N.N.; Ou, C.X.; Wang, X.Y.; Zhang, L.; Jiang, F. Removal of heavy metals using a novel sulfidogenic AMD treatment system with sulfur reduction: Configuration, performance, critical parameters and economic analysis. Environ. Int. 2020, 136, 105457. [Google Scholar] [CrossRef]
- Fischer, S.; Jarsjö, J.; Rosqvist, G.; Mörth, C.M. Catchment-scale microbial sulfate reduction (MSR) of acid mine drainage (AMD) revealed by sulfur isotopes. Environ. Pollut. 2022, 292, 118478. [Google Scholar] [CrossRef]
- Hedderich, R.; Klimmek, O.; Kröger, A.; Dirmeier, R.; Keller, M.; Stetter, K.O. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 1998, 22, 353–381. [Google Scholar] [CrossRef]
- Wang, J.T.; Zhang, L.; Kang, Y.; Chen, G.H.; Jiang, F. Long-term feeding of elemental sulfur alters microbial community structure and eliminates mercury methylation potential in sulfate-reducing bacteria abundant activated sludge. Environ. Sci. Technol. 2018, 52, 4746–4753. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Parks, J.M.; Johs, A.; Podar, M.; Bridou, R.; Hurt, R.A., Jr.; Smith, S.D.; Tomanicek, S.J.; Qian, Y.; Brown, S.D.; Brandt, C.C.; et al. The genetic basis for bacterial mercury methylation. Science 2013, 339, 1332–1335. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Jia, F.J.; Wang, L.; Jalalah, M.; Al-Assiri, M.S.; Gao, T.; Harraz, F.A.; Li, G. Fabrication of an artificial ionic gate inspired by mercury-resistant bacteria for simple and sensitive detection of mercury ion. Sens. Actuators B. Chem. 2021, 326, 128976. [Google Scholar] [CrossRef]
- Zhang, H.D.; Ma, Y.L.; Zhou, Y.H.; Liu, H.C.; Nie, Z.Y.; Pan, X.; Fan, X.-L.; Xia, J.-L. The differential inhibitive effects and fates of As(III) and As(V) mediated by Sulfobacillus thermosulfidooxidans grown on S0, Fe2+ and FeS2. Ecotoxicol. Environ. Safe. 2021, 222, 112502. [Google Scholar] [CrossRef]
- Cheng, J.P.; Zhao, W.C.; Liu, Y.Y.; Wu, C.; Liu, C.; Wang, W.H. Adsorption properties and gaseous mercury transformation rate of natural biofilm. Bull. Environ. Contam. Toxicol. 2008, 81, 516–520. [Google Scholar] [CrossRef]
- Dash, H.R.; Das, S. Interaction between mercuric chloride and extracellular polymers of biofilm-forming mercury resistant marine bacterium Bacillus thuringiensis PW-05. RSC Adv. 2016, 6, 109793–109802. [Google Scholar] [CrossRef]
- Liu, H.C.; Xia, J.L.; Nie, Z.Y.; Peng, A.A.; Ma, C.Y.; Zheng, L.; Zhao, Y.-D. Comparative study of sulfur utilization and speciation transformation of two elemental sulfur species by thermoacidophilic Archaea Acidianus manzaensis YN-25. Process. Biochem. 2013, 48, 1855–1860. [Google Scholar] [CrossRef]
- Payne, B.P.; Biesinger, M.C.; McIntyre, N.S. X-ray photoelectron spectroscopy studies of reactions on chromium metal and chromium oxide surfaces. J. Electron. Spectros. Relat. Phenomena. 2011, 184, 29–37. [Google Scholar] [CrossRef]
- Viltres, H.; Odio, O.F.; Lartundo-Rojas, L.; Reguera, E. Degradation study of arsenic oxides under XPS measurements. Appl. Surf. Sci. 2020, 511, 145606. [Google Scholar] [CrossRef]
- Corkhill, C.L.; Wincott, P.L.; Lloyd, J.R. The oxidative dissolution of arsenopyrite (FeAsS) and enargite (Cu3AsS4) by Leptospirillum ferrooxidans. Geochim. Cosmochim. Acta 2008, 72, 5616–5633. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, C.; Liu, H.C.; Nie, Z.Y.; Zhou, Y.H.; Xia, J.L. Research progresses on occurrence forms and microbial transformation of mercury. Chinese J. Appl. Environ. Biology. 2022, 28, 1–10. (In Chinese) [Google Scholar]
