Microbacterium Cluster EA Bacteriophages: Phylogenomic Relationships and Host Range Predictions
Abstract
1. Introduction
2. Materials and Methods
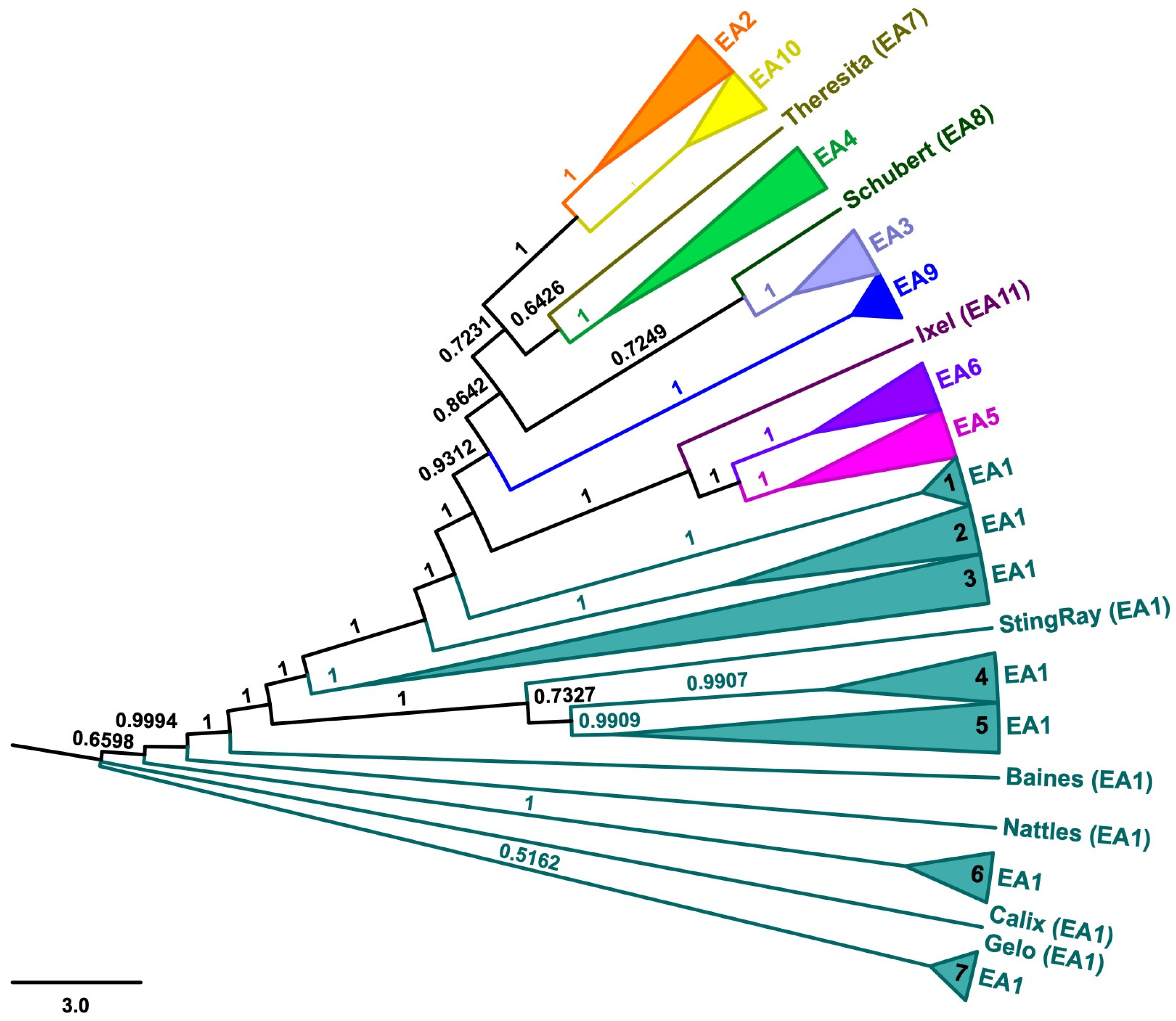
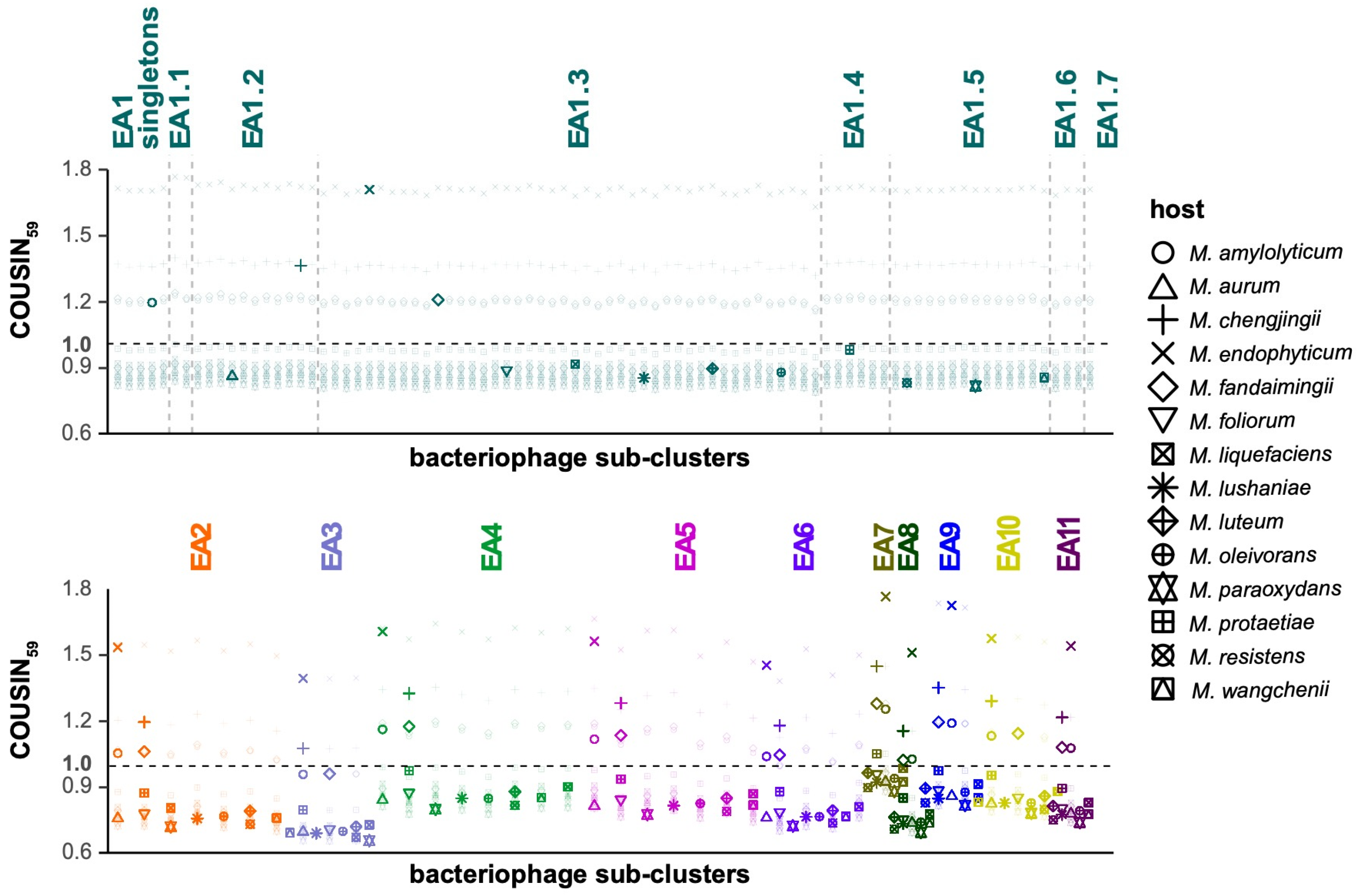
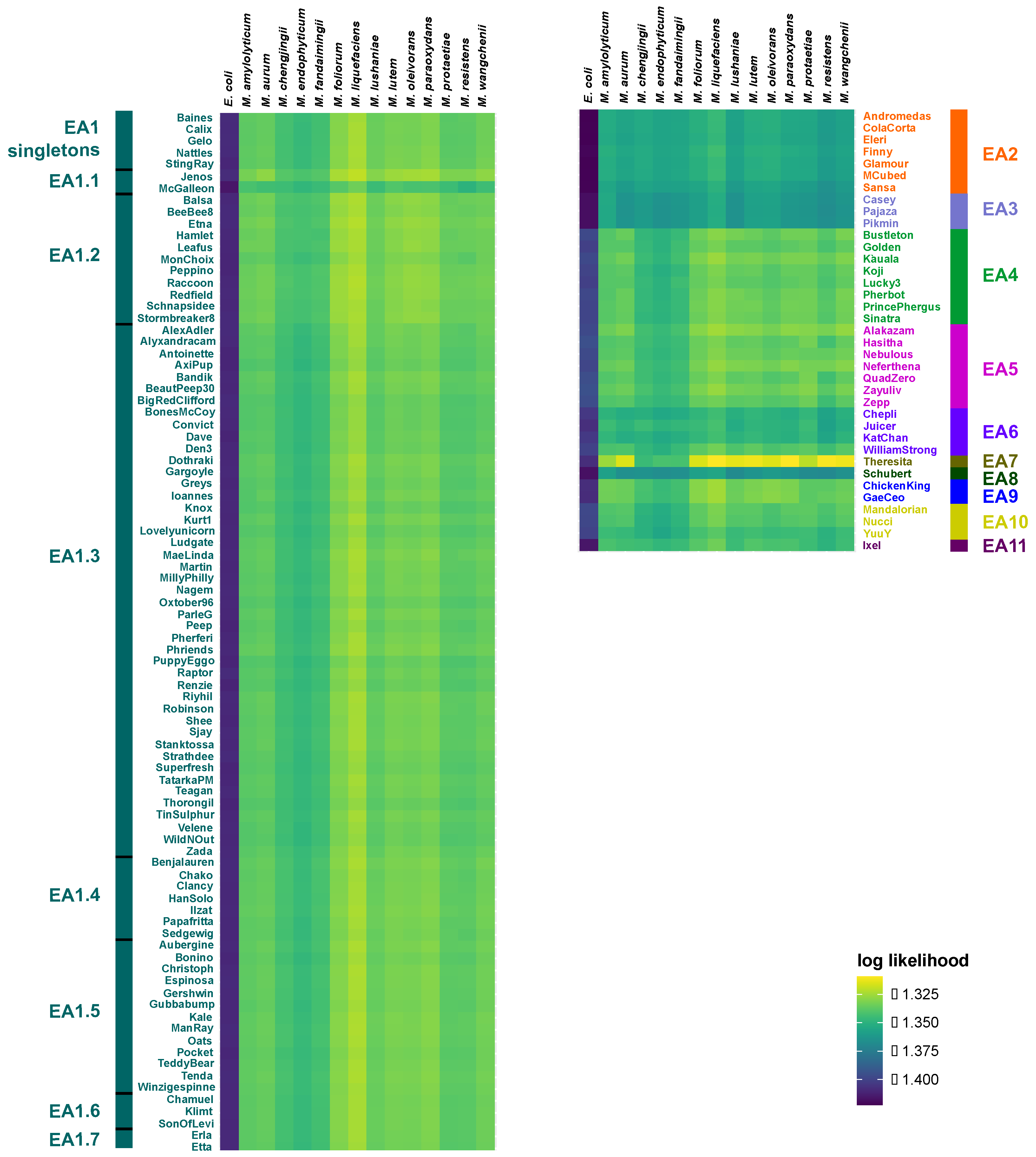
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Monk, A.B.; Rees, C.D.; Barrow, P.; Hagens, S.; Harper, D.R. Bacteriophage applications: Where are we now? Lett. Appl. Microbiol. 2010, 51, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Dyson, Z.A.; Tucci, J.; Seviour, R.J.; Petrovski, S. Lysis to Kill: Evaluation of the lytic abilities, and genomics of nine bacteriophages infective for Gordonia spp. and their potential use in activated sludge foam biocontrol. PLoS ONE 2015, 10, e0134512. [Google Scholar] [CrossRef]
- Heller, D.; Sivanathan, V. Publishing student-led discoveries in genetics. G3 Genes Genomes Genet. 2022, 12, jkac141. [Google Scholar] [CrossRef]
- Gneiding, K.; Frodl, R.; Funke, G. Identities of Microbacterium spp. encountered in human clinical specimens. J. Clin. Microbiol. 2008, 46, 3646–3652. [Google Scholar] [CrossRef]
- McLean, R.A.; Sulzbacher, W.L. Microbacterium thermosphactum, spec: nov.; a nonheat resistant bacterium from fresh pork sausage. J. Bacteriol. 1953, 65, 428–433. [Google Scholar] [CrossRef]
