Growth and Volatile Organic Compound Production of Pseudomonas Fish Spoiler Strains on Fish Juice Agar Model Substrate at Different Temperatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Bacterial Strains
2.2. Preparation, Inoculation and Storage of Model Substrate
2.3. Enumeration of Bacterial Population
2.4. Determination of Volatile Organic Compounds Profile
2.5. Modeling and Statistical Analysis
3. Results
3.1. Microbial Populations and Kinetics
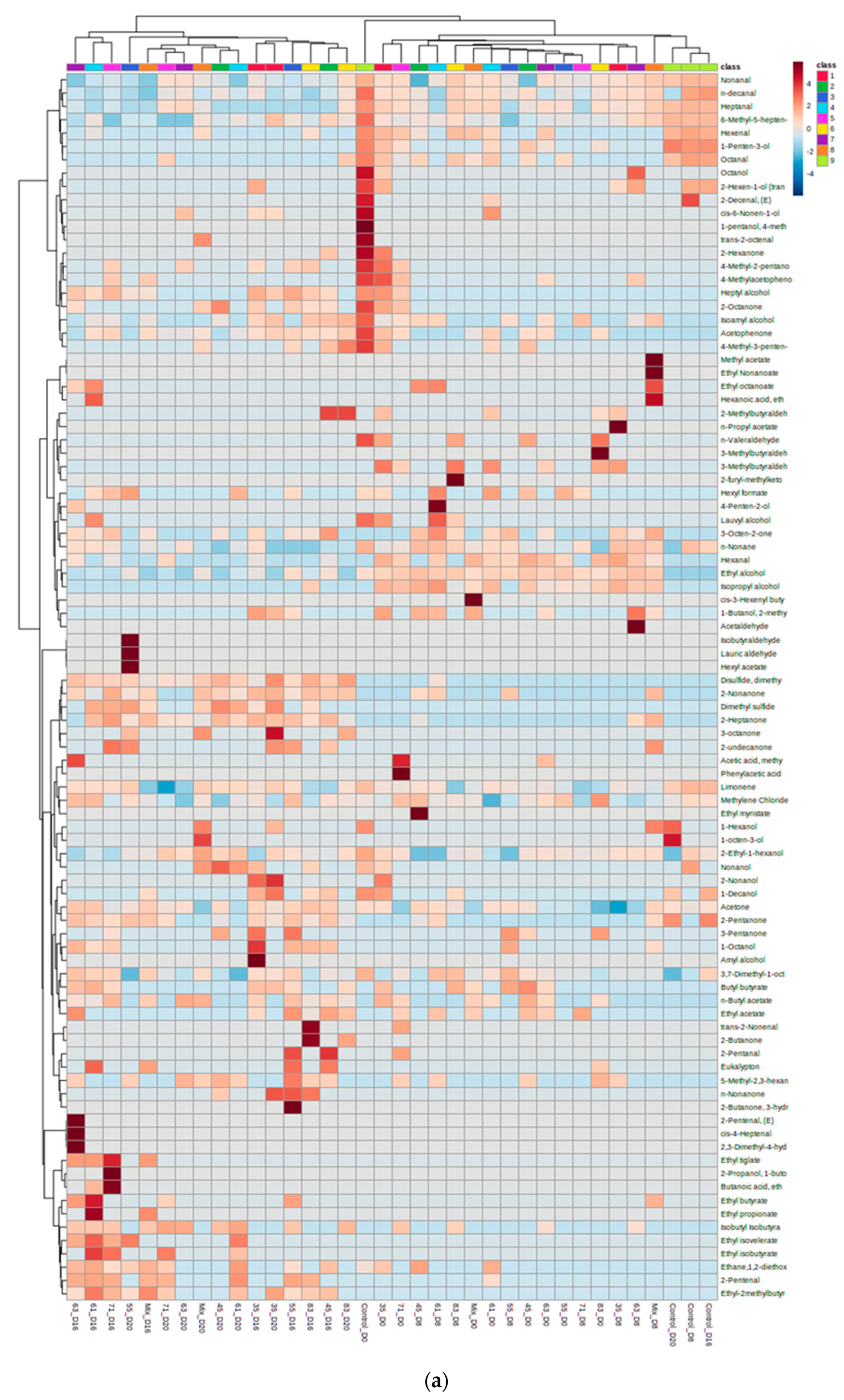
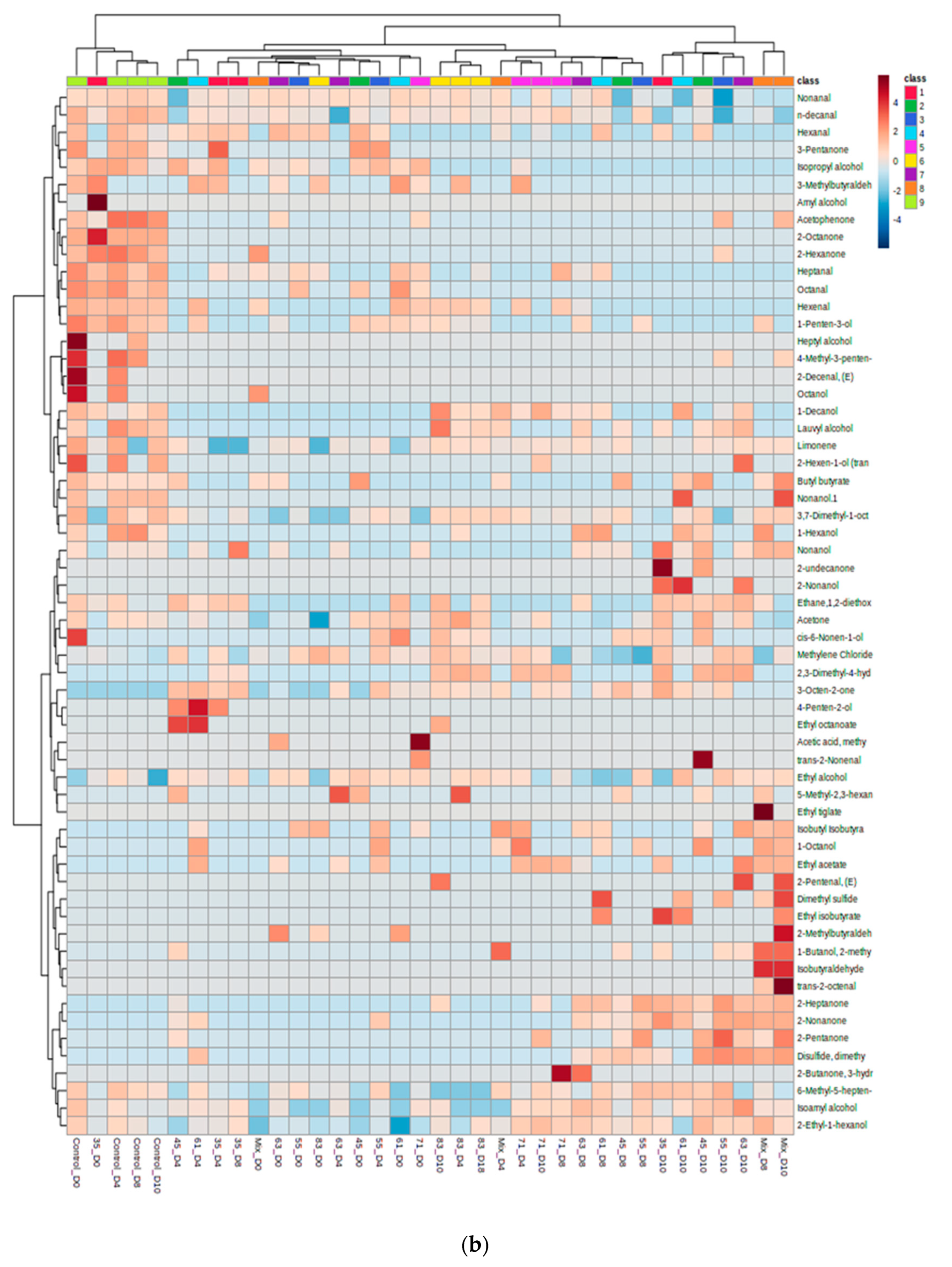
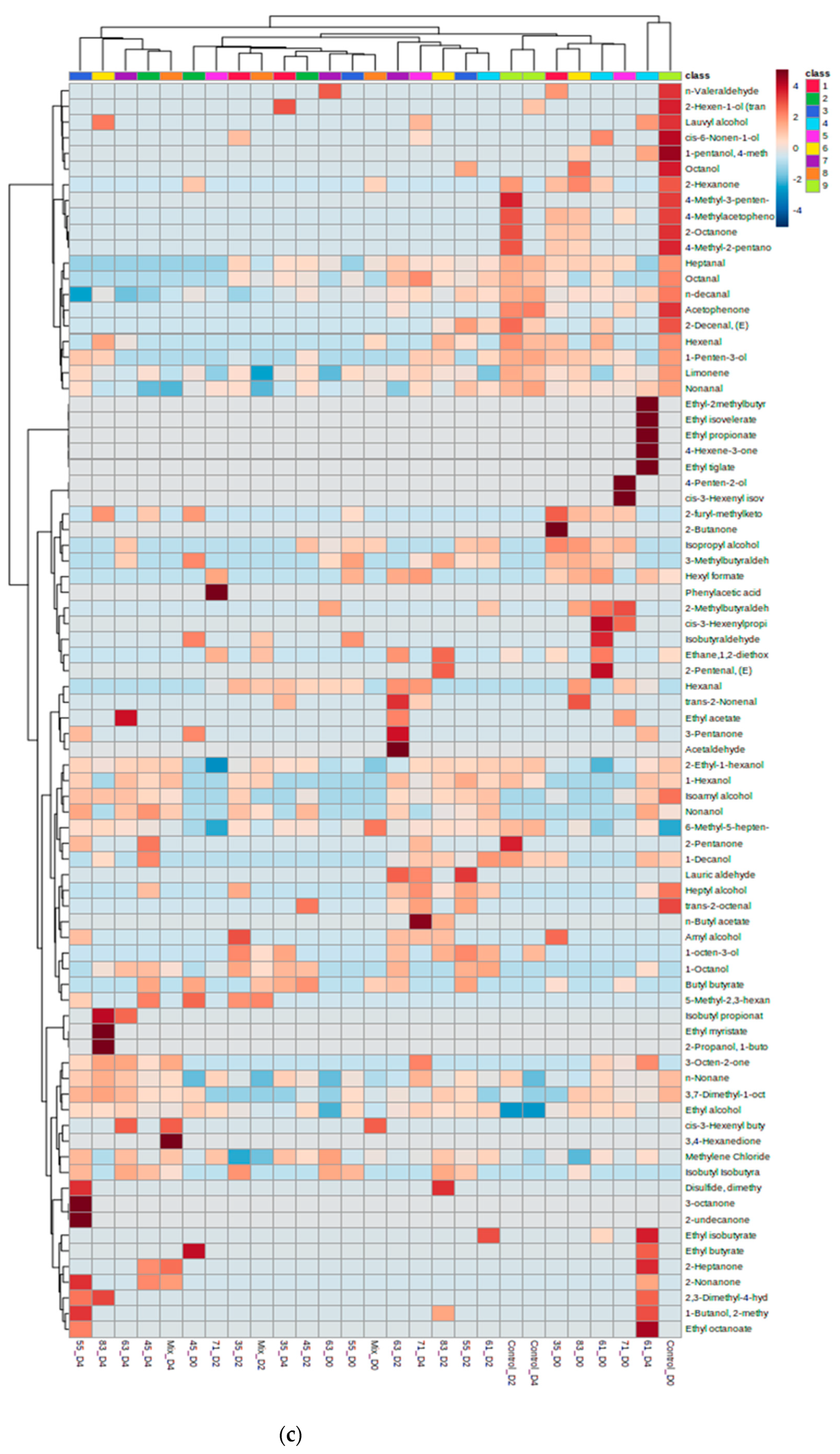
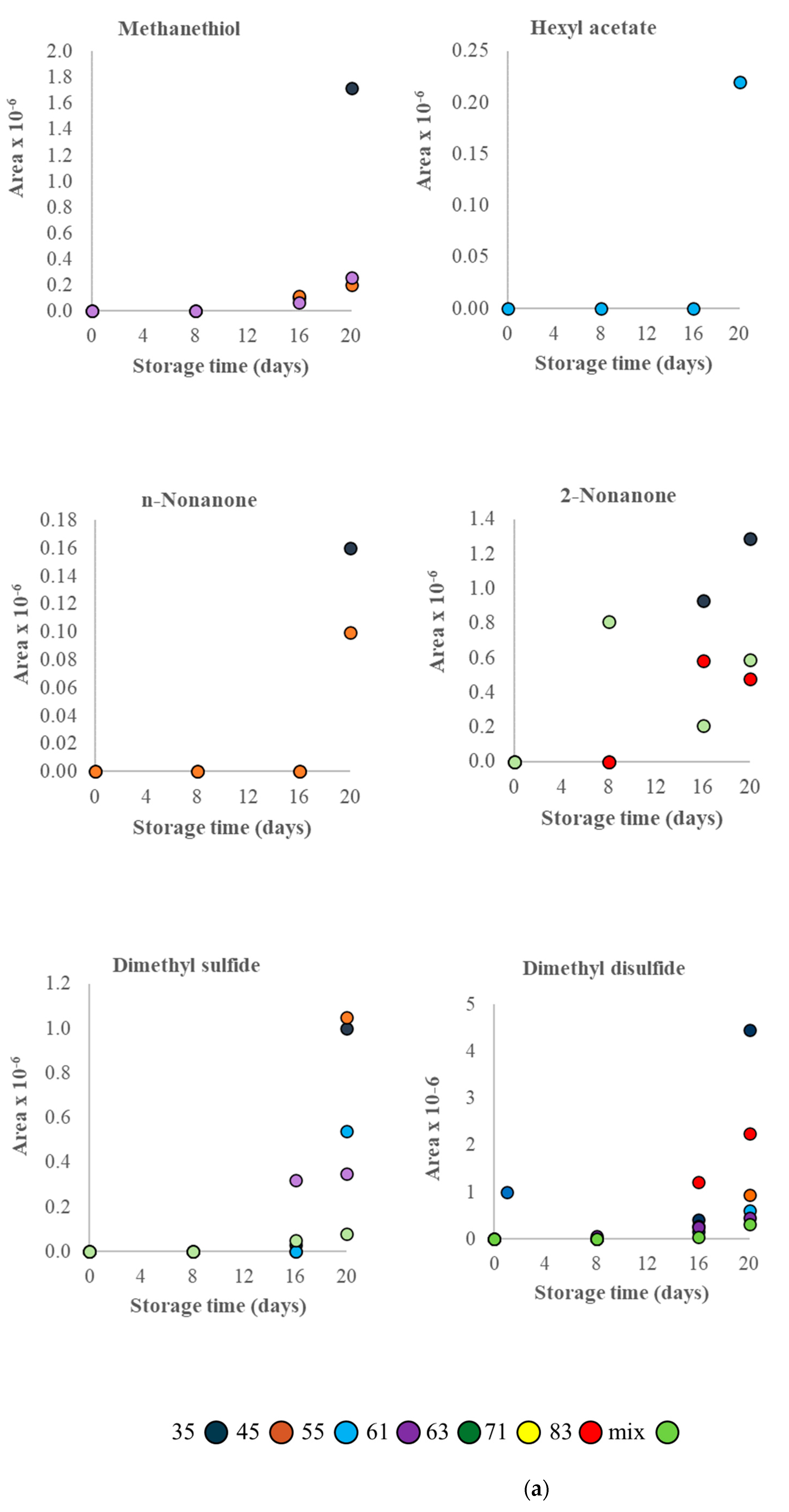
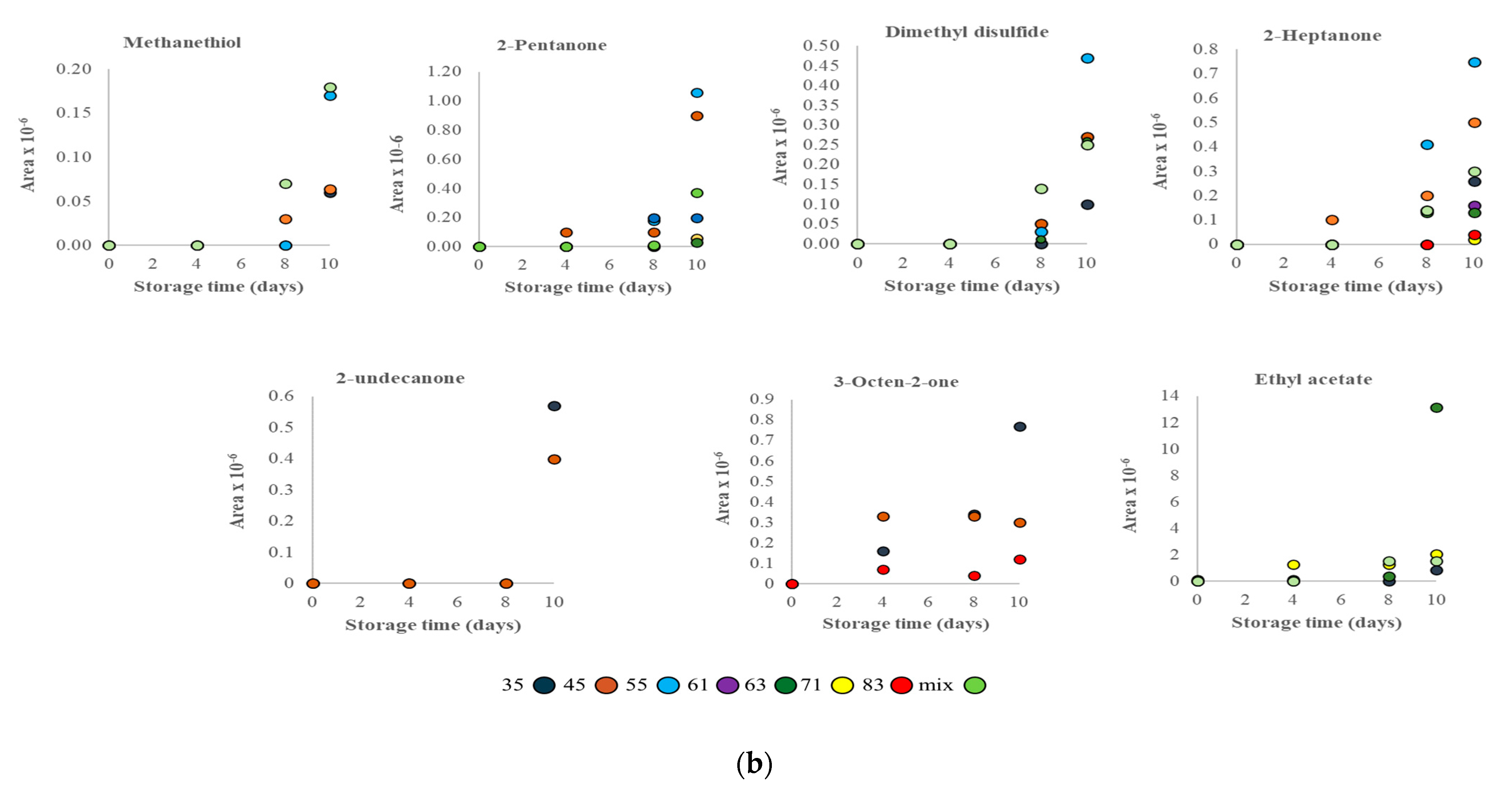
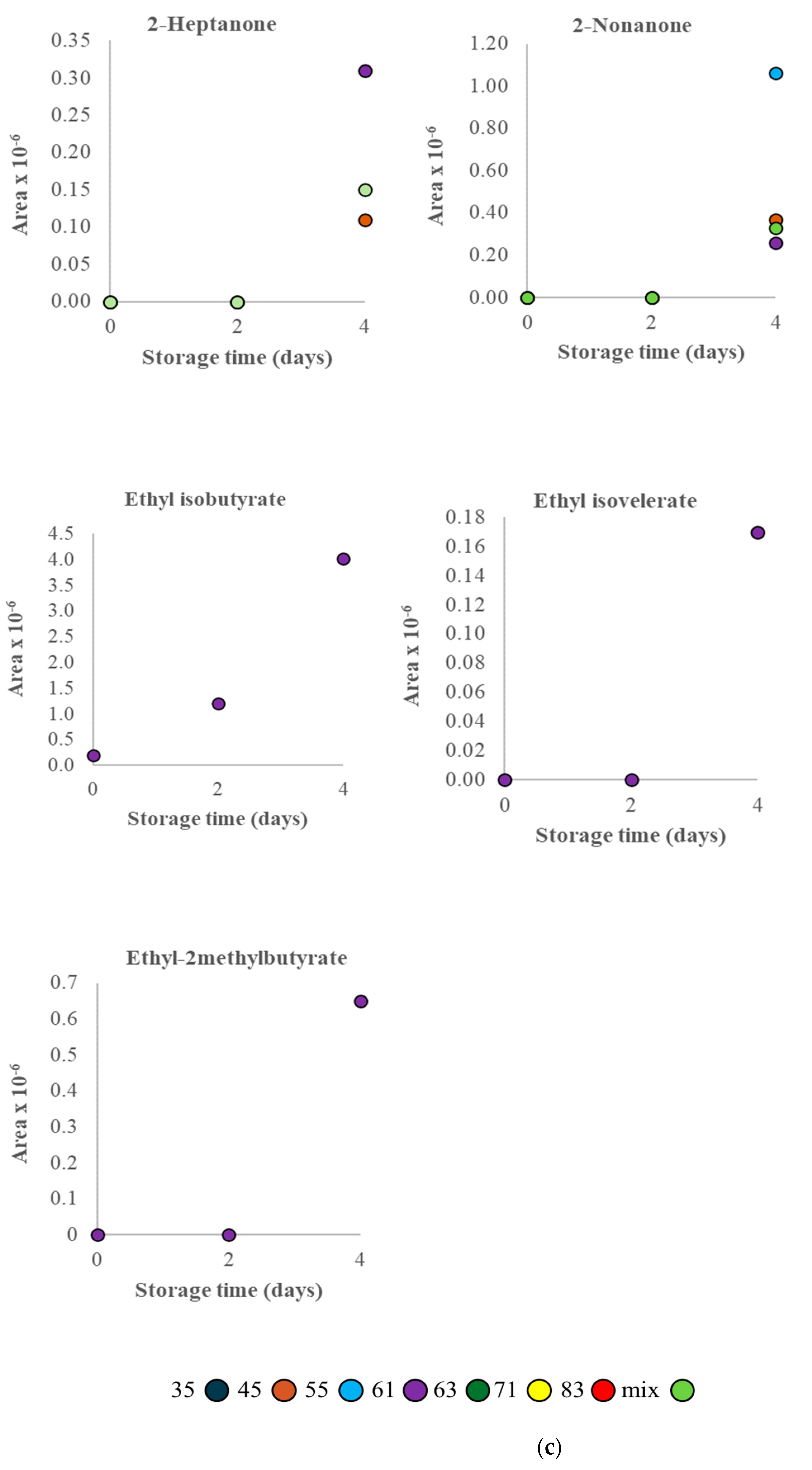
3.2. VOC Profile and Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gram, L.; Huss, H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Boziaris, I.S.; Parlapani, F.F. Specific Spoilage Organisms (SSOs) in Fish. In Microbiological Quality of Food; Woodhead Publishing: Sawston, UK, 2017; pp. 61–98. [Google Scholar] [CrossRef]
- Gram, L.; Dalgaard, P. Fish spoilage bacteria—Problems and solutions. Curr. Opin. Biotechnol. 2002, 13, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Nychas, G.J.E.; Marshall, D.L.; Sofos, J.N. Food microbiology: Fundamentals and frontiers. In Meat, Poultry and Seafood; CABI: Wallingford, UK, 2007; pp. 105–140. [Google Scholar]
