Inter-Species Redox Coupling by Flavin Reductases and FMN-Dependent Two-Component Monooxygenases Undertaking Nucleophilic Baeyer–Villiger Biooxygenations
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Maintenance, and Growth Conditions
2.2. Extract Preparation
2.3. Purification of 2,5-DKCMO, 3,6-DKCMO, and LuxAB Luciferase
2.4. Purification of Frp1, Frp2, FreEc, FRGVf, FreVf, and FRDAa
2.5. Single-Enzyme Kinetic Studies
2.6. Coupled-Enzyme Kinetic Studies
2.7. Longer-Term (120 min) Biocatalytic Reactions with Combinations of Highly Purified Enzymes
2.8. Reproducibility
3. Results and Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willetts, A. The isoenzymic diketocamphane monooxygenases of Pseudomonas putida ATCC 17453—An episodic history and still mysterious after 60 years. Microorganisms 2021, 9, 2592. [Google Scholar] [CrossRef] [PubMed]
- Zenno, S.; Saigo, K.; Kanoh, H.; Inouye, S. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bio-luminescent bacterium Vibrio fischeri ATCC 7744. J. Bacteriol. 1994, 176, 3536–3543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Willetts, A. Structural studies and biosynthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 1997, 15, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Leisch, H.; Morley, K.; Lau, P.C.K. Baeyer-Villiger monooxygenases: More than just green chemistry. Chem. Rev. 2011, 111, 4165–4222. [Google Scholar] [CrossRef]
- Ellis, H.R. The FMN-dependent two-component monooxygenase systems. Arch. Biochem. Biophys. 2010, 497, 1–12. [Google Scholar] [CrossRef]
- Wu, J.; Weiss, B. Two-stage induction of the soxRS regulon in Escherichia coli. J. Bacteriol. 1992, 174, 3915–3920. [Google Scholar] [CrossRef]
- Manchado, M.; Michen, C.; Pueyo, C. Hydrogen peroxide activates the soxRS regulon in vivo. J. Bacteriol. 2000, 182, 6842–6844. [Google Scholar] [CrossRef][Green Version]
- Woodmansee, A.N.; Imlay, J.A. Reduced flavins promote oxidative damage in non-respiring Escherichia coli by delivering electrons to intracellular iron. J. Biol. Chem. 2002, 277, 34055–34066. [Google Scholar] [CrossRef]
- Imlay, J.A. Cellular defences against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Willetts, A.; Kelly, D.R. Flavin-dependent redox transfers by two-component diketocamphane monooxygenases of camphor-grown Pseudomonas putida NCIMB 10007. Microorganisms 2016, 4, 38. [Google Scholar] [CrossRef]