- Liu, J.; Nakayama, T.; Hemmi, H.; Asano, Y.; Tsuruoka, N.; Shimomura, K.; Nishijima, M.; Nishino, T. Microbacterium natoriense sp. nov., a novel d-aminoacylase-producing bacterium isolated from soil in natori, Japan. Int. J. Syst. Evol. Microbiol. 2005, 55, 661–665. [Google Scholar] [CrossRef]
- Behrendt, U.; Ulrich, A.; Schumann, P. Description of Microbacterium foliorum sp. nov. and Microbacterium phyllosphaerae sp. nov., isolated from the phyllosphere of grasses and the surface litter after mulching the sward, and reclassification of Aureobacterium resistens (Funke et al. 1998) as Microbacterium resistens comb. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1267–1276. [Google Scholar] [CrossRef]
- Park, M.-J.; Kim, M.K.; Kim, H.-B.; Im, W.-T.; Yi, T.-H.; Kim, S.-Y.; Soung, N.-K.; Yang, D.-C. Microbacterium ginsengisoli sp. nov., a glucosidase-producing bacterium isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2008, 58, 429–433. [Google Scholar] [CrossRef]
- Russell, D.A.; Hatfull, G.F. PhagesDB: The actinobacteriophage database. Bioinformatics 2017, 33, 784–786. [Google Scholar] [CrossRef]
- Jacobs-Sera, D.; Abad, L.A.; Alvey, R.M.; Anders, K.R.; Aull, H.G.; Bhalla, S.S.; Blumer, L.S.; Bollivar, D.W.; Bonilla, J.A.; Butela, K.A.; et al. Genomic diversity of bacteriophages infecting Microbacterium spp. PLoS ONE 2020, 15, e0234636. [Google Scholar] [CrossRef]