- Parlapani, F.F.; Boziaris, I.S. Monitoring of spoilage and determination of microbial communities based on 16S rRNA gene sequence analysis of whole sea bream stored at various temperatures. LWT 2016, 66, 553–559. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Verdos, G.I.; Haroutounian, S.A.; Boziaris, I.S. The dynamics of Pseudomonas and volatilome during the spoilage of gutted sea bream stored at 2 °C. Food Control 2015, 55, 257–265. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Haroutounian, S.A.; Nychas, G.J.E.; Boziaris, I.S. Microbiological spoilage and volatiles production of gutted European sea bass stored under air and commercial modified atmosphere package at 2 °C. Food Microbiol. 2015, 50, 44–53. [Google Scholar] [CrossRef]
- Syropoulou, F.; Parlapani, F.F.; Kakasis, S.; Nychas, G.J.E.; Boziaris, I.S. Primary Processing and Storage Affect the Dominant Microbiota of Fresh and Chill-Stored Sea Bass Products. Foods 2021, 10, 671. [Google Scholar] [CrossRef]
- Tryfinopoulou, P.; Tsakalidou, E.; Nychas, G.J.E. Characterization of Pseudomonas spp. associated with spoilage of gilt-head sea bream stored under various conditions. Appl. Environ. Microbiol. 2002, 68, 65–72. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Pasentsis, K.; Argiriou, A.; Krey, G.; Boziaris, I.S. A meta-barcoding approach to assess and compare the storage temperature-dependent bacterial diversity of gilt-head sea bream (Sparus aurata) originating from fish farms from two geographically distinct areas of Greece. Int. J. Food Microbiol. 