Abstract
Antimicrobial resistance and tolerance are natural phenomena that arose due to evolutionary adaptation of microorganisms against various xenobiotic agents. These adaptation mechanisms make the current treatment options challenging as it is increasingly difficult to treat a broad range of infections, associated biofilm formation, intracellular and host adapted microbes, as well as persister cells and microbes in protected niches. Therefore, novel strategies are needed to identify the most promising drug targets to overcome the existing hurdles in the treatment of infectious diseases. Furthermore, discovery of novel drug candidates is also much needed, as few novel antimicrobial drugs have been introduced in the last two decades. In this review, we focus on the strategies that may help in the development of innovative small molecules which can interfere with microbial resistance mechanisms. We also highlight the recent advances in optimization of growth media which mimic host conditions and genome scale molecular analyses of microbial response against antimicrobial agents. Furthermore, we discuss the identification of antibiofilm molecules and their mechanisms of action in the light of the distinct physiology and metabolism of biofilm cells. This review thus provides the most recent advances in host mimicking growth media for effective drug discovery and development of antimicrobial and antibiofilm agents.
1. Introduction
Antimicrobial agents are produced by almost all organisms, including bacteria, fungi, and humans [1,2,3]. These compounds are synthesized by microorganisms since they exist, not only as protecting agents against other microorganisms, but also as signaling molecules as well as nutrients [4,5,6]. Antimicrobial resistance has only been observed to arise concomitantly locally, excluding mainly resistance against clinically relevant antibiotics in major human pathogens. The massive anthropogenic use of antimicrobial agents in different fields, such as medicine, agriculture, and husbandry including aquafarming has, however, promoted a global spread of resistance against those antimicrobial agents. Emergence of resistance includes the resistance against last resort drugs, such as colistin [7]. Mimicking nature by application of a panel of diverse antimicrobial agents targeting different essential pathways, applied as combinatorial antimicrobial therapy, has been one way to restrict the wider spread of antimicrobial resistance.
The massive anthropogenic use of antimicrobial agents, detergents, disinfectants, and heavy metals in mono-application contributes significantly to the alteration of the human microflora. Equally has their use led to the emergence of resistance and tolerance phenotypes, which can already arise at subinhibitory concentration of the antimicrobial agent (Figure 1).
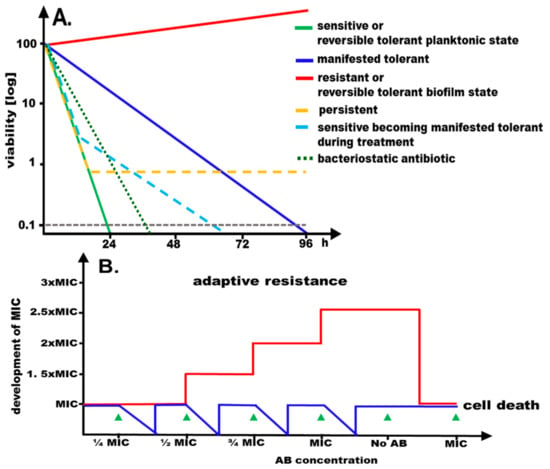
Figure 1.
Consequences of exposure to antimicrobials. (A) Graphical representation of time-dependent development of viability of a sensitive, resistant, persistent, manifested tolerant and reversible tolerant microbial population to a bactericidal antimicrobial at the minimal inhibitory concentration (MIC). A time course of a hypothetical antibiotic experiments is shown with bacterial cells subjected to the antibiotic at MIC with cellular viability monitored over time. While the majority of cells of a sensitive population are readily killed (>99.9% killing after 24 h; green line), cells of the resistant population continue to grow (red line). Cells of a manifested tolerant population display prolonged viability as compared to sensitive cells (blue line). Upon exposure of a sensitive population with a large fraction of persister cells to the antimicrobial (yellow line), the majority of the cells are killed, but a subpopulation remains viable for an extended period. Upon exposure of a sensitive population to an antimicrobial, manifested tolerance can emerge (light blue line). Biofilms display a reversible tolerance phenotype, showing apparent resistance at the MIC of the planktonic state. The decrease in viability for a bacteriostatic antibiotic is shown for comparison (90–99% killing after 24 h; dotted dark green line). (B) Repeated exposure to increasing concentrations of antibiotic below the MIC followed by regrowth can lead to adaptive resistance, which is reversed upon removal of the antibiotic. Red line, development of the minimal inhibitory concentration; blue line, development of viability.