- Frost, V.J.; Westover, K.M. Complete genome sequences of Microbacterium liquefaciens phages Mercedes, Leafus, Nebulous, and Ixel. Microbiol. Resour. Announc. 2021, 10, e00068-21. [Google Scholar] [CrossRef]
- Versoza, C.J.; Pfeifer, S.P. Computational prediction of bacteriophage host ranges. Microorganisms 2022, 10, 149. [Google Scholar] [CrossRef]
- Węgrzyn, G. Should bacteriophages be classified as parasites or predators? Pol. J. Microbiol. 2022, 71, 3–9. [Google Scholar] [CrossRef]
- Esposito, L.A.; Gupta, S.; Streiter, F.; Prasad, A.; Dennehy, J.J. Evolutionary interpretations of mycobacteriophage biodiversity and host-range through the analysis of codon usage bias. Microb. Genom. 2016, 2, e000079. [Google Scholar] [CrossRef]
- Thankeswaran Parvathy, S.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef]
- Galiez, C.; Siebert, M.; Enault, F.; Vincent, J.; Söding, J. WIsH: Who is the host? Predicting prokaryotic hosts from metagenomic phage contigs. Bioinformatics 2017, 33, 3113–3114. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Dalgaard, P.; R Development Core Team. R: A Language and Environment for Statistical Computing. 2010. Available online: https://www.r-project.org/ (accessed on 29 November 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 9783319242774. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Krumsiek, J.; Arnold, R.; Rattei, T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007, 23, 1026–1028. [Google Scholar] [CrossRef]
- Bourret, J.; Alizon, S.; Bravo, I.G. COUSIN (COdon Usage Similarity INdex): A normalized measure of codon usage preferences. Genome Biol. Evol. 2019, 11, 3523–3528. [Google Scholar] [CrossRef] [PubMed]
- Howell, A.A.; Versoza, C.J.; Cerna, G.; Johnston, T.; Kakde, S.; Karuku, K.; Kowal, M.; Monahan, J.; Murray, J.; Nguyen, T.; et al. Phylogenomic analyses and host range prediction of cluster P mycobacteriophages. G3 Genes Genomes Genet. 2022, 12, jkac244. [Google Scholar] [CrossRef] [PubMed]
- Baláž, A.; Kajsík, M.; Budiš, J.; Szemeš, T.; Turňa, J. PHERI—Phage Host Exploration Pipeline. bioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.05.13.093773v3 (accessed on 20 December 2022).
- Gibb, B.; Hyman, P.; Schneider, C. The many applications of engineered bacteriophages—An overview. Pharmaceuticals 2021, 14, 634. [Google Scholar] [CrossRef]
- Milhaven, M.; Cai, L.; Cherian, S.; Johnson, K.; Salas Perez, K.; Blanco, M.; Garg, A.; Lobatos, J.; Mitra, C.; Strasser, M.; et al. Complete genome sequence of the Microbacterium bacteriophage Chako. Microbiol. Resour. Announc. in press.