2018, 278, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Syropoulou, F.; Parlapani, F.F.; Anagnostopoulos, D.A.; Stamatiou, A.; Mallouchos, A.; Boziaris, I.S. Spoilage Investigation of Chill Stored Meagre (Argyrosomus regius) Using Modern Microbiological and Analytical Techniques. Foods 2021, 10, 3109. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ianniello, R.G.; De Filippis, F.; Ricciardi, A. Dynamics of bacterial communities and interaction networks in thawed fish fillets during chilled storage in air. Int. J. Food Microbiol. 2019, 293, 102–113. [Google Scholar] [CrossRef]
- Nychas, G.J.E.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Jay, J.M. Microbial spoilage indicators and metabolites. In Food-Borne Microorganisms and Their Toxins: Developing Methodology; Pierson, M.D., Sterm, N.J., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 1986; pp. 219–240. [Google Scholar]
- Parlapani, F.F.; Mallouchos, A.; Haroutounian, S.A.; Boziaris, I.S. Volatile organic compounds of microbial and non-microbial origin produced on model fish substrate un-inoculated and inoculated with gilt-head sea bream spoilage bacteria. LWT 2017, 78, 54–62. [Google Scholar] [CrossRef]
- Chinivasagam, H.N.; Bremner, H.A.; Thrower, S.J.; Nottingham, S.M. Spoilage pattern of five species of australian prawns: Deterioration is influenced by environment of capture and mode of storage. J. Aquat. Food Prod. Technol. 1996, 5, 25–50. [Google Scholar] [CrossRef]
- Miller, A.I.; Scanlan, R.A.; Lee, J.S.; Libbey, L.M. Volatile Compounds Produced in Sterile Fish Muscle (Sebastes melanops) by Pseudomonas putrefaciens, Pseudomonas fluorescens, and an Achromobacter Species. Appl. Microbiol. 1973, 26, 18–21. [Google Scholar] [CrossRef]