Tolerance is distinct from adaptive and acquired resistance that are defined as enhanced resistance upon exposure to gradually increasing concentrations of the antimicrobials and the acquisition of resistance by mutations and antibiotic resistance genes, respectively. Both modes of resistance significantly alter the minimal inhibitory concentration (MIC) temporarily and permanently, respectively (Figure 1). Reversible tolerance can be displayed by a distinct physiological, yet reversible, state of the organisms such as slow growth or biofilm formation. On the other hand, slowed down killing by the antimicrobial agent at MIC, thereby maintaining significant cell viability, is defined as a manifested mode of tolerance (Figure 1; ref. [8]). A bactericidal effect is only observed at higher concentration of the antimicrobial agent (>32-fold higher as the minimal inhibitory concentration).
In a wider perspective, the composition and phenotypes of microbial populations in the environment are equally altered, which can be accompanied by the emergence of pandemic clones and enhanced biofilm formation [9,10,11]. Thus, treatment of microbial infections continues to be hampered by major challenges, such as antimicrobial resistance and tolerance. For instance, microbial biofilms, as well as metabolically silent persister cells, a subfraction of cells in a population found predominantly in biofilms, are examples of metabolically altered and downregulated cells, which display extended tolerance (Figure 1; refs. [12,13,14,15]). An immune and antibiotic protected niche for microbes inside host cells and host-matrix embedded biofilms can further lead to recurrent and refractory infections [16,17,18]. Thereby, dissemination of antimicrobial resistance is facilitated by mobile elements, including plasmids, transposons, and integrons. These mobile elements can rapidly thrive within mixed populations consisting of multiple microbial species which are promoted by biofilm formation [10,19]. However, to what extent mutated genomic targets of antibiotic action are horizontally transferred via extracellular matrix DNA (eDNA) or high frequency of recombination status of isolates remains to be determined [20,21]. The formation of biofilms, the build-up of multicellular matrix embedded communities, makes organisms reversibly tolerant towards antimicrobial agents. This tolerance triggers chronic infections which are refractory to diverse antimicrobial treatments. Interestingly, cells forming biofilms are seemingly susceptible to antimicrobials in conventional testing using standard parameters which usually monitors the planktonic state of the organism (Figure 1). Intensive research activities are currently underway to identify novel antimicrobial and antibiofilm agents [22,23,24,25]. These efforts already led to the approval of several novel effective analogs of established and novel classes of antibacterial agents, as well as novel β-lactam/β-lactamase inhibitor combinations, to treat even multi-drug resistant bacterial infections [26,27]. We discuss in this article complementary aspects that might be important for the identification and characterization of novel antimicrobial and antibiofilm agents and their targets.
