Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Environment and Animals
2.2. Sample Isolation, Isolate Recovery, and Species Identification
2.3. Antimicrobial Susceptibility Testing
2.4. DNA Extraction and Whole Genome Sequencing (WGS)
2.5. Phylogenetic Analyses
2.6. Genotypic Antibiotic Resistance Analyses
2.7. Statistics
3. Results
3.1. Species Detection and Distribution
3.2. Phenotypic Resistance Profiles of NAS Isolates
3.3. Genome Sequencing and Phylogenetic Analyses
3.4. Genotypic Resistance Profiles of Isolates
3.5. Multiple Resistance Determinant (MRD) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Review on Antimicrobial Resistance: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; UK government and Welcome Trust: London, UK, 2016. [Google Scholar]
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Rolo, J.; Worning, P.; Nielsen, J.B.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Tomasz, A.; Westh, H.; et al. Evolutionary Origin of the Staphylococcal Cassette Chromosome mec (SCCmec). Antimicrob. Agents Chemoth. 2017, 61, 6. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, N.; Maeusli, M.; Papafotis, D.; Miller, S.; Lee, B.; Theologidis, I.; Luna, B. Environmental Spread of Antibiotic Resistance. Antibiotics 2021, 10, 640. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Waste; UK government and Welcome Trust: London, UK, 2015; pp. 1–41. [Google Scholar]
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 2021, 74, 21. [Google Scholar] [CrossRef] [PubMed]
- Goerge, T.; Lorenz, M.B.; van Alen, S.; Hubner, N.O.; Becker, K.; Kock, R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: Prevalence, preventive options and evidence. Vet. Microbiol. 2017, 200, 6–12. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fevre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Association, A.V.M. One Health: A New Professional Imperative. Maced. J. Med. Sci. 2008, 3, 229–232. [Google Scholar] [CrossRef]
- Nationales Forschungsnetz zoonotische Infektionskrankheiten. #1Health-PREVENT—One Health Interventionen zur Prävention der Zoonotischen Verbreitung von Antibiotikaresistenten Erregern. Available online: https://www.gesundheitsforschung-bmbf.de/de/1health-prevent-one-health-interventionen-zur-pravention-der-zoonotischen-verbreitung-von-7131.php (accessed on 31 October 2022).
- Schollenbruch, H.; Kobusch, I.; Schroter, I.; Mellmann, A.; Kock, R.; Boelhauve, M. Pilot Study on Alteration of LA-MRSA Status of Pigs during Fattening Period on Straw Bedding by Two Types of Cleaning. Antibiotics 2021, 10, 521. [Google Scholar] [CrossRef]
- Kobusch, I.; Schroter, I.; Linnemann, S.; Schollenbruch, H.; Hofmann, F.; Boelhauve, M. Prevalence of LA-MRSA in pigsties: Analysis of factors influencing the (De)colonization process. Sci. Rep. 2022, 12, 18000. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 11.0; European Society of Clinical microbiology and Infectious Diseases: Basel, Switzerland, 2021. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carrico, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic. Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2018. Available online: https://www.R-project.org/ (accessed on 14 December 2022).
- Heilmann, C.; Ziebuhr, W.; Becker, K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 2019, 25, 1071–1080. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Andam, C.P. Extensive Horizontal Gene Transfer within and between Species of Coagulase-Negative Staphylococcus. Genome Biol. Evol. 2021, 13, evab206. [Google Scholar] [CrossRef] [PubMed]
- Landbau, B. Bioland-Richtlinien. Tierhaltung. 2022. Available online: www.bioland.de (accessed on 15 October 2022).
- Naturland Standards on Production. 2022. Available online: www.naturland.de (accessed on 15 October 2022).
