1. Introduction
The current era shows great awareness of environmental problems. Microbial diversity can help in sustaining a clean and balanced ecosystem. Plant-associated microorganisms, especially endophytes, represent a hopeful tool for ecological maintenance and sustainable development. To do so, beneficial microorganisms have several mechanisms, e.g., acting as biofertilizers, bio-stimulants, and increasing resistance against abiotic and biotic stresses. Economically, applying plant-associated microorganisms is expected to drop production budgets [
1].
Besides the ecological role, plant growth-promoting fungi (PGPF) is useful fungi that have dual uses; they stimulate the growth of plants and protect plants from pathogens. The group of PGPF contains diverse species of fungi, e.g.,
Trichoderma spp., non-pathogenic species of
Aspergillus, and
Penicillium, however, more investigations to elucidate the biological role of PGPF are encouraged [
2].
Trichoderma spp. contains diverse fungi that are present in different environments, mostly in soils.
Trichoderma spp. can be linked with plants such as epiphytes, endophytes, and/or in the rhizosphere region, acting as promotors of plant growth and/or bioagents against various plant diseases.
Trichoderma species secrete diverse volatile and nonvolatile compounds that display antibiotic activity [
3]. Additionally,
Trichoderma spp. produces many compounds that improve the plant’s defense, growth, and resistance to numerous biotic and abiotic stresses like heavy metals, salinity drought, cold, etc. Several biocontrol modes were reported by
Trichoderma spp., i.e., (i) the production of hydrolytic enzymes, antibiotics, and many bioactive metabolites, (ii) competition with the pathogen for nutrients, and space, (iii) mycoparasitism, and/or (iv) induction of defense-related genes and systemic resistance in plants [
4].
The bioactive metabolites, especially lytic enzymes squirted by
Trichoderma spp., hinder the growth of plants’ pathogens. For example, proteases, glucanases, and chitinases have been stated to hydrolyze the cell wall of the pathogen. Also, extracellular proteins produced by
Trichoderma spp. have a vital task in the inhibition of phytopathogens, as well as the enhancement of plants’ immune response [
5]. These modes of action may indirectly or directly suppress the phytopathogens’ growth and pathogenicity.
Furthermore, as bio-stimulants,
Trichoderma spp. improves plant health and productivity through the production of peptides, hormones, volatiles, and nonvolatile metabolites that enhance the growth, and rooting system [
6].
Trichoderma spp. induces the plant to produce defense-related enzymes, such as polyphenol oxidase and peroxidase, which improve defense response against fungal, viral, and bacterial plant pathogens [
7,
8,
9]. Ecologically, the presence of
Trichoderma spp. in soil improves soil microbial communities leading to improvement of plant growth and yield [
10].
Pea (
Pisum sativum L.) is grown in numerous regions in the world and is consumed by animals and humans. It is known as a low-cost, readily available source of protein, minerals, vitamins, and complex carbohydrates [
11]. The world cultivated area of pea reached 2.5 million ha, yielding 19.8 million tons [
12]. Pea is exposed to the attack by several fungal phytopathogens, causing significant decreases in quality and yield. For instance, the damping-off disease, caused by different abiotic or biotic stresses, leads to an obvious decline in seed germination and seedling emergence, or development. Plant pathogenic fungi have been stated as the most vital biotic stress.
Fusarium spp.,
Pythium spp.,
Phytophthora spp., and
Rhizoctonia spp. are the greatest commonly damping-off pathogens [
13]. Thus, this disease is one of the most significant yield constraints in fields and nurseries.
To the authors’ information, the existing pioneer work is the first state of domiciliation of a new Trichoderma sp., isolated from pea seeds, into pea plant tissue as a new habitat for the bioagent fungus. The growth and health-promoting action, as well as the biocontrol of pea damping-off disease, were investigated. Evidence of the domiciliation process of the endophytic fungus was followed up by ultrastructural and biochemical studies. Additional investigation on the changes in the rhizosphere microbial biodiversity was carried out.
4. Discussion
Owing to their biodiversity in various ecosystems, beneficial fungi must be considered when evolving plant protection strategies. Unfortunately, public awareness, mostly, neglected fungal biodiversity and its critical role in this respect. Therefore, there is a great need for investigating the diverse fungal communities for new fungal strains, having new roles, instead of the already available ones.
This study was carried out to draw attention to the possibility of domiciliating a new beneficial microbiome into the plant tissue. In this connection, a pioneer isolate of T. asperellum ZNW, an endophytic fungus isolated from pea seeds, was domiciliated into the pea plant, representing a case study for the settlement of the bioagent fungus into plant tissue.