2. Assessment of Antimicrobial Resistance Mimicking the Host Milieu
Assessment of antimicrobial resistance is conventionally performed by using standardized antimicrobial susceptibility testing [28]. However, these assessment processes are limited as they do not necessarily mimic the antimicrobial susceptibility under host conditions [29]. Comparison of the results of standardized antimicrobial susceptibility testing in vitro with the treatment outcome in patients represent different scenarios with incongruent results, such as in vitro inactive molecules that turn out to be effective in vivo, and vice versa [29,30,31]. For example overcomes a horizontally acquired folate transporter susceptibility of group A streptococci to sulfamethoxazole in vivo due to the acquisition of host folate. This resistance phenotype has been overlooked in conventional susceptibility testing due to the low folate concentration in the growth medium [32]. On the other hand, antibiotics were found effective against multidrug resistant bacteria in animal studies, while no in vitro effect was observed [33]. Furthermore, human biotransformation of clarithromycin produces more potent molecules [34,35], which cannot be predicted by conventional antimicrobial assays. It can thus be concluded that host conditions can alter antimicrobial resistance. Recently, it has been observed that some of these limitations in the assessment of the antimicrobial activity can be overcome by supplementation of the growth medium with bicarbonate which is present in host blood and tissues. Bicarbonate is growth inhibitory towards microorganisms when applied as an individual component at high concentrations. Furthermore, bicarbonate effectively enhanced the susceptibility towards aminoglycoside and macrolide antibiotics at subinhibitory concentrations [36,37,38,39]. Interestingly, enhanced susceptibility was observed even in the presence of an antimicrobial resistance cassette. In the same line, growth of Escherichia coli, resistant to the beta-lactam mecillinam, in urine medium partially or fully reverted the antimicrobial resistance of a clinically relevant mutation in the cysB gene [40]. Moreover, testing a range of antibiotics under standardized conditions by using Mueller–Hinton Broth versus host-like conditions demonstrated that susceptibility can be substantially different. For example, tissue-dependent high level of antimicrobial resistance in the gastrointestinal pathogen and model organism for typhoid fever Salmonella typhimurium was reflected only by the host-like medium [41].
3. Assessment of Antimicrobial Tolerance Mimicking the Host Milieu
Another limitation of standardized antimicrobial susceptibility testing arises as usually planktonic cell cultures are assessed for susceptibility. This conventional evaluation method identifies antimicrobial susceptible isolates even if a treatment refractory biofilm infection persists. However, the observed resistance towards treatment is based on tolerance of a microbial biofilm which develops specifically under host conditions in vivo [42]. In order to more closely mimic the relevant host environment, including the altered physiological and metabolic state of microorganisms, more complex semisynthetic (also named as synthetic, artificial, or simulated) growth media were developed (Table 1). Such growth media enable us to assess biofilm formation and to understand the effect of individual medium components on microbial biofilm behavior, including antimicrobial tolerance. The impact and interactions among individual members of a microbial consortium can be investigated in an in vitro set up using a host-simulating growth medium.

Table 1.
Growth media closely simulating host conditions.
Cystic fibrosis is a genetically inherited disease which is characterized by enhanced lung sputum production with altered composition prone to be colonized by microbes [58]. Persistent P. aeruginosa lung infection is one of the key factors which determines morbidity and mortality in cystic fibrosis patients. Enhanced biofilm formation of the microorganisms, due to overproduction of the exopolysaccharide alginate, is the key determinant of persistent infection. Thus, this phenotypic and/or genotypic development represents another example of developed tolerance alternatively resistance towards antimicrobial treatment. Growth of P. aeruginosa in artificial sputum medium resembling the cystic fibrosis lung environment (Table 1), including the sputum components, such as mucin and host DNA, leads to a rapid microbial adaptation. These metabolic and physiological adaptations include emergence of small colony variants and enhanced biofilm formation, and thus the microbe displaying enhanced tolerance against antimicrobial agents [45,46,59,60,61]. Interestingly, the biofilm displays mainly as tight cellular aggregates in the artificial sputum medium with few cells attached to the abiotic surface. A similar situation is observed in the cystic fibrosis lung [62], where the microbes reside in the sputum, but do not adhere to the epithelial lining. Sputum components, including amino acids which are present in high concentrations under deteriorating health conditions of the patient, contribute to an enhanced biofilm formation. Thus, a vicious cycle between disease severity and the difficulty to eradicate the microorganisms can be emulated in vitro, as it has been observed in vivo [63]. Consequently, with the toolbox to modulate individual components of an artificial medium in hand, different disease stages and conditions in individual patients, sputum composition reflecting disease severity, as well as environmental conditions, including the contribution of the accompanying microflora, can be mimicked [61,64,65]. Furthermore, the artificial sputum medium can help in understanding the genetic components of the bacterial isolates required for growth [66,67]. Therefore, use of host-like conditions in microbial susceptibility testing, including biofilm-related tolerance, supports in identifying specific drug targets, as well as unraveling the bacterial physiology, along with physiological and metabolic basis of biofilm formation [61,68]. By using this approach, growth, biofilm formation, and antimicrobial tolerance have not only been assessed for P. aeruginosa, but also for other microorganisms colonizing the cystic fibrosis lung, including Stenotrophomonas maltophilia, Mycobacterium abscessus, and black yeast [69,70,71]. It is worth mentioning that the antimetabolite oxythiamine was initially identified as an antimicrobial agent in artificial sputum medium, showing synergistic effects in combination with tetracycline for which P. aeruginosa is intrinsically resistant [72]. It remains to be shown whether the recent pharmacological restoration of functionality of the cystic fibrosis transmembrane conductance regulator CFTR, that leads to diminished growth of pathogens and re-establishment of a conventional microflora, can be mimicked by altering the composition of the artificial sputum medium. Artificial medium use reflecting distinctly similar host conditions can also be applied to specifically monitor bacterial colonization in primary ciliary dyskinesia and chronic obstructive pulmonary diseases (COPD) [73,74].