- Hamel, J.; Zhang, Y.; Wente, N.; Kromker, V. Non-S. aureus staphylococci (NAS) in milk samples: Infection or contamination? Vet. Microbiol. 2020, 242, 108594. [Google Scholar] [CrossRef] [PubMed]
- Stepien-Pysniak, D.; Wilczynski, J.; Marek, A.; Smiech, A.; Kosikowska, U.; Hauschild, T. Staphylococcus simulans associated with endocarditis in broiler chickens. Avian. Pathol. 2017, 46, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Pyorala, S.; Taponen, S. Coagulase-negative staphylococci-emerging mastitis pathogens. Vet. Microbiol. 2009, 134, 3–8. [Google Scholar] [CrossRef]
- Shields, B.E.; Tschetter, A.J.; Wanat, K.A. Staphylococcus simulans: An emerging cutaneous pathogen. JAAD Case Rep. 2016, 2, 428–429. [Google Scholar] [CrossRef]
- Mallet, M.; Loiez, C.; Melliez, H.; Yazdanpanah, Y.; Senneville, E.; Lemaire, X. Staphylococcus simulans as an authentic pathogenic agent of osteoarticular infections. Infection 2011, 39, 473–476. [Google Scholar] [CrossRef]
- Lal, A.; Akhtar, J.; Ullah, A.; Abraham, G.M. First Case of Pleural Empyema Caused by Staphylococcus simulans: Review of the Literature. Case Rep. Infect. Dis. 2018, 2018, 7831284. [Google Scholar] [CrossRef]
- Go, J.R.; Corsini Campioli, C.; DeSimone, D.; Sohail, M.R. Staphylococcus simulans bloodstream infection following CIED extraction. BMJ Case Rep. 2021, 14, e240309. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Fessler, A.T.; Hanke, D.; Sanz, S.; Olarte, C.; Mama, O.M.; Eichhorn, I.; Schwarz, S.; Torres, C. Coagulase-negative staphylococci carrying cfr and PVL genes, and MRSA/MSSA-CC398 in the swine farm environment. Vet. Microbiol. 2020, 243, 108631. [Google Scholar] [CrossRef]
- Khazandi, M.; Al-Farha, A.A.; Coombs, G.W.; O’Dea, M.; Pang, S.; Trott, D.J.; Aviles, R.R.; Hemmatzadeh, F.; Venter, H.; Ogunniyi, A.D.; et al. Genomic characterization of coagulase-negative staphylococci including methicillin-resistant Staphylococcus sciuri causing bovine mastitis. Vet. Microbiol. 2018, 219, 17–22. [Google Scholar] [CrossRef]
- De Visscher, A.; Supre, K.; Haesebrouck, F.; Zadoks, R.N.; Piessens, V.; Van Coillie, E.; Piepers, S.; De Vliegher, S. Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet. Microbiol. 2014, 172, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.M.; Jimenez, L.; Perea, J.M.; Arias, R.; Palop, M.L. Farming Practices Influence Antibiotic Resistance and Biogenic Amine Capacity of Staphylococci from Bulk Tank Ewe’s Milk. Animals 2020, 10, 1622. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.M.; Dong, Y.; Fessler, A.T.; Schwarz, S.; Schoen, C.; Kock, R.; Ziebuhr, W. Antibiotic resistance profiles of coagulase-negative staphylococci in livestock environments. Vet. Microbiol. 2017, 200, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Rissi, D.R.; Elsmo, E.J.; Sanchez, S. Cystitis and peritonitis caused by Staphylococcus xylosus infection in a calf. Braz. J. Vet. Pathol. 2015, 8, 99–101. [Google Scholar]
- Akhaddar, A.; Elouennass, M.; Naama, O.; Boucetta, M. Staphylococcus xylosus isolated from an otogenic brain abscess in an adolescent. Surg. Infect. 2010, 11, 559–561. [Google Scholar] [CrossRef]
- Klibi, A.; Maaroufi, A.; Torres, C.; Jouini, A. Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 930–935. [Google Scholar] [CrossRef]
- Yuan, Z.; Wang, J.; Che, R.; God’spower, B.O.; Zhou, Y.; Dong, C.; Li, L.; Chen, M.; Eliphaz, N.; Liu, X.; et al. Relationship between L-lactate dehydrogenase and multidrug resistance in Staphylococcus xylosus. Arch. Microbiol. 2021, 204, 91. [Google Scholar] [CrossRef]
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef]
- Kloos, W.E.; Zimmerman, R.J.; Smith, R.F. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 1976, 31, 53–59. [Google Scholar] [CrossRef]
- De la Rosa-Ramos, M.A.; Salcedo-Hernandez, R.; Sarmiento-Silva, R.E.; Aguilera-Arreola, M.G.; Alcantar-Curiel, M.D.; Betanzos-Cabrera, G.; Rodriguez-Martinez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Non-epidermidis coagulase-negative Staphylococcus isolated from farm animals can inhibit the hemagglutinating activity of Newcastle disease virus and bovine parainfluenza virus type 3. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101649. [Google Scholar] [CrossRef]