- Miller, A.I.; Scanlan, R.A.; Lee, J.S.; Libbey, L.M. Identification of the Volatile Compounds Produced in Sterile Fish Muscle (Sebastes melanops) by Pseudomonas fragi. Appl. Microbiol. 1973, 25, 952–955. [Google Scholar] [CrossRef]
- Miller, A.I.; Scanlan, R.A.; Lee, J.S.; Libbey, L.M.; Morgan, M.E. Volatile Compounds Produced in Sterile Fish Muscle (Sebastes melanops) by Pseudomonas perolens. Appl. Microbiol. 1973, 25, 257–261. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Kormas, K.A.; Boziaris, I.S. Microbiological changes, shelf life and identification of initial and spoilage microbiota of sea bream fillets stored under various conditions using 16S rRNA gene analysis. J. Sci. Food Agric. 2015, 95, 2386–2394. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Mallouchos, A.; Haroutounian, S.A.; Boziaris, I.S. Microbiological spoilage and investigation of volatile profile during storage of sea bream fillets under various conditions. Int. J. Food Microbiol. 2014, 189, 153–163. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Koutsoumanis, K. Predictive Modeling of the Shelf Life of Fish under Nonisothermal Conditions. Appl. Environ. Microbiol. 2001, 67, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Dalgaard, P.; Gram, L.; Huss, H.H. Spoilage and shelf-life of cod fillets packed in vacuum or modified atmospheres. Int. J. Food Microbiol. 