- Wilson, T.; Hastings, J.W. Bioluminescence. Ann. Rev. Cell Dev. 1998, 14, 197–230. [Google Scholar] [CrossRef] [PubMed]
- Willetts, A. Characterised flavin-dependent two-component mono-oxygenases from the CAM plasmid of Pseudomonas putida ATCC 17453 (NCIMB 10007): Ketolactonases by another name. Microorganisms 2019, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Willetts, A. Oxidations by microbial NADH plus FMN-dependent luciferases from Photobacterium phosphoreum and Vibrio fischeri. J. Mol. Catal. B Enzymatic 1997, 2, 193–197. [Google Scholar] [CrossRef]
- Konigsburger, K.; Griengl, H. Microbisl Baeyer-Villiger reactions of bicyclo[3.2.0]heptan-6-ol–a novel approach of sarkomycin A. Bioorg. Med. Chem. 1994, 2, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.; Furstoss, R. Synthesis of (1S,5R)-2,8-dioxabicyclo[3.3.0]oct-3-one from its enantiomer: A subunit of clerodane derivatives. Synthesis 1995, 27, 1514–1521. [Google Scholar]
- Adger, B.; Bes, T.; Grogan, G.; McCague, R.; Pedragosa-Moreau, S.; Roberts, S.M.; Villa, R.; Wan, P.H.; Willetts, A. Applications of enzymic Baeyer-Villiger oxidations of 2-substituted cycloalkanones to the total synthesis of R-(+)-lipoic acid. J. Chem. Soc. Chem. Commun. 1995, 1563–1564. [Google Scholar] [CrossRef]
- Bes, T.M.; Villa, R.; Roberts, S.M.; Wan, P.W.H.; Willetts, A. Oxidative biotransformations by microorganisms: Production of chiral synthons by cyclopentanone monooxygenase from Pseudomonas sp. NCIMB 9872. J. Mol. Catal. B Enzymatic 1996, 1, 127–134. [Google Scholar] [CrossRef]
- Gagnon, R.; Grogan, G.; Roberts, S.M.; Villa, R.; Willetts, A. Enzymatic Baeyer-Villiger oxidation of some bicyclo[2.2.1]heptan-2-ones using monooxygenases from Pseudomonas putida NCIMB 10007: Enantioselective preparation of a precursor of azadirachtin. J. Chem. Soc. Perkin Trans. 1995, 1, 1505–1511. [Google Scholar] [CrossRef]
- Levitt, M.S.; Newton, R.F.; Roberts, S.M.; Willetts, A. Preparation of optically active 6-fluorocarbocyclic nucleosides using enantioselective enzyme-catalysed Baeyer-Villiger oxidations. J. Chem. Soc. Chem. Commun. 1990, 619–620. [Google Scholar] [CrossRef]
- Alphand, V.; Furstoss, R. Microbial transformations 22. Microbiologically mediated Baeyer-Villiger reactions: A unique route to several bicyclic-γ-lactones in high enanatiomeric purity. J. Org. Chem. 1992, 57, 1306–1309. [Google Scholar] [CrossRef]