- Frey, Y.; Rodriguez, J.P.; Thomann, A.; Schwendener, S.; Perreten, V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 2013, 96, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- De Visscher, A.; Piepers, S.; Haesebrouck, F.; Supre, K.; De Vliegher, S. Coagulase-negative Staphylococcus species in bulk milk: Prevalence, distribution, and associated subgroup- and species-specific risk factors. J. Dairy Sci. 2017, 100, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Garland-Lewis, G.; Trufan, S.; Meschke, S.J.; Fowler, H.; Shean, R.C.; Greninger, A.L.; Rabinowitz, P.M. Distribution of Staphylococcus species in dairy cows, workers and shared farm environments. FEMS Microbiol. Lett. 2018, 365, fny146. [Google Scholar] [CrossRef] [PubMed]
- Sizemore, C.; Buchner, E.; Rygus, T.; Witke, C.; Gotz, F.; Hillen, W. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon. Mol. Gen. Genet. 1991, 227, 377–384. [Google Scholar] [CrossRef]
- Sizemore, C.; Wieland, B.; Gotz, F.; Hillen, W. Regulation of Staphylococcus xylosus xylose utilization genes at the molecular level. J. Bacteriol. 1992, 174, 3042–3048. [Google Scholar] [CrossRef][Green Version]
- Kloos, W.E.; Wolfshohl, J.F. Identification of Staphylococcus species with the API STAPH-IDENT system. J. Clin. Microbiol. 1982, 16, 509–516. [Google Scholar] [CrossRef]
- Marples, R.R.; Richardson, J.F. Evaluation of a micromethod gallery (API Staph) for the identification of staphylococci and micrococci. J. Clin. Pathol. 1982, 35, 650–656. [Google Scholar] [CrossRef]
- BioMérieux, S. API® STAPH—Manual 2021/04. Available online: www.biomerieux.com (accessed on 1 October 2022).
- Marincola, G.; Liong, O.; Schoen, C.; Abouelfetouh, A.; Hamdy, A.; Wencker, F.D.R.; Marciniak, T.; Becker, K.; Kock, R.; Ziebuhr, W. Antimicrobial Resistance Profiles of Coagulase-Negative Staphylococci in Community-Based Healthy Individuals in Germany. Front. Public Health 2021, 9, 684456. [Google Scholar] [CrossRef]
- Reynaga, E.; Navarro, M.; Vilamala, A.; Roure, P.; Quintana, M.; Garcia-Nunez, M.; Figueras, R.; Torres, C.; Lucchetti, G.; Sabria, M. Prevalence of colonization by methicillin-resistant Staphylococcus aureus ST398 in pigs and pig farm workers in an area of Catalonia, Spain. BMC Infect. Dis. 2016, 16, 716. [Google Scholar] [CrossRef]
- Argudin, M.A.; Roisin, S.; Nienhaus, L.; Dodemont, M.; de Mendonca, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic Diversity among Staphylococcus aureus Isolates Showing Oxacillin and/or Cefoxitin Resistance Not Linked to the Presence of mec Genes. Antimicrob. Agents Chemother. 2018, 62, e00091-18. [Google Scholar] [CrossRef]
- McDougal, L.K.; Thornsberry, C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J. Clin. Microbiol. 1986, 23, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Ishikawa, H.; Itoh, T.; Arano, M.; Hirata, K.; Ueshiba, H. Penicillin-Binding Proteins and Associated Protein Mutations Confer Oxacillin/Cefoxitin Tolerance in Borderline Oxacillin-Resistant Staphylococcus aureus. Microb. Drug Resist. 2021, 27, 590–595. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Chen, L.; Gilbert, A.; da Costa, T.M.; Nair, V.; Datta, S.K.; Kreiswirth, B.N.; Chambers, H.F. PBP4 Mediates beta-Lactam Resistance by Altered Function. Antimicrob. Agents Chemother. 2017, 61, e00932-17. [Google Scholar] [CrossRef] [PubMed]
- Sommer, A.; Fuchs, S.; Layer, F.; Schaudinn, C.; Weber, R.E.; Richard, H.; Erdmann, M.B.; Laue, M.; Schuster, C.F.; Werner, G.; et al. Mutations in the gdpP gene are a clinically relevant mechanism for beta-lactam resistance in meticillin-resistant Staphylococcus aureus lacking mec determinants. Microb. Genom. 2021, 7, 623. [Google Scholar] [CrossRef]
- European Medicines Agency Science Medicines Health. Categorisation of Antibiotics in the European Union. Available online: www.ema.europa.eu (accessed on 12 December 2019).