1993, 19, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Ravn, L.; Rasch, M.; Bruhn, J.B.; Christensen, A.B.; Givskov, M. Food spoilage—Interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002, 78, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Chinivasagam, H.N.; Bremner, H.A.; Wood, A.F.; Nottingham, S.M. Volatile components associated with bacterial spoilage of tropical prawns. Int. J. Food Microbiol. 1998, 42, 45–55. [Google Scholar] [CrossRef]
- Laursen, B.G.; Leisner, J.J.; Dalgaard, P. Carnobacterium Species: Effect of metabolic activity and interaction with Brochothrix thermosphacta on sensory characteristics of modified atmosphere packed shrimp. J. Agric. Food Chem. 2006, 54, 3604–3611. [Google Scholar] [CrossRef]
- Duflos, G.; Coin, V.M.; Cornu, M.; Antinelli, J.F.; Malle, P. Determination of volatile compounds to characterize fish spoilage using headspace/mass spectrometry and solid-phase microextraction/gas chromatography/mass spectrometry. J. Sci. Food Agric. 2006, 86, 600–611. [Google Scholar] [CrossRef]
- Iglesias, J.; Medina, I. Solid-phase microextraction method for the determination of volatile compounds associated to oxidation of fish muscle. J. Chromatogr. A 2008, 1192, 9–16. [Google Scholar] [CrossRef]
- Selli, S.; Cayhan, G.G. Analysis of volatile compounds of wild gilthead sea bream (Sparus aurata) by simultaneous distillation–extraction (SDE) and GC–MS. Microchem. J. 2009, 93, 232–235. [Google Scholar] [CrossRef]
| Strain | GenBank Accession Number | Strain | Origin | Reference |
|---|---|---|---|---|