- Milhaven, M.; Hastings, E.; Brister, D.; Cevallos, L.; Chilukuri, S.; Diyya, A.; Garg, A.; Hernandez, R.; Kelly, D.; Lazo, K.; et al. Complete genome sequence of Microbacterium bacteriophage Erla. Microbiol. Resour. Announc. 2021, 10, e01354-20. [Google Scholar] [CrossRef]
- El Yaman, R.; Anderson, J.S.; Anderson, T.M.; Avalos, M.A.; Bangurah, S.K.; Dada, K.B.; Degener, K.R.; Hadeed, M.N.; Issa, L.H.; Jaeran, A.S.; et al. Genome sequences of microbacteriophages Zada and Ioannes. Microbiol. Resour. Announc. 2020, 9, e01012–e01020. [Google Scholar] [CrossRef]
- Nguyen Quach, A.M.; Varghese, A.E.; Siddiq, K.; Pappano, I.G.; Thaha, A.; Marcelis, T.A.; Feruku, B.; Vindyala, A.; Mohamed, R.; Johnson, K.A.; et al. Complete genome sequences of seven EA cluster microbacteriophages, Bustleton, MillyPhilly, Riyhil, Phriends, Pherbot, PrincePhergus, and TinSulphur. Microbiol. Resour. Announc. 2019, 8, e01193-19. [Google Scholar] [CrossRef]
- Lee, T.; Aguirre, M.; Andrews, S.; Bahr, K.; Ballard, A.; Bristerpostma, M.; Cox, F.; Dowell, L.; Kiker, D.; Lujan, T.; et al. Complete genome sequence of bacteriophage Finny, isolated from a Microbacterium foliorum culture. Microbiol. Resour. Announc. 2019, 8, e01039-19. [Google Scholar] [CrossRef]
- Lin, H.; Reeves, M.; Acevedo, M.; Bass, K.; Chau, E.; Ching, B.; Enriquez, E.; Evans, S.; Mamora, K.; Pang, C.; et al. Genome sequences of bacteriophages ClearAsMud and Kauala, isolated from Microbacterium foliorum. Microbiol. Resour. Announc. 2020, 9, e01026-20. [Google Scholar] [CrossRef] [PubMed]
- Cook-Easterwood, J.; Bethel, D.J.; Kurikose, I.; Katsanos, J. The genomic characterization of two Microbacterium foliorum–specific bacteriophages, QuadZero and AnnaLie. Microbiol. Resour. Announc. 2022, 11, e00208–e00222. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Paul, E.S.; Diaz, A. Characterization of phages YuuY, KaiHaiDragon, and OneinaGillian isolated from Microbacterium foliorum. Int. J. Mol. Sci. 2022, 23, 6609. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Bala, K.; Saxena, A.; Schumann, P.; Lal, R. Microbacterium amylolyticum sp. nov., isolated from soil from an industrial waste site. Int. J. Syst. Evol. Microbiol. 2012, 62, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Takeuchi, M.; Weiss, N. Proposal of two new species in the genus Microbacterium: Microbacterium dextranolyticum sp. nov. and Microbacterium Aurum sp. nov. Int. J. Syst. Evol. Microbiol. 1993, 43, 549–554. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, Y.; Yang, J.; Lai, X.-H.; Jin, D.; Lu, S.; Pu, J.; Zhang, S.; Zhu, W.; Xu, M.; et al. Microbacterium chengjingii sp. nov. and Microbacterium fandaimingii sp. nov., isolated from bat faeces of Hipposideros and Rousettus species. Int. J. Syst. Evol. Microbiol. 2021, 71, 004858. [Google Scholar] [CrossRef]
- Alves, A.; Correia, A.; Igual, J.M.; Trujillo, M.E. Microbacterium endophyticum sp. nov. and Microbacterium halimionae sp. nov., endophytes isolated from the salt-marsh plant Halimione portulacoides and emended description of the genus Microbacterium. Sys. Appl. Microbiol. 2014, 37, 474–479. [Google Scholar] [CrossRef]
- Russell, D.A.; Garlena, R.A.; Hatfull, G.F. Complete genome sequence of Microbacterium foliorum NRRL B-24224, a host for bacteriophage discovery. Microbiol. Resour. Announc. 2019, 8, e01467-18. [Google Scholar] [CrossRef]