- Schwarz, S.; Shen, J.; Kadlec, K.; Wang, Y.; Brenner Michael, G.; Fessler, A.T.; Vester, B. Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a027037. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef] [PubMed]
- Schaekel, F.; May, T.; Seiler, J.; Hartmann, M.; Kreienbrock, L. Antibiotic drug usage in pigs in Germany-Are the class profiles changing? PLoS ONE 2017, 12, e0182661. [Google Scholar] [CrossRef]
- Marosevic, D.; Kaevska, M.; Jaglic, Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage—A review. Ann. Agric. Environ. Med. 2017, 24, 338–344. [Google Scholar] [CrossRef]
- Agency, E.M. Questions and Answers on Veterinary Medicinal Products Containing a Combination of Lincomycin and Spectinomycin to Be Administered Orally to Pigs and/or Poultry: Outcome of a Referral Procedure under Article 35 of Directive 2001/82/EC (EMEA/V/A/110). 2016. Available online: https://www.ema.europa.eu/en/medicines/veterinary/referrals/veterinary-medicinal-products-containing-combination-lincomycin-spectinomycin-be-administered-orally (accessed on 15 October 2022).
- Falagas, M.E.; Athanasaki, F.; Voulgaris, G.L.; Triarides, N.A.; Vardakas, K.Z. Resistance to fosfomycin: Mechanisms, Frequency and Clinical Consequences. Int. J. Antimicrob. Agents 2019, 53, 22–28. [Google Scholar] [CrossRef]
- Michalopoulos, A.S.; Livaditis, I.G.; Gougoutas, V. The revival of fosfomycin. Int. J. Infect. Dis. 2011, 15, e732–e739. [Google Scholar] [CrossRef]
- Werner, A.H.; Russell, A.D. Mupirocin, fusidic acid and bacitracin: Activity, action and clinical uses of three topical antibiotics. Vet. Derm. 1999, 10, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Frosini, S.M.; Bond, R.; Loeffler, A.; Larner, J. Opportunities for topical antimicrobial therapy: Permeation of canine skin by fusidic acid. BMC Vet. Res. 2017, 13, 345. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Muller, E.; Braun, S.D.; Armengol-Porta, M.; Bes, M.; Boswihi, S.; El-Ashker, M.; Engelmann, I.; Gawlik, D.; Gwida, M.; et al. Characterisation of S. aureus/MRSA CC1153 and review of mobile genetic elements carrying the fusidic acid resistance gene fusC. Sci. Rep. 2021, 11, 8128. [Google Scholar] [CrossRef] [PubMed]