| 35 | KJ411280 | Bacterium SBF-B5-d0 (Pseudomonas fluorescens) | Sea bream fillets stored at 0 °C | [20] |
| 45 | KJ411285 | Bacterium SBF-B19-dsp (Pseudomonas fragi) | Sea bream fillets stored at 0 °C | [20] |
| 55 | KJ411284 | Bacterium SBF-B18-dsp (Pseudomonas fragi) | Sea bream fillets stored at 0 °C | [20] |
| 61 | KJ411289 | Bacterium SBF-A12-dsp (Pseudomonas fragi) | Sea bream fillets stored at 5 °C | [20] |
| 63 | KR778800 | Pseudomonas fragi strain b67 | Whole sea bream stored at 0 and 5 °C | [5] |
| 71 | KR778803 | Pseudomonas fluorescens strain b84 | Whole sea bream stored at 5 and 15 °C | [5] |
| 83 | KP010026 | Pseudomonas sp. GSB-b11 | Gutted sea bream stored at 2 °C | [6] |
| T °C | Culture | y0 (log cfu/g) | ymax (log cfu/g) | lag (days) | μmax (day −1) |
|---|---|---|---|---|---|
| 0 °C | 35 | 2.85 ± 0.18 (2.78 ± 0.22) | 9.52 ± 0.05 e (9.49 ± 0.09) | 0.00 | 0.515 ± 0.015 b |
| 45 | 2.86 ± 0.65 (2.86 ± 0.83) | 9.40 ± 0.07 c,d,e (9.44 ± 0.13) | 2.22 ± 0.80 | 0.755 ± 0.070 c | |
| 55 | 2.83 ± 0.12 (2.90 ± 0.00) | 9.46 ± 0.08 d.e (9.51 ± 0.07) | 1.21 ± 0.47 | 0.650 ± 0.087 b,c | |
| 61 | 2.72 ± 0.12 (2.85 ± 0.17) | 9.24 ± 0.10 b,c (9.34 ± 0.08) | 1.54 ± 0.02 | 0.572 ± 0.065 b,c | |
| 71 | 3.01 ± 0.14 (2.90 ± 0.05) | 9.11 ± 0.02 b (9.12 ± 0.06) | 1.50 ± 0.98 | 0.740 ± 0.155 c | |
| 83 | 2.66 ± 0.29 (2.79 ± 0.20) | 9.29 ± 0.03 b,c,d (9.23 ± 0.08) | 0.76 ± 0.00 | 0.484 ± 0.044 b | |
| 63 | 2.88 ± 0.20 (2.79 ± 0.26) | 9.22 ± 0.09 b,c (9.25 ± 0.22) | 3.65 ± 0.13 | 0.775 ± 0.010 c | |
| Mix | 2.89 ± 0.28 (2.96 ± 0.23) | 9.13 ± 0.05 b (9.19 ± 0.04) | 1.41 ± 0.15 | 0.641 ± 0.071 b,c | |