Trichoderma spp. is a diverse group of wonderful bioagents widely used in sustainable agriculture owing to their ability to enhance plant growth and productivity, and further alleviate abiotic and biotic stresses [
37].
Trichoderma spp. are used in various remedies (foliar application, seed treatment, and/or soil remedy) for the management of diseases caused by bacteria, fungi, and nematodes [
38].
Cultural, morphological, and microscopic (light, and SEM) identification of fungal isolate ZNW declares that the current isolate ZNW resembles the morphology of
T. asperellum RMCK01 [
39]. The morphology of
T. asperellum ZNW growing on a PDA plate did not have the same morphology as observed in previous investigations, suggesting a new variant of
T. asperellum. The conidia of
T. asperellum ZNW are slightly globose in shape and the conidiophore is branched, which resembles
T. asperellum Ta13 [
40]. The morphological characters of
T. asperellum ZNW have followed the classification of
T. asperellum [
18]. This data supports the claim that myco-diversity needs more studies to explore the undiscovered area of seed-borne fungi.
Identification of the damping-off phytopathogen,
G. ultimum NZW, declared that its mycelia resemble those of
Pythium ultimum var.
ultimum associated with soybean damping-off [
41]. SEM micrographs of
G. ultimum NZW illustrated that its hyphae had a swelling end with a slightly globose shape, which was confirmed by other studies on
G. ultimum strains [
19,
42,
43]. All these works confirm the classification of the current fungus.
The pathogenicity of the current strain was confirmed by the pathogenicity test [
44]. However,
Globisporangium ultimum (Trow) (syn.
G. ultimum Trow, syn.
G. ultimum Trow var.
ultimum) is an oomycete species that causes damping-off and/or root rot on a vast range of plants throughout the world [
42,
43,
45].
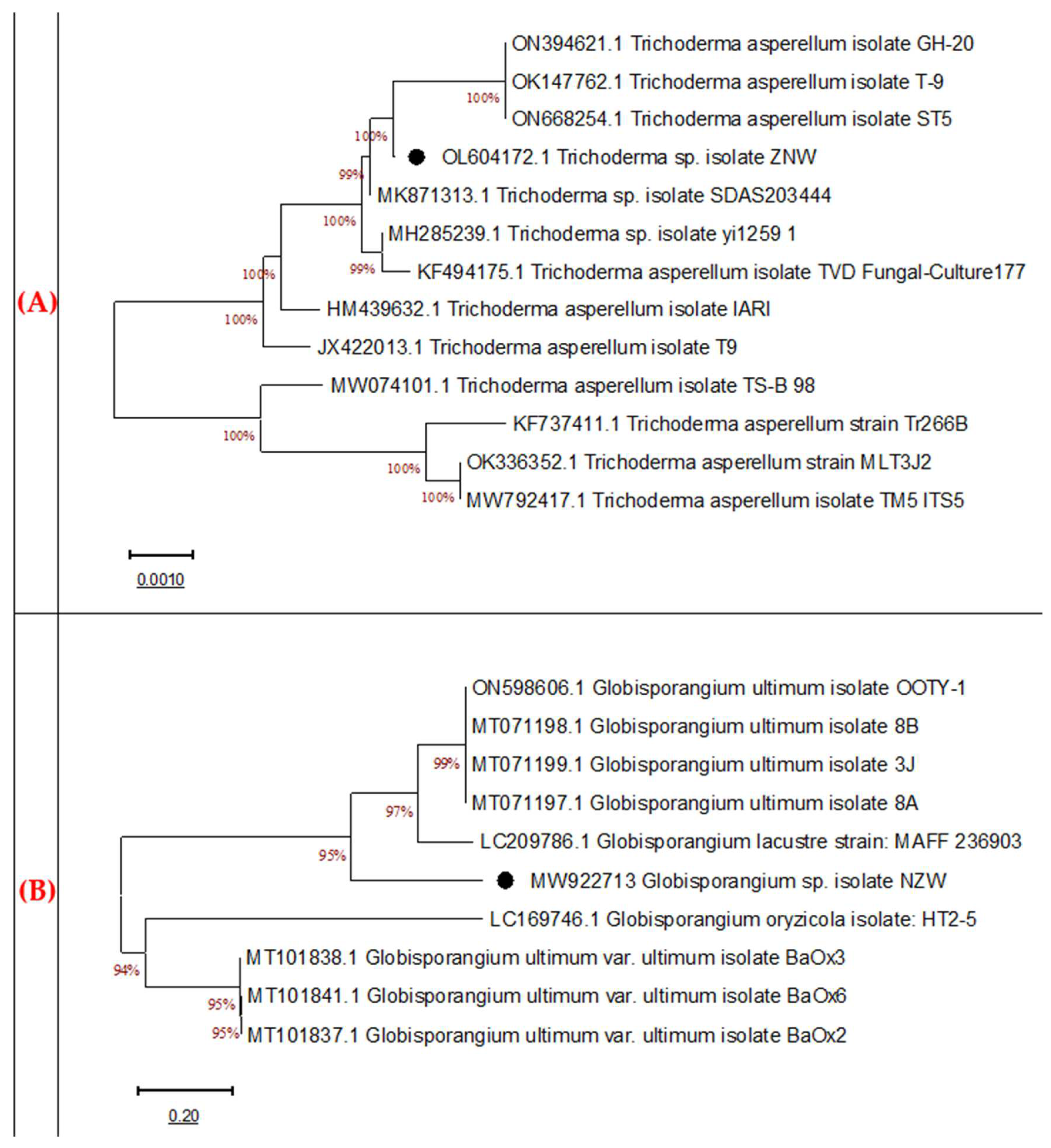
The previous identification of both fungi was further confirmed. Molecular identification is frequently used due to its high specificity and sensitivity for the quick identification of numerous microorganisms [
46]. The sequences of the tested fungi were related to similar strains in the GenBank. Both the bioagent and pathogenic fungi were identified as
T. asperellum ZNW (OL604172) and
G. ultimum NZW (MW922713), respectively, which came in line with the morphological identification features.
Molecular identification is highly specific and sensitive and is frequently used for fast identification. The nucleotide sequencing of the ITS region is similar in broad fungal classes, thus helping in revealing the interspecific and intraspecific differences among microorganisms. This region has non-functional sequences and is very variable among species, therefore, it can be accepted for the identification of very wide groups of fungal species. Technically, the multi-copy or repeated feature of the rDNA facilitates the amplification of the ITS regions from a tiny part of DNA, thus, providing fast and accurate identification compared to the other markers [