- Song, J.W.; Lee, J.H.; Bornscheuer, U.T.; Park, J.B. Microbial synthesis of medium chain α.ω-dicarboxylic acids and ω-aminocarboxylic acids from renewable long-chain fatty acids. Adv. Synth. Catal. 2014, 356, 1782–1788. [Google Scholar] [CrossRef]
- Sattler, J.H.; Fuchs, M.; Mutti, F.G.; Grischek, B.; Engel, P.; Pfeffer, J.; Woodley, J.M.; Kroutil, W. Introducing an in-situ capping strategy in systems catalysis to access 6-aminohexanoic acid. Angew. Chem. Int. Ed. 2014, 53, 14153–14157. [Google Scholar] [CrossRef]
- Milker, S.; Fink, M.J.; Rudroff, F.; Mihovilovic, M.D. Non-hazadous biocatalytic oxidation in Nylon 9 monomer synthesis on a 40 g scale with efficient downstream processing. Biotechnol. Bioeng. 2017, 114, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Zenno, S.; Saigo, K. Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J. Bacteriol. 1994, 176, 3544–3551. [Google Scholar] [CrossRef][Green Version]
- Gaudu, P.; Touati, D.; Niviere, V.; Fontecave, M. The NAD(P)H: Flavin oxidoreductase from Escherichia coli as a source of superoxide radicals. J. Biol. Chem. 1994, 269, 8182–8188. [Google Scholar] [CrossRef] [PubMed]
- Fieschi, F.; Niviere, V.; Frier, C.; Decout, J.-L. The mechanism and substrate specificity of the NADPH: Flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 1995, 270, 30392–30400. [Google Scholar] [CrossRef]
- Willetts, A.; Kelly, D.R. Multiple native flavin reductases in camphor-utilising Pseudomonas putida NCIMB 10007: Functional interaction with two-component diketocamphane monooxygenase isoenzymes. Microbiology 2014, 160, 1784–1794. [Google Scholar] [CrossRef]
- Niviere, V.; Fieschi, F.; Decout, J.-L.; Fontecave, M. The NAD(P)H:flavin oxidoreductase from Escherichia coli. Evidence for a new mode of binding for reduced pyridine nucleotides. J. Biol. Chem. 1999, 274, 18252–18260. [Google Scholar] [CrossRef]
- Willetts, A. Functional Studies of Type II Baeyer-Villiger Monooxygenases. In Annual Research Report: Faculty of Science; University of Exeter: Exeter, UK, 1998; pp. 68–74. [Google Scholar]
- Liu, Y.; Golden, S.S.; Kondo, T.; Ishiura, M.; Johnson, C.H. Bacterial luciferase as a reporter of circadian gene expression in cyano-bacteria. J. Bacteriol. 1995, 177, 2080–2086. [Google Scholar] [CrossRef]
- Waldman, M.S.; Fenja, S.; Bieichrodt, T.L.; Riedel, C.U. Bacterial luciferase reporters: The Swiss army knife of molecular biology. Bioeng. Bugs 2011, 2, 8–16. [Google Scholar] [CrossRef]