- Boswihi, S.S.; Udo, E.E.; Mathew, B.; Noronha, B.; Verghese, T.; Tappa, S.B. Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Patients Admitted to Kuwait Hospitals in 2016-2017. Front. Microbiol. 2019, 10, 2912. [Google Scholar] [CrossRef]
- O’Neill, A.J.; McLaws, F.; Kahlmeter, G.; Henriksen, A.S.; Chopra, I. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 2007, 51, 1737–1740. [Google Scholar] [CrossRef]
- Castanheira, M.; Watters, A.A.; Mendes, R.E.; Farrell, D.J.; Jones, R.N. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). J. Antimicrob. Chemoth. 2010, 65, 1353–1358. [Google Scholar] [CrossRef]
- Chen, H.J.; Hung, W.C.; Lin, Y.T.; Tsai, J.C.; Chiu, H.C.; Hsueh, P.R.; Teng, L.J. A novel fusidic acid resistance determinant, fusF, in Staphylococcus cohnii. J. Antimicrob. Chemoth. 2015, 70, 416–419. [Google Scholar] [CrossRef]
- McKenna, M. Antibiotic resistance: The last resort. Nature 2013, 499, 394–396. [Google Scholar] [CrossRef]
- Bayer, A.S.; Schneider, T.; Sahl, H.G. Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 2013, 1277, 139–158. [Google Scholar] [CrossRef]
- Schwarz, S.; Werckenthin, C.; Kehrenberg, C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemoth. 2000, 44, 2530–2533. [Google Scholar] [CrossRef]
- Liu, B.G.; Yuan, X.L.; He, D.D.; Hu, G.Z.; Miao, M.S.; Xu, E.P. Research progress on the oxazolidinone drug linezolid resistance. Eur. Rev. Med. Pharm. Sci. 2020, 24, 9274–9281. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemoth. 2018, 73, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Gostev, V.; Leyn, S.; Kruglov, A.; Likholetova, D.; Kalinogorskaya, O.; Baykina, M.; Dmitrieva, N.; Grigorievskaya, Z.; Priputnevich, T.; Lyubasovskaya, L.; et al. Global Expansion of Linezolid-Resistant Coagulase-Negative Staphylococci. Front. Microbiol. 2021, 12, 661798. [Google Scholar] [CrossRef] [PubMed]