47,
48,
49].
SEM investigation of the dual culture showed the ability of
T. asperellum ZNW to mycoparasite and coil around
G. ultimum NZW, causing damage, fractionation, and breakdown of
G. ultimum NZW hyphae. Mycoparasitism is an effective method through which
Trichoderma spp. physically contact and coil their hyphae around the hyphae of the pathogenic fungi, then the bioagent obtains nutrients from the dead host biomass after hydrolysis by the secretion of lytic enzymes and other metabolites [
50,
51].
However, there is no antagonistic activity of
T. asperellum ZNW volatile organic compounds against
G. ultimum NZW. This result means that
T. asperellum ZNW may not produce volatile compounds or produce volatile compounds that did not inhibit
G. ultimum NZW, the latter assumption may be the more accepted one. Conversely to our results,
T. asperellum produced volatile compounds that had antifungal activity against other plant pathogens (
Curvularia aeria and
Corynespora cassiicola), enhanced the defense, and improved the growth of lettuce [
52]. Moreover,
T. asperellum T76-14 produced volatile organic compounds that inhibited
Fusarium incarnatum, which causes fruit rot of muskmelons [
53].
The biochemical feature of the bioagent was assessed as a possible role in the biological control process. Lytic activity of T. asperellum ZNW was searched on pea straw. This pea straw is hard to disintegrate due to its cellulose, hemicellulose, protein, and pectin content. Generally, such structures hinder fungal invasion into plant tissues. These enzymes are known as cell wall-degrading enzymes and play a role in the biocontrol process against the phytopathogen. Another suggested role is the facilitation of hyphal penetration to plant cell-wall tissue during the domiciliation process.
There was an obvious lytic activity of xylanase and cellulase that hydrolyzes hemicellulose (xylan) and cellulose, leading to the release of xylose and glucose units, respectively [
47]. Whereas pectinase and proteases degrade pectin polymer and protein into galacturonic acid [
47] and amino acids [
28], respectively. The breakdown of plant tissues by these enzymes eases microbial entrance into plants [
54], thus facilitating the domiciliation process.
Chitinase analyzes 1–4 β-glycoside linkage of N-acetyl-D-glucosamine in the fungal chitin; thus, acting as a bio-fungicide by antagonizing the other phytopathogens on host plants [
55]. The growth medium was free from the chitin substrate, demonstrating that chitinase is produced constitutively by the
Trichoderma asperellum ZNW.