- Kadow, M.; Sass, S.; Schmidt, M.; Bornschueur, U. Recombinant expression and purification of the 2,5-diketocamphane 1,2-monooxgenase from the camphor-metabolising Pseudomonas putida strain NCIMB 10007. AMB Express 2011, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Kadow, M.; Loschinski, K.; Sass, S.; Schnidt, M.; Bornschueur, U. Completing the series of BVMOs involved in camphor metabolism of Pseudomonas putida NCIMB 10007 by identification of two missing genes, their functional expression in E. coli, and biochemical characteristics. Appl. Microbiol. Biotechnol. 2012, 96, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Kadow, M.; Balke, K.; Willetts, A.; Bornschueur, U.T.; Backwall, J.E. Baeyer-Villiger monooxygenases with a flavin reductase from E. coli. Appl. Microbiol. Biotechnol. 2014, 98, 3975–3986. [Google Scholar] [CrossRef] [PubMed]
- Campbell, Z.T.; Baldwin, T.O. Fre is the major flavin reductase supporting bioluminescence from Vibrio harveyi luciferase in Escherichia coli. J. Biol.Chem. 2009, 284, 8322–8328. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Mortimer, M.W.; Fisher, T.S.; Kahn, M.L.; Brockman, F.J.; Xun, L. Cloning, sequencing, and analysis of a gene cluster from Chelo-bacterium heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J. Bacteriol. 1997, 179, 1112–1116. [Google Scholar] [CrossRef]
- Thibaut, D.; Ratet, N.; Bisch, D.; Faucher, D.; Debussch, L.; Blanche, F. Purification of the two-enzyme system catalysing the oxidation of the D-proline residue of pristamycin IIB during the last step of pristamycin IIA biosynthesis. J. Bacteriol. 1995, 177, 5199–5205. [Google Scholar] [CrossRef]
- Witschel, M.; Nagel, S.; Egli, T. Identification and characterisation of the two-enzyme system catalysing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J. Bacteriol. 1997, 179, 6937–6943. [Google Scholar] [CrossRef]
- Rollig, R.; Paul, C.E.; Claeys-Bruno, M.; Duquesne, K.; Kara, S.; Alphand, V. Divorce in the two-component BVMO family:the single oxygenase for enantioselective chemo-enzymatic Baeyer-Villiger oxidations. Org. Biomol. Chem. 2021, 19, 3441–3450. [Google Scholar] [CrossRef]
- Uetz, T.; Schneider, R.; Snozzi, M.; Egli, T. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from ‘Chelatobacter’ strain ATCC 29600. J. Bacteriol. 1992, 174, 1179–1188. [Google Scholar] [CrossRef]
- Louie, T.M.; Xie, S.; Xun, L. Coordinated production and utilization of FADH2 by NAD(P)H-flavin oxidoreductase and 4-hydroxyphenylacetate 3-monooxygenase. Biochemistry 2003, 42, 7509–7517. [Google Scholar] [CrossRef]