| 4 °C | 35 | 3.04 ± 0.13 (2.79 ± 0.22) | 9.32 ± 0.09 (9.27 ± 0.12) | 1.33 ± 0.09 | 1.083 ± 0.038 e |
| 45 | 2.89 ± 0.42 (2.86 ± 0.34) | 9.40 ± 0.13 (9.41 ± 0.15) | 0.40 ± 0.70 | 0.914 ± 0.060 c,d | |
| 55 | 2.82 ± 0.06 (2.90 ± 0.01) | 9.57 ± 0.13 (9.47 ± 0.20) | 0.0 | 0.896 ± 0.048 b,c | |
| 61 | 2.85 ± 0.22 (2.85 ± 0.17) | 9.28 ± 012 (9.32 ± 0.01) | 0.67 ± 0.69 | 0.985 ± 0.158 d | |
| 71 | 2.79 ± 0.12 (2.90 ± 0.05) | 8.76 ± 0.33 (8.73 ± 0.19) | 0.11 ± 0.19 | 0.967 ± 0.055 d | |
| 83 | 2.68 ± 0.09 (2.79 ± 0.21) | 9.46 ± 0.26 (9.24 ± 0.21) | 0 | 0.733 ± 0.044 b | |
| 63 | 2.89 ± 0.36 (2.79 ± 0.26) | 9.08 ± 0.18 (9.21 ± 0.12) | 1.10 ± 0.47 | 1.208 ± 0.081 e,f | |
| Mix | 2.92 ± 0.42 (2.96 ± 0.24) | 9.55 ± 0.07 (9.54 ± 0.17) | 0.93 ± 0.46 | 1.040 ± 0.044 e | |
| 8 °C | 35 | 2.93 ± 0.27 (2.78 ± 0.22) | 8.33 ± 0.32 b (8.50 ± 0.44) | 1.22 ± 0.23 | 1.434 ± 0.110 b,c |
| 45 | 2.93 ± 0.95 (2.86 ± 033) | 8.63 ± 0.18 b,c (8.90 ± 0.03) | 0.59 ± 0.19 | 1.872 ± 0.211 c | |
| 55 | 2.85 ± 017 (2.90 ± 0.00) | 8.69 ± 026 c (8.83 ± 010) | 0.74 ± 0.00 | 1.653 ± 0.317 b,c | |
| 61 | 2.84 ± 0.40 (2.85 ± 0.17) | 9.16 ± 0.16 c,d (9.03 ± 0.08) | 0.65 ± 0.53 | 1.492 ± 0.213 b,c | |
| 71 | 2.83 ± 0.04 (2.90 ± 0.05) | 9.07 ± 0.22 c,d (9.00 ± 0.08) | 0 | 1.409 ± 0.012 b | |
| 83 | 2.79 ± 0.20 (2.79 ± 0.20) | 9.22 ± 0.29 c,d (9.06 ± 0.02) | 0.46 ± 0.35 | 1.374 ± 0.140 a,b | |
| 63 | 2.73 ± 0.38 (2.79 ± 026) | 8.10 ± 0.21 b (8.22 ± 0.21) | 0.60 ± 0.21 | 1.473 ± 0.034 b,c | |