Furthermore,
Trichoderma spp. can biosynthesize many metabolites, which have antimicrobial activity against various phytopathogens, induce disease resistance, and enhance plant growth [
56]. Besides many components, 9-octadecenoic acid (peak area-36.32%) was the main secondary metabolite detected in the filtrate of
T.
asperellum ZNW by GC-MS analysis. This component is presented in different
Trichoderma strains in other investigations at lower concentrations in
T.
asperellum (peak area-1.57%) [
57] and
T. hamatum (peak area-1.77%) that inhibited
Candida albicans [
58]. The current compound, 9-octadecenoic acid, was reported to be belonging to the unsaturated fatty acids that have antimicrobial activity [
59] and plays an obvious task in the plant’s defense against biotic and abiotic stresses [
60]. Another fatty acid (9-hexadecenoic acid or palmitic acid) was present in the filtrate (peak area-3.96%) of this investigation, which was previously detected in the filtrate of
T. hamatum [
58] with antimicrobial activity [
61]. Moreover, palmitic acid is found in the filtrate of
T.
asperellum and is also used for disease management and as a plant nutrient [
62]. The possible role of palmitic acid is the ability to alter the rhizosphere microbial structure, control fungal disease, and encourage plant growth [
63].
Oleamide, a TMS derivative (peak area-29.89%) present in the filtrate of
T. asperellum ZNW, was previously reported as an amide derivative of the fatty acid oleic acid [
64]. Fatty acids and their derivatives have vital roles in plant defense against bacterial and fungal pathogens [
65]. Also, the detected 1,2-15,16-diepoxyhexadecane (peak area-8.13%) in this study may have antioxidant, anti-cancer, and anti-inflammatory activities [
66]. Moreover, 1-Monopalmitin, 2TMS derivative (peak area-5.74%) detected in the filtrate of
T.
asperellum ZNW is a secondary metabolite in most active actinomycetes, having anticancer and antimicrobial activities [
67]. In general, the antimicrobial effect of
T. asperellum ZNW may be due to the synergism among the diverse chemicals, especially the main components, however, the synergistic effects between the main and the minor components exceed the expected individual outcome of each component alone [
54,
68].
Under greenhouse conditions, pea seeds were treated by the bioagent through biopriming with
T.
asperellum ZNW. Beneficial microbial inoculants are a vital method for improving the germination of seeds and/or enhancing growth, as well as the resistance of plants against several abiotic and biotic stresses. Moreover, seed biopriming permits the microorganisms, such as
Trichoderma spp., to adhere to and enter the tissues [
69,
70,
71].
Seed biopriming in
T. asperellum ZNW increased the survival percentage and decreased percentages of rotted seeds and infected seedlings in the absence or presence of
G. ultimum. The promotion of growth may be due to the action of the metabolites and the ability of fungal inoculation to reduce the disease (root rot and damping-off) and also to enhance the survival percentage of the plant [
72]. The use of
T. asperellum for biopriming of chili seeds reduced disease severity caused by
Fusarium solani and enhanced seedling health [
70].
T. asperellum ZNW enhanced the vegetative growth of pea plants that were planted in infested or non-infested soil due to its ability to promote plant growth, antagonize
G. ultimum NZW, and its ability to induce the defense-related enzymes, and improve the concentration of photosynthetic pigment. These improvements resemble those reported on the growth parameter of tomato due to
T. asperellum, which is used as a bioagent against damping-off disease [
73].
Ultrastructural alterations were noticed by SEM in the cross sections of the root of the pea plant.
T.
asperellum ZNW could domiciliate and grow as an endophytic fungus in pea roots and lead to the thickened cell wall of vessels (xylem). Moreover,
T.
asperellum ZNW prevented
G. ultimum NZW from invading the region of the root, in which
T. asperellum ZNW is present. Furthermore, no traces of the pathogen could be observed in the region of
T. asperellum ZNW. According to the authors’ information, this is the first trial that indicates the ability of domiciliation of
Trichoderma spp. into plant tissue so that it can grow as an endophytic fungus. The growth of
T.
asperellum ZNW as an endophytic fungus gave a high opportunity for plant protection from phytopathogens and allow the plant to get several profits from
T.
asperellum ZNW metabolites.
T.
asperellum ZNW led to an increase in thickness of the xylem of pea roots, this may be due to the lignification, i.e., lignin accumulation in the cell wall of the root that is accompanied by low methyl-esterified pectin in intercellular spaces, which increases in cell wall thickness improved wheat protection against pathogens [
74].