- Gunsalus-Miguel, A.; Meighen, E.A.; Nicholi, M.Z.; Nealson, K.H. Purification and properties of bacterial luciferases. J. Biol. Chem. 1972, 247, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Duane, W.; Hastings, J.W. Flavin mononucleotide reductase of luminous bacteria. Mol. Cell. Biochem. 1975, 6, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Carnell, A.; Willetts, A. Biotransformations by fungi. Regio- plus stereoselective Baeyer-Villiger oxidations by dematiaceous fungi. Biotechnol. Lett. 1992, 14, 17–21. [Google Scholar] [CrossRef]
- Grogan, G.; Roberts, S.M.; Wan, P.; Willetts, A. Camphor-grown Pseudomonas putida: A multifunctional biocatalysts for oxygenations. Biotechnol. Lett. 1993, 15, 913–918. [Google Scholar] [CrossRef]
- Gagnon, R.; Grogan, G.; Levitt, M.S.; Roberts, S.M.; Wan, P.W.H.; Willetts, A. Biological Baeyer-Villiger oxidation of some monocyclic and bicyclic ketones using monooxygenases from Acinetobacter calcoaceticus NCIMB 9871 and Pseudomonas putida NCIMB 10007. J. Chem. Soc. Perkin Trans. 1994, 1, 2537–2543. [Google Scholar] [CrossRef]
- Lei, B.; Tu, S.-C. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry 1998, 37, 14529–14623. [Google Scholar] [CrossRef]
- Ottolina, G.; Carrea, G.; Colonna, S.; Ruchemann. A predictive active site model for cyclohexanone monooxygense catalysed Baeyer-Villiger oxidations. Tetrahedron Asymm. 1996, 7, 1123–1136. [Google Scholar] [CrossRef]
- Kelly, D.R. A proposal for the origin of stereoselectivity in enzyme catalysed Baeyer-Villiger reactions. Tetrahedron Asymm. 1996, 7, 1149–1152. [Google Scholar] [CrossRef]
- Tanase, C.; Pintilie, L.; Tanase, R.E. Lactones in the synthesis of prostaglandins and prostaglandin analogues. Int. J. Mol. Sci. 2021, 22, 1572. [Google Scholar] [CrossRef]
- Balke, K.; Baumgen, M.; Bornscheuer, U.T. Controlling the regioselectivity of Baeyer-Villiger monooxygenases by mutations of active site residues. ChemBioChem. 2017, 18, 1627–1638. [Google Scholar] [CrossRef]
- Alphand, V.; Archelas, A.; Furstoss, R. Microbial transformations 16. One-step synthesis of the pivotal prostaglandin chiral synthon via a highly enantioselective microbiological Baeyer-Villiger type reaction. Tetrahedron Lett. 1989, 30, 3663–3664. [Google Scholar] [CrossRef]