- Gómez-Ramírez, M.; Montero-Álvarez, L.A.; Tobón-Avilés, A.; Fierros-Romero, G.; Rojas-Avelizapa, N.G. Microbacterium oxydans and Microbacterium liquefaciens: A biological alternative for the treatment of Ni-V-containing wastes. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 2015, 50, 602–610. [Google Scholar] [CrossRef]
- Tian, Z.; Yang, J.; Lai, X.-H.; Pu, J.; Jin, D.; Luo, X.; Huang, Y.; Li, J.; Zhang, G.; Wang, S.; et al. Microbacterium caowuchunii sp. nov. and Microbacterium lushaniae sp. nov., isolated from Plateau Pika (Ochotona Curzoniae) on the Qinghai–Tibet plateau of PR China. Int. J. Syst. Evol. Microbiol. 2021, 71, 004662. [Google Scholar] [CrossRef]
- Xie, F.; Niu, S.; Lin, X.; Pei, S.; Jiang, L.; Tian, Y.; Zhang, G. Description of Microbacterium luteum sp. nov., Microbacterium cremeum sp. nov., and Microbacterium atlanticum sp. nov., three novel C50 carotenoid producing bacteria. J. Microbiol. 2021, 59, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Ortet, P.; Gallois, N.; Piette, L.; Long, J.; Berthomieu, C.; Armengaud, J.; Barakat, M.; Chapon, V. Draft genome sequence of Microbacterium oleivorans strain A9, a bacterium isolated from Chernobyl radionuclide-contaminated soil. Genome Announc. 2017, 5, e00092-17. [Google Scholar] [CrossRef] [PubMed]
- Laffineur, K.; Avesani, V.; Cornu, G.; Charlier, J.; Janssens, M.; Wauters, G.; Delmeée, M. Bacteremia due to a novel Microbacterium species in a patient with leukemia and description of Microbacterium paraoxydans sp. nov. J. Clin. Microbiol. 2003, 41, 2242–2246. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Cho, H.; Kim, M.A.; Hamada, M.; Tamura, T.; Saitou, S.; Kim, S.-J.K.; Kwon, S.-W. 2020 Microbacterium protaetiae sp. nov. isolated from gut of larva of Protaetia brevitarsis seulensis. Int. J. Syst. Evol. Microbiol. 2020, 70, 2226–2232. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Yang, J.; Lu, S.; Pu, J.; Lai, X.-H.; Jin, D.; Li, J.; Zhang, G.; Wang, X.; Zhang, S.; et al. Microbacterium wangchenii sp. nov., isolated from faeces of Tibetan gazelles (Procapra picticaudata) on the Qinghai-Tibet plateau. Int. J. Syst. Evol. Microbiol. 2019, 70, 1307–1314. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milhaven, M.; Versoza, C.J.; Garg, A.; Cai, L.; Cherian, S.; Johnson, K.; Salas Perez, K.; Blanco, M.; Lobatos, J.; Mitra, C.; et al. Microbacterium Cluster EA Bacteriophages: Phylogenomic Relationships and Host Range Predictions. Microorganisms 2023, 11, 170. https://doi.org/10.3390/microorganisms11010170
Milhaven M, Versoza CJ, Garg A, Cai L, Cherian S, Johnson K, Salas Perez K, Blanco M, Lobatos J, Mitra C, et al. Microbacterium Cluster EA Bacteriophages: Phylogenomic Relationships and Host Range Predictions. Microorganisms. 2023; 11(1):170. https://doi.org/10.3390/microorganisms11010170
Chicago/Turabian StyleMilhaven, Mark, Cyril J. Versoza, Aman Garg, Lindsey Cai, Sanjana Cherian, Kamalei Johnson, Kevin Salas Perez, Madison Blanco, Jackelyn Lobatos, Corinne Mitra, and et al. 2023. "Microbacterium Cluster EA Bacteriophages: Phylogenomic Relationships and Host Range Predictions" Microorganisms 11, no. 1: 170. https://doi.org/10.3390/microorganisms11010170
APA StyleMilhaven, M., Versoza, C. J., Garg, A., Cai, L., Cherian, S., Johnson, K., Salas Perez, K., Blanco, M., Lobatos, J., Mitra, C., Strasser, M., & Pfeifer, S. P. (2023). Microbacterium Cluster EA Bacteriophages: Phylogenomic Relationships and Host Range Predictions. Microorganisms, 11(1), 170. https://doi.org/10.3390/microorganisms11010170