| Mix | 2.88 ± 0.29 (2.96 ± 0.23) | 9.34 ± 0.19 d (9.14 ± 0.02) | 0.46 ± 0.00 | 1.435 ± 0.056 b,c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parlapani, F.F.; Anagnostopoulos, D.A.; Karamani, E.; Mallouchos, A.; Haroutounian, S.A.; Boziaris, I.S. Growth and Volatile Organic Compound Production of Pseudomonas Fish Spoiler Strains on Fish Juice Agar Model Substrate at Different Temperatures. Microorganisms 2023, 11, 189. https://doi.org/10.3390/microorganisms11010189
Parlapani FF, Anagnostopoulos DA, Karamani E, Mallouchos A, Haroutounian SA, Boziaris IS. Growth and Volatile Organic Compound Production of Pseudomonas Fish Spoiler Strains on Fish Juice Agar Model Substrate at Different Temperatures. Microorganisms. 2023; 11(1):189. https://doi.org/10.3390/microorganisms11010189
Chicago/Turabian StyleParlapani, Foteini F., Dimitrios A. Anagnostopoulos, Evangelia Karamani, Athanasios Mallouchos, Serkos A. Haroutounian, and Ioannis S. Boziaris. 2023. "Growth and Volatile Organic Compound Production of Pseudomonas Fish Spoiler Strains on Fish Juice Agar Model Substrate at Different Temperatures" Microorganisms 11, no. 1: 189. https://doi.org/10.3390/microorganisms11010189
APA StyleParlapani, F. F., Anagnostopoulos, D. A., Karamani, E., Mallouchos, A., Haroutounian, S. A., & Boziaris, I. S. (2023). Growth and Volatile Organic Compound Production of Pseudomonas Fish Spoiler Strains on Fish Juice Agar Model Substrate at Different Temperatures. Microorganisms, 11(1), 189. https://doi.org/10.3390/microorganisms11010189