- Willetts, A.; Knowles, C.J.; Levitt, M.S.; Roberts, S.M.; Sandey, H.; Shipston, N.F. Biotransformation of endo-bicyclo[2.2.1]heptanol and endo-bicyclo[3.2.0]hept-2-en-6-ol into corresponding lactones. J. Chem. Soc. Perkin Trans. 1991, 1, 1608–1610. [Google Scholar] [CrossRef]
- Baldwin, C.V.F.; Wohlgemuth, R.; Woodley, J.M. Reactor operation and scale-up for whole-cell Baeyer-Villiger catalysed lactone synthesis. Org. Process. Res. Dev. 2008, 12, 660–665. [Google Scholar] [CrossRef]
- Ohshiro, T.; Aoi, Y.; Torii, K.; Izumi, Y. Flavin reductase coupling with two monooxygenases involved in dibenzothiophene desulfurization: Characterization of a non-desulfurizing bacterium Paenibacillus polymxya A-1. Appl. Microbiol. Biotechnol. 2002, 59, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Squires, C.H.; Ji, W.; Xi, L.; Ortego, B.; Pogrebinsky, O.S.; Gray, K.A. Method of Desulfurization of Fossil Fuel with Flavoprotein. US Patent no. 5,733,773, 31 March 1998. [Google Scholar]
- Rambosek, L.; Paddington, C.S.; Kovacevich, B.R.; Young, K.D.; Denome, S. Recombinant DNA Encoding a Desulfurization Biocatalyst. US Patent no. 5,879, 914, 12 December 1999. [Google Scholar]
- Reichmuth, D.S.; Hittle, J.L.; Blanch, H.W.; Keasling, J.D. Bio-desulfurization of dibenzothiophene in Escherichia coli enhanced by expression of a Vibrio harveyi oxidoreductase gene. Biotechnol. Bioeng. 2000, 67, 72–79. [Google Scholar] [CrossRef]
- Bornschueur, U.T.; Kazlauskas, R.J. Catalytic promiscuity in biocatalysis: Using old enzymes to form new bonds and follow new pathways. Angew. Chem. Int. Ed. 2004, 43, 6032–6041. [Google Scholar] [CrossRef]
- Kazlauskas, R.J. Enhancing catalytic promiscuity for biocatalysis. Curr. Opinion Chem. Biol. 2005, 9, 195–210. [Google Scholar] [CrossRef]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and application. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef]
- Meng, J.; Feng, R.; Zheng, G.; Mast, Y.; Wohleben, W.; Gao, J.; Jiang, W.; Lu, Y. Improvements of pristamycin production in Streptomyces pristinaespiralis by metabolic engineering approach. Synth. Syst. Biotechnol. 2017, 2, 130–136. [Google Scholar] [CrossRef]
- Galan, B.; Diaz, E.; Garcia, J.L. Enhancing desulfurization by engineering a flavin reductase-encoding gene cassette in recombinant biocatalysis. Environ. Microbiol. 2000, 2, 687–694. [Google Scholar] [CrossRef]
- Alcin, A.; Santos, V.E.; Martin, A.B.; Yustos, P.; Garcia-Ochoa, F. Biodesulfurization of DBT with Pseudomonas putida CECT5279 by resting cells: Influence of cell growth time on reducing equivalent concentration and HpaC activity. Biochem. Eng. 2005, 26, 168–175. [Google Scholar] [CrossRef]