- Liu, X.; Ma, A.Z.; Zhuang, G.Q.; Zhuang, X.L. Diversity of microbial communities potentially involved in mercury methylation in rice paddies surrounding typical mercury mining areas in China. MicrobiologyOpen 2018, 7, e577. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskaya, T.A.; Hu, H.Y.; Van Nostrand, J.D.; Wymore, A.M.; Xu, X.H.; Qiu, G.L.; Feng, X.; Zhou, J.; Brown, S.D.; Brandt, C.C.; et al. Microbial community structure with trends in methylation gene diversity and abundance in mercury-contaminated rice paddy soils in Guizhou, China. Environ. Sci. Proc. Imp. 2018, 20, 673–685. [Google Scholar] [CrossRef]
- Rasmussen, L.D.; Zawadsky, C.; Binnerup, S.J.; Øregaard, G.; Sørensen, S.J.; Kroer, N. Cultivation of hard-to-culture subsurface mercury-resistant bacteria and discovery of new merA gene sequences. Appl. Environ. Microbiol. 2008, 74, 3795–3803. [Google Scholar] [CrossRef]
- Allen, R.C.; Tu, Y.K.; Nevarez, M.J.; Bobbs, A.S.; Friesen, J.W.; Lorsch, J.R.; McCauley, J.A.; Voet, J.G.; Hamlett, N.V. The mercury resistance (mer) operon in a marine gliding flavobacterium, Tenacibaculum discolor 9A5. FEMS Microbiol. Ecol. 2013, 83, 135–148. [Google Scholar] [CrossRef]
- Azaroff, A.; Urriza, M.G.; Gassie, C.; Monperrus, M.; Guyoneaud, R. Marine mercury-methylating microbial communities from coastal to Capbreton Canyon sediments (North Atlantic Ocean). Environ. Pollut. 2020, 262, 114333. [Google Scholar] [CrossRef]
- McDaniel, E.A.; Peterson, B.D.; Stevens, S.L.R.; Tran, P.Q.; Anantharaman, K.; McMahon, K.D. Expanded phylogenetic diversity and metabolic flexibility of mercury-methylating microorganisms. mSystems 2020, 5, e00299-20. [Google Scholar] [CrossRef]
- Kerin, E.J.; Gilmour, C.C.; Roden, E.; Suzuki, M.T.; Coates, J.D.; Mason, R.P. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 2006, 72, 7919–7921. [Google Scholar] [CrossRef]
- Fatimawali; Kepel, B.; Tallei, T.E. Potential of organic mercury-resistant bacteria isolated from mercury contaminated sites for organic mercury remediation. Pak. J. Biol. Sci. 2019, 22, 45–50. [Google Scholar]
- Fong, J.C.; De Guzman, B.E.; Lamborg, C.H.; Sison-Mangus, M.P. The mercury-tolerant microbiota of the zooplankton Daphnia aids in host survival and maintains fecundity under mercury stress. Environ. Sci. Technol. 2019, 53, 14688–14699. [Google Scholar] [CrossRef] [PubMed]
- Nealson, K.H.; Saffarini, D. Iron and manganese in anaerobic respiration: Environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 1994, 48, 311–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, D.; Liang, J.L.; Wang, L.; Zhou, Y. Triclosan transformation and impact on an elemental sulfur-driven sulfidogenic process. Chem. Eng. J. 2021, 421, 129634. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, Y.; Liu, H.; Nie, Z.; Wang, Y.; Chen, L. The Transformation of Hg2+ during Anaerobic S0 Reduction by an AMD Environmental Enrichment Culture. Microorganisms 2023, 11, 72. https://doi.org/10.3390/microorganisms11010072
Zhou Y, Liu Y, Liu H, Nie Z, Wang Y, Chen L. The Transformation of Hg2+ during Anaerobic S0 Reduction by an AMD Environmental Enrichment Culture. Microorganisms. 2023; 11(1):72. https://doi.org/10.3390/microorganisms11010072
Chicago/Turabian StyleZhou, Yuhang, Yue Liu, Hongchang Liu, Zhenyuan Nie, Yirong Wang, and Lu Chen. 2023. "The Transformation of Hg2+ during Anaerobic S0 Reduction by an AMD Environmental Enrichment Culture" Microorganisms 11, no. 1: 72. https://doi.org/10.3390/microorganisms11010072
APA StyleZhou, Y., Liu, Y., Liu, H., Nie, Z., Wang, Y., & Chen, L. (2023). The Transformation of Hg2+ during Anaerobic S0 Reduction by an AMD Environmental Enrichment Culture. Microorganisms, 11(1), 72. https://doi.org/10.3390/microorganisms11010072