- Argudin, M.A.; Lauzat, B.; Kraushaar, B.; Alba, P.; Agerso, Y.; Cavaco, L.; Butaye, P.; Porrero, M.C.; Battisti, A.; Tenhagen, B.A.; et al. Heavy metal and disinfectant resistance genes among livestock-associated methicillin-resistant Staphylococcus aureus isolates. Vet. Microbiol. 2016, 191, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Hoefer, A.; Butaye, P. Heavy metal resistance in bacteria from animals. Res. Vet. Sci. 2019, 122, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Su, J.-Q.; An, X.-L.; Huang, F.-Y.; Rensing, C.; Brandt, K.K.; Zhu, Y.-G. Feed additives shift gut microbiota and enrich antibiotic resistance in swine gut. Sci. Total Environ. 2018, 621, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends. Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020’. (EMA/58183/2021). 2021. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2019-2020-trends-2010-2020-eleventh_en.pdf (accessed on 15 October 2022).
- (EIP-AGRI). Final Report: Focus group Reducing Antibiotics In Pig Farming. 2013. Available online: https://ec.europa.eu/eip/agriculture/sites/default/files/eip-agri_fg3_pig_antibiotics_final_report_2014_en_0.pdf (accessed on 15 October 2022).
- Alt, K.; Fetsch, A.; Schroeter, A.; Guerra, B.; Hammerl, J.A.; Hertwig, S.; Senkov, N.; Geinets, A.; Mueller-Graf, C.; Braeunig, J.; et al. Factors associated with the occurrence of MRSA CC398 in herds of fattening pigs in Germany. BMC Vet. Res. 2011, 7, 69. [Google Scholar] [CrossRef]
- van Duijkeren, E.; Ikawaty, R.; Broekhuizen-Stins, M.J.; Jansen, M.D.; Spalburg, E.C.; de Neeling, A.J.; Allaart, J.G.; van Nes, A.; Wagenaar, J.A.; Fluit, A.C. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 2008, 126, 383–389. [Google Scholar] [CrossRef]