- Li, G.Q.; Li, S.S.; Zhang, M.L.; Wang, J.; Zhu, L.; Liang, F.L.; Liu, R.L.; Ma, T. Genetic rearrangement strategy for optimising the dibenzo-thiophene biodesulfurization pathway in Rhodococcus erythropolis. Appl. Environ. Microbiol. 2008, 74, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Mohebali, G.; Ball, A.S. Biocatalytic desulfurization (BDS) of petrodeisel fuels. Microbiology 2008, 154, 2169–2183. [Google Scholar] [CrossRef]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology. J. Amer. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef] [PubMed]
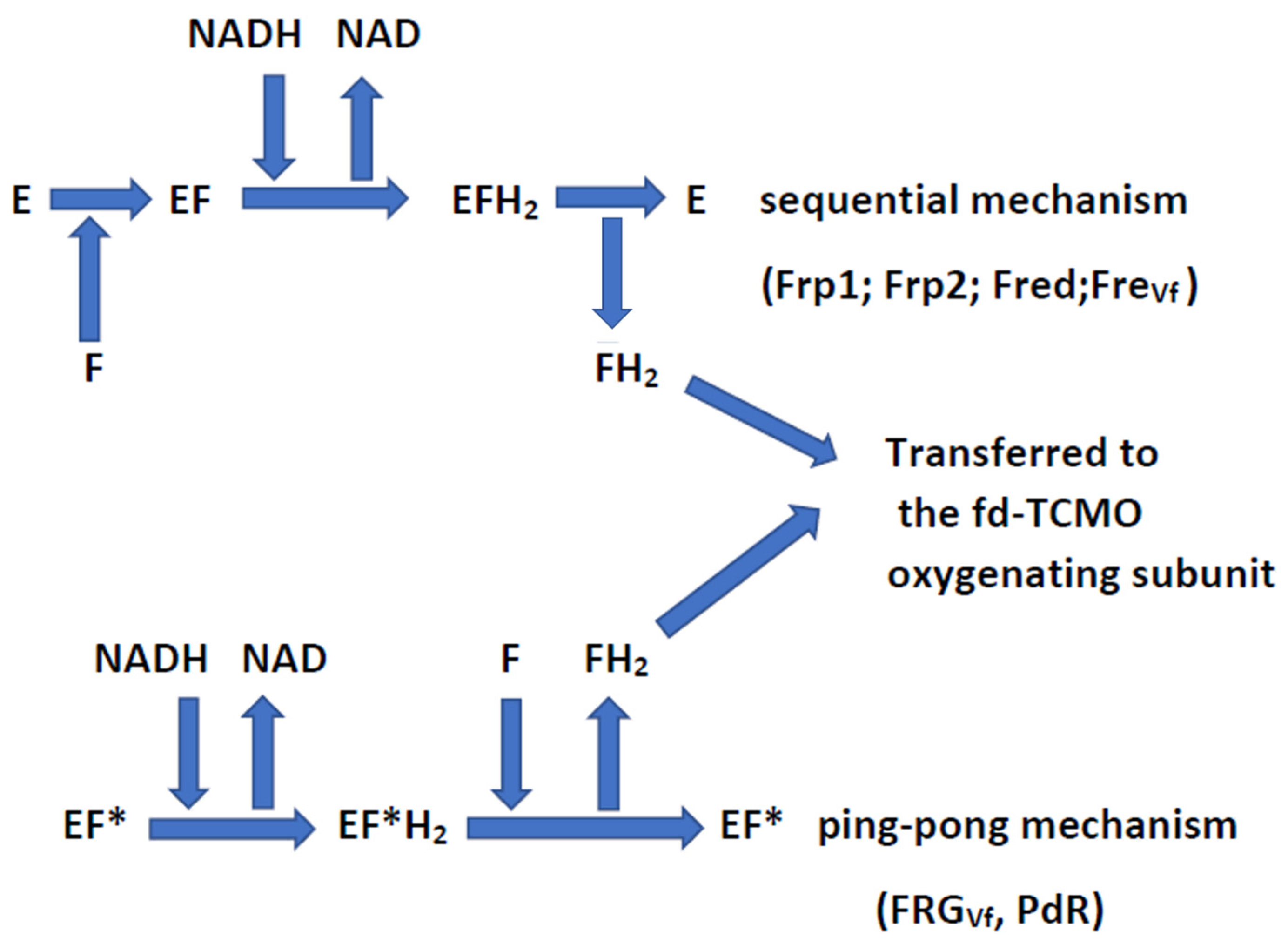
| Flavin Reductase: Nomenclature and Reaction Mechanism | Microbial Source | MW and Structure | Bound or Unbound FMN |
|---|---|---|---|
| Frp1 Sequential | P. putida | 26.0 kDa | Unbound |
| Frp2 Sequential | P. putida | 27.0 kDa | Unbound |
| Fred Sequential | P. putida | 2 × 18.0 kDa | Unbound |
| PdR Ping-pong | P. putida | 48.5 kDa | Bound |
| FreEc (FR-II) Sequential | E. coli | 28.5 kDa | Unbound |
| FRGVf (FRG-I) Ping-pong | V. fischeri | 24.6 kDa | Bound |
| FreVf Sequential | V. fischeri | 25.5 kDa | Unbound |
| FRDVh Sequential | V. harveyi | 26.7 kDa | Unbound |
| FRPVh Ping-pong | V. harveyi | 26.3 kDa | Bound |
| ActVB (FRD-II) Ping-pong | Streptomyces coelicolor | 2 × 18.0 kDa | Bound |
| SnaC (FRD-I, PIIB) Ping-pong | S. pristinaspiralis | 28.0 kDa | Bound |
| EmoB (cB’) Ping-pong | Chelatovorans multitrophicus | 2 × 25.0 kDa | Bound |
| DszD (FRD-III) Sequential | Rhodococcus erythropolis | 4 × 22.5 kDa | Unbound |
| FRDAa (cB) Sequential | Aminobacter aminovorans | 2 × 44.0 kDa | Bound |
| Flavin Reductase and Source | Apparent KmFMN (μM) | Source of Hydride Ion | Mechanism of Hydride Ion Transfer |
|---|---|---|---|
| Frp1 P. putida | 2.5 (S) 2.0 (C + 2,5-MO) 2.5 (C + 3,6-MO) | NADH | Sequential |
| Frp2 P. putida | 4.2 (S) 3.6 (C + 2,5-MO) 4.1 (C + 3,6-MO) | NADH | Sequential |
| FRGVf (V. fischeri) | 4.3 (S) 4.0 (C + LuxAB) | NADH | Ping-pong |
| FreVf (V. fischeri) | 2.5 (S) 2.6 (C + LuxAB) | NADH | Sequential |
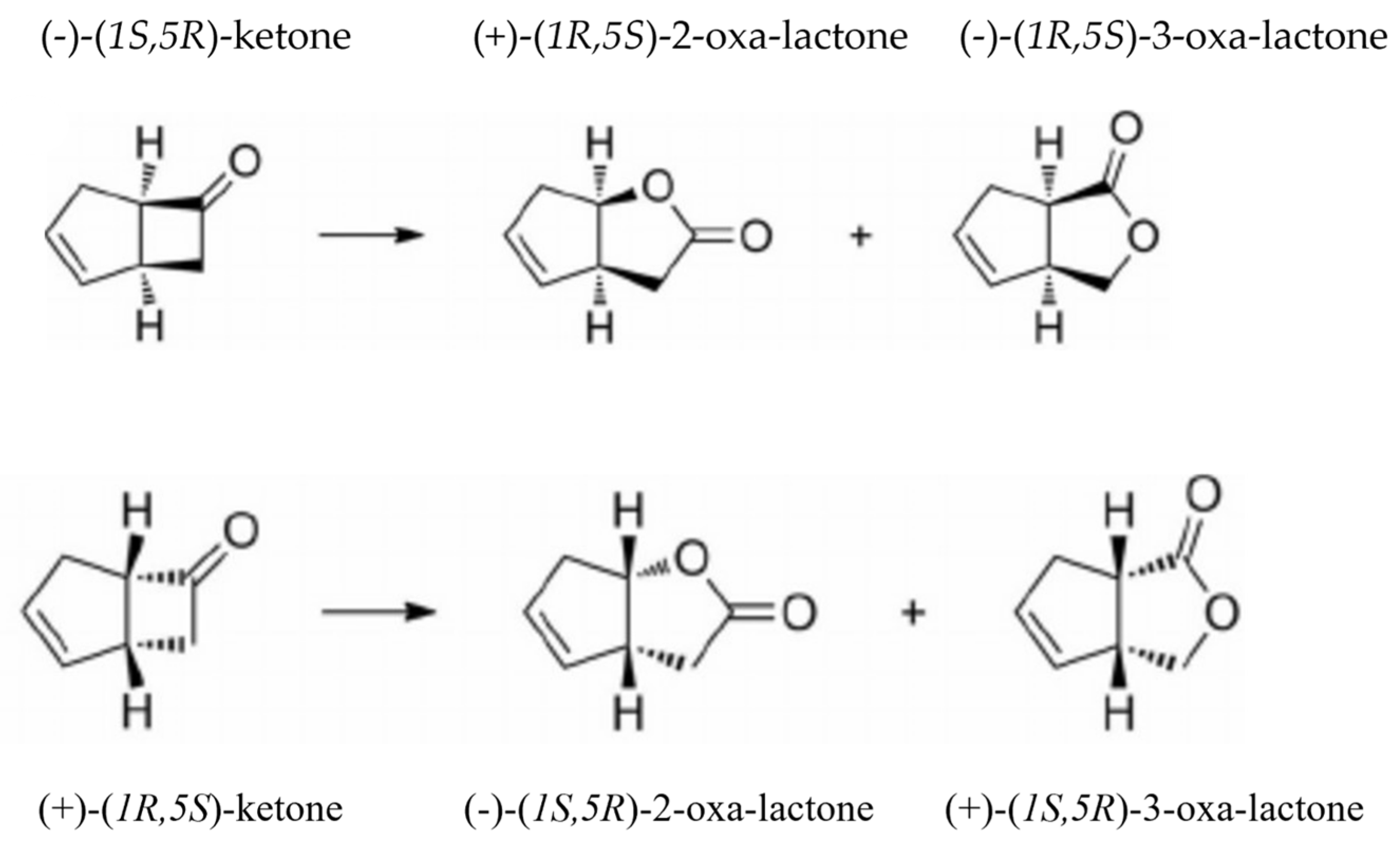
| fd-TCMO and Partner FR in Coupled-enzyme Reaction | Residual Ketone (mM) Remaining after 120 min | Percentage and Ratio of the Ketone (k) Enantiomers Biotransformed | Predominant Regioisomeric Lactones (l) Formed Expressed as ee% | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (+)k | (−)k | (+):(−) | (+)2l | (−)2l | (+)3l | (−)3l | |||