Additionally, the enzymatic system of T. asperellum ZNW enables partial maceration of plant tissue that allows the smooth penetration, and domiciliation of T. asperellum ZNW into plant tissue without causing any pathogenic symptoms. Generally, Trichoderma spp. do not have any virulence factor and, consequently, no disease development occurs.
T. asperellum ZNW enhanced the induced resistance and increased the production of total phenols and improved the activities of peroxidase and polyphenol oxidase in pea planted. This, in turn, leads to an increase in defense response against viral, bacterial, and fungal pathogens [
7,
8,
9]. Phenolic compounds have significant roles in plant health through increasing colonization of beneficial microorganisms and inhibition of insect attack and/or pathogen infection [
75]. Phenolic compounds have antioxidant and antimicrobial properties and protect the plant tissues from poisonous effects, resulting from reactive oxygen species [
76]. Under adverse conditions, phenolics accumulate in the plant tissues and play a vital role in the regulation of different environmental stresses, such as nutrient deficiency, low temperatures, and high light [
77]. Peroxidase and polyphenol oxidase are defense-related enzymes that enhance the defense against phytopathogens infections and improve plant resistance against biotic and abiotic stress [
78].
T. asperellum ZNW enhanced chlorophylls and carotenoid content in pea plants.
T. asperellum could enhance the pea plant’s health and enhance the activity of the photosynthesis process [
79]. The improvement of chlorophyll concentration refers to the enhancement of the plant’s defense against abiotic and biotic stresses [
80].
T. asperellum enhanced chlorophylls and carotenoids content when used as biocontrol of plant pathogens [
81]. Furthermore,
T. asperellum enhanced the concentration of photosynthetic pigments and helped to overcome drought stress [
82].
The changes in the biological features in the rhizosphere soil of pea were explored. The activity of the soil dehydrogenase was enhanced due to the application of
T. asperellum.
Trichoderma spp. was reported to improve the activity of dehydrogenase and positively amend the soil properties [
83]. Dehydrogenase is one of the most important soil enzymes, which is an indicator of soil quality due to different stress, or management practices. Enzyme activity in the soil can be changed as a result of the activities of the soil’s microbes [
84]. Since dehydrogenases are present intracellularly in the living microbial cells and do not accumulate out of the microbial cells, it is considered an indicator of the microbial activity in the soil; furthermore, dehydrogenases play a vital role in the biological oxidation of the organic matter in the soil by transferring hydrogen from organic substrates to inorganic acceptors [
85]. The enhancement of dehydrogenase is a good sign of the improvement of microbial population and physicochemical soil properties [
86,
87].
T. asperellum ZNW led to an alteration in the bacterial and fungal communities in the rhizosphere, where both microbial counts decreased greater after 90 days than after 60 days. This effect of
T. asperellum ZNW on the rhizosphere microbial count may be directly and/or indirectly related to plant physiology, and root exudates in soil. This effect may be due to the competition of
Trichoderma with other microbes, leading to an alteration in the population of the soil microbiome. Also, the activity of soil enzymes may have a vital role in changing the microbial community [
88]. The relative abundance of
Trichoderma spp. in the soil causes increases in the availability of soil nutrients, and changes the rhizosphere chemical structure, thus altering soil microbial communities [
10].
T. asperellum causes an increase in the total count of beneficial microbes and a decrease in the total count of phytopathogens in the plants’ rhizosphere that help control plant diseases and promote growth [
89].
Trichoderma spp. can boost the physiology of plants and can modulate root exudates that cause a change in nature and kind of the plant-microbe interaction [
90]. Root exudates are secretions produced by roots that are considered a basis of nutrition for beneficial soil microbial communities [
91]. Root exudates enhance soil respiration and positively change the configuration of the microbial community, which effectively enhances the growth of plants [
92]. The root exudates decrease near the end of plant age leading to decreasing in the microbial count of rhizosphere microbial communities [
93].
The biopriming of pea seeds in
T. asperellum ZNW led to an increase in the productivity of pea plants and yield parameters. This is an expected outcome due to the enhancement of plant growth and physiology, as well as the management of phytopathogen, leading to an enhancement in induced resistance and also an enhancement of the rhizosphere microbial community.
T. asperellum could promote different plants and control diseases, leading to an increase in yield, as occurs when controlling late-wilt disease, promoting growth, and increasing the yield of maize [
94]. Furthermore,
T. asperellum could reduce
Pythium root rot, promote growth, and improve the productivity of lettuce [
95].