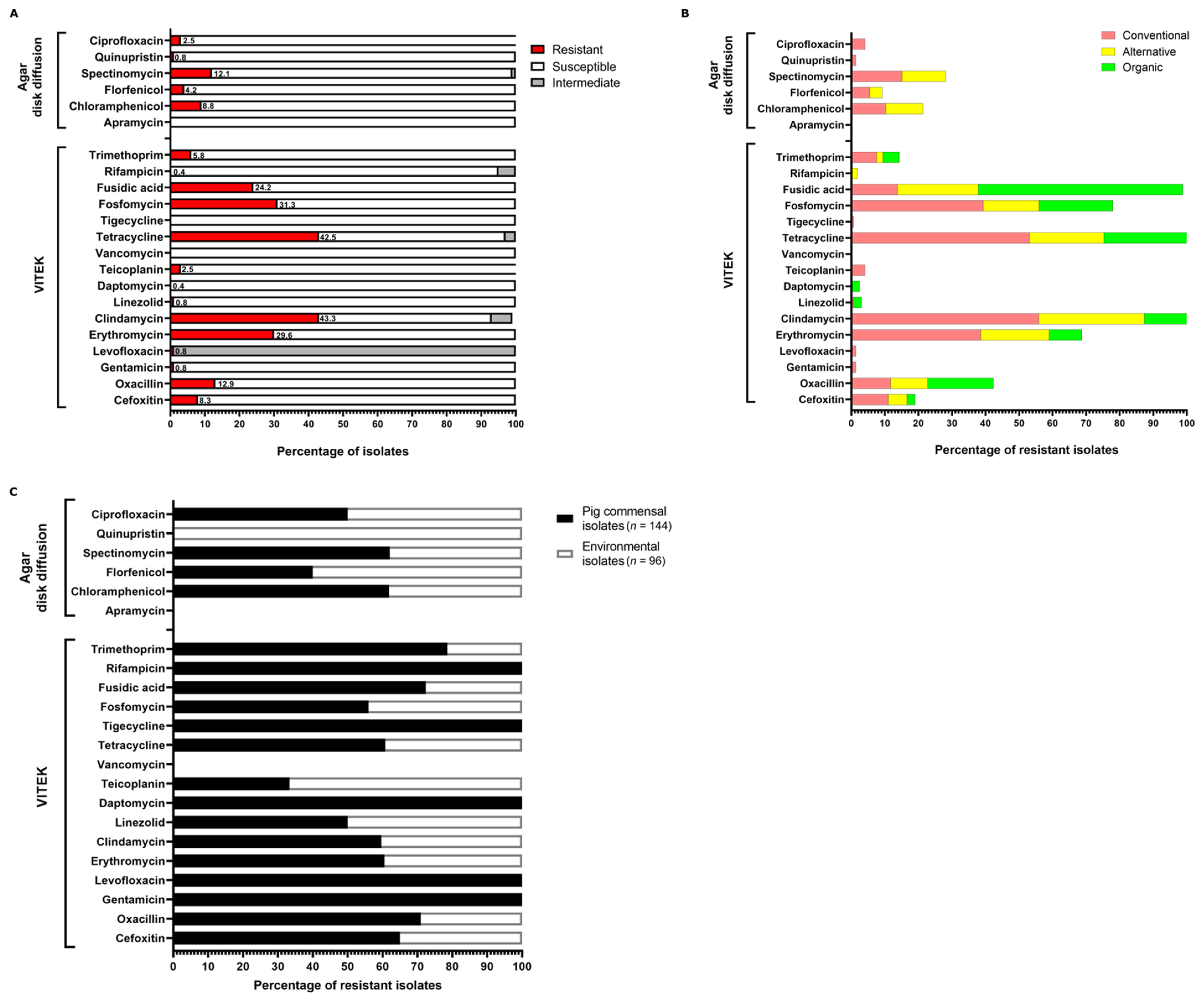

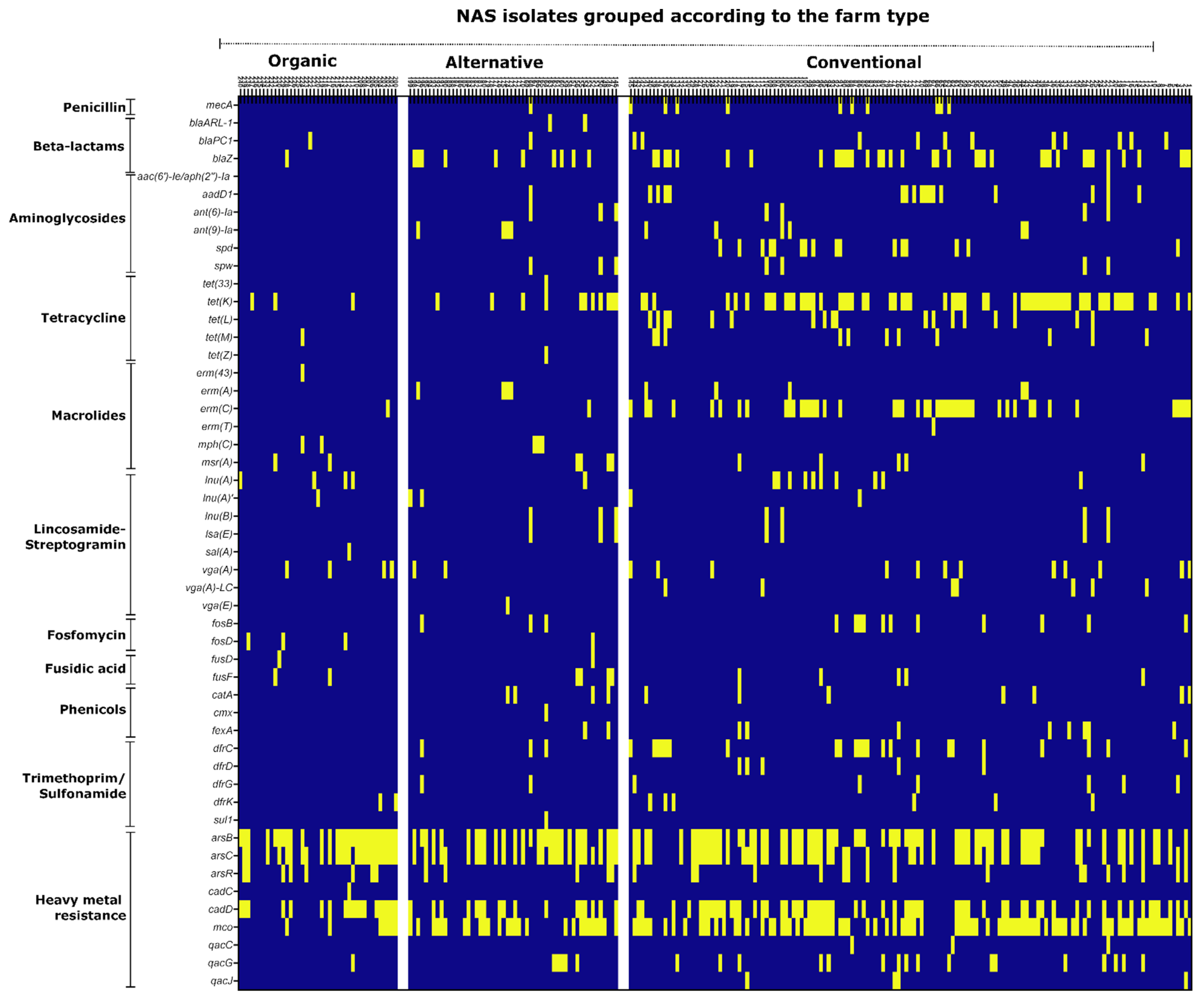

| Farm Type | Total Number of Isolates in the Farm-Type | Number of MRD Isolates | Percentage of Isolates that Were MRD |
|---|---|---|---|
| Conventional | 145 | 74 | 51 |
| Alternative | 54 | 18 | 33.3 |
| Organic | 41 | 10 | 24.4 |
| Farm type | Total Number of Isolates in the Farm-Type | Number of MRD Isolates | Percentage of Isolates that Were MRD |
|---|---|---|---|
| Conventional | 145 | 74 | 51 |
| Alternative | 54 | 15 | 27.8 |
| Organic | 41 | 4 | 9.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soundararajan, M.; Marincola, G.; Liong, O.; Marciniak, T.; Wencker, F.D.R.; Hofmann, F.; Schollenbruch, H.; Kobusch, I.; Linnemann, S.; Wolf, S.A.; et al. Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries. Microorganisms 2023, 11, 31. https://doi.org/10.3390/microorganisms11010031
Soundararajan M, Marincola G, Liong O, Marciniak T, Wencker FDR, Hofmann F, Schollenbruch H, Kobusch I, Linnemann S, Wolf SA, et al. Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries. Microorganisms. 2023; 11(1):31. https://doi.org/10.3390/microorganisms11010031
Chicago/Turabian StyleSoundararajan, Manonmani, Gabriella Marincola, Olivia Liong, Tessa Marciniak, Freya D. R. Wencker, Franka Hofmann, Hannah Schollenbruch, Iris Kobusch, Sabrina Linnemann, Silver A. Wolf, and et al. 2023. "Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries" Microorganisms 11, no. 1: 31. https://doi.org/10.3390/microorganisms11010031
APA StyleSoundararajan, M., Marincola, G., Liong, O., Marciniak, T., Wencker, F. D. R., Hofmann, F., Schollenbruch, H., Kobusch, I., Linnemann, S., Wolf, S. A., Helal, M., Semmler, T., Walther, B., Schoen, C., Nyasinga, J., Revathi, G., Boelhauve, M., & Ziebuhr, W. (2023). Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries. Microorganisms, 11(1), 31. https://doi.org/10.3390/microorganisms11010031