| LuxAB luciferase | |||||||||
| Frp1 (n.n) | 0.70 | 41.6 | 18.4 | 2.26:1 | – | >98 | – | 16 | |
| Frp2 (n.n) | 0.69 | 43.0 | 18.4 | 2.33:1 | – | >98 | – | 18 | |
| FRGVf (n) | 0.75 | 34.2 | 15.0 | 2.28:1 | – | 96 | – | 18 | |
| FreVf (n) | 0.79 | 42.8 | 18.8 | 2.28:1 | – | >99 | – | 16 | |
| FRDA (n.n) | 0.62 | 52.2 | 22.8 | 2.29:1 | – | >98 | – | 16 | |
| FreEc (n.n) | 0.61 | 54.2 | 24.0 | 2.25:1 | – | >99 | – | 18 | |
| 2,5-DKCMO | |||||||||
| Frp1 (n) | 0.24 | 81.8 | 69.7 | 1.17:1 | 84 | – | >99 | – | |
| Frp2 (n) | 0.20 | 84.3 | 74.8 | 1.13:1 | 86 | – | >98 | – | |
| FRGVf (n.n) | 0.26 | 78.8 | 68.4 | 1.15:1 | 82 | – | >98 | – | |
| FreVf (n.n) | 0.23 | 82.5 | 71.7 | 1.15:1 | 84 | – | >98 | – | |
| FRDAa (n.n) | 0.06 | 100 | 88.6 | 1.13:1 | 88 | – | >99 | – | |
| FreEc (n.n) | 0.04 | 100 | 92.0 | 1.09:1 | 90 | – | >98 | – | |
| 3,6-DKCMO | |||||||||
| Frp1 (n) | 0.53 | 58.8 | 35.0 | 1.68:1 | 30 | – | 80 | – | |
| Frp2 (n) | 0.53 | 59.8 | 35.2 | 1.70:1 | 30 | – | 82 | – | |
| FRGVf (n.n) | 0.55 | 57.0 | 33.0 | 1.72:1 | 32 | – | 86 | – | |
| FreVf (n.n) | 0.52 | 61.0 | 35.4 | 1.72:1 | 32 | – | 84 | – | |
| FRDAa (n.n) | 0.38 | 77.6 | 45.0 | 1.72:1 | 34 | – | 82 | – | |
| FreEc (n.n) | 0.37 | 80.0 | 45.8 | 1.74:1 | 32 | – | 88 | – | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willetts, A. Inter-Species Redox Coupling by Flavin Reductases and FMN-Dependent Two-Component Monooxygenases Undertaking Nucleophilic Baeyer–Villiger Biooxygenations. Microorganisms 2023, 11, 71. https://doi.org/10.3390/microorganisms11010071
Willetts A. Inter-Species Redox Coupling by Flavin Reductases and FMN-Dependent Two-Component Monooxygenases Undertaking Nucleophilic Baeyer–Villiger Biooxygenations. Microorganisms. 2023; 11(1):71. https://doi.org/10.3390/microorganisms11010071
Chicago/Turabian StyleWilletts, Andrew. 2023. "Inter-Species Redox Coupling by Flavin Reductases and FMN-Dependent Two-Component Monooxygenases Undertaking Nucleophilic Baeyer–Villiger Biooxygenations" Microorganisms 11, no. 1: 71. https://doi.org/10.3390/microorganisms11010071
APA StyleWilletts, A. (2023). Inter-Species Redox Coupling by Flavin Reductases and FMN-Dependent Two-Component Monooxygenases Undertaking Nucleophilic Baeyer–Villiger Biooxygenations. Microorganisms, 11(1), 71. https://doi.org/10.3390/microorganisms11010071