Biofilms formed under conditions simulating the host environment are not only morphologically different, but also follow different genetic programs [75]. Artificial urine medium, simulated synovial fluid, chronic wound medium, and simulated colonic environment medium form a panel of growth media developed to mimic specific infectious disease situations more closely (Table 1; refs. [48,54,76]). For example, in chronic wound medium, biofilms exhibit enhanced resistance to disinfectants and integrated anaerobes have been recovered from chronic wounds [53,54]. In such a medium, the physiology of microorganisms is significantly altered and relevant wound microbes, such as P. aeruginosa and S. aureus, have been found to be less virulent [77]. These in vitro observations are consistent with the fact that sepsis does not develop from chronic wound infections. Growth in the chronic wound medium also demonstrated a beneficial effect of co-infection of S. aureus and P. aeruginosa, including enhanced antimicrobial tolerance. Furthermore, distinct disease situations such as urinary tract and cystic fibrosis lung infection, provide particular growth conditions for the pathogen. Consequently, upon growth in different artificial media, perhaps not surprisingly, specific mutations can be selected upon exposure to antimicrobials. Upon selection, mutations leading to resistance towards fosfomycin, an epoxid-based cell wall inhibiting antibiotic, could be distinct in S. maltophilia grown either in urine or synthetic sputum medium [78]. Undoubtedly, the growth of microbial isolates in urine, sputum, saliva, or synovial fluid, derived directly from patients or animals, is an excellent approach [79,80,81]. The use of synthetic media has, however, the advantage that the effect of individual components in the medium on biofilm formation and antimicrobial tolerance can be assessed and correlated with different patients’ conditions. Besides the alteration of the growth medium, the properties of the biotic and abiotic surfaces can be determinants of the type of biofilm formed. For example, initial bacterial adhesion as a first step to develop dental plaque biofilm on a biological apatite surface is not affected by electrostatic repulsive forces [82]. Biofilm formation in osteomyelitis has been mimicked using bone blocks from bovine femur [83]. Multiple microbes adhere to central venous and urinary catheters made of silicone to develop a biofilm that eventually contains pathogens [84]. Dentures made of acrylic resins select for Candida albicans biofilm formation [85]. Surface roughness was identified as one parameter which is a determinant of initiation of biofilm formation [86]. Furthermore, static and dynamic (continuous flow with constant renewal of the medium) biofilm models have also been developed to reflect differential access to nutrients and the removal of waste products in different systems [87].
4. Innovative Strategies to Overcome Antimicrobial Resistance
Many of the established antimicrobials target core functions essential for viability, such as DNA (quinolones), RNA, and protein (aminoglycosides) synthesis, cell wall biosynthesis (β-lactams, fosfomycin and vancomycin), and outer membrane integrity, whereas a few are targeting central metabolism. In order to overcome antimicrobial resistance, effective antibiotics are isolated from natural resources [88,89]. This approach is complemented by the discovery of novel antimicrobial agents based on metabolic or virulence targets, host-adapted screening approaches, biotransformation, as well as machine learning approaches, along with genetic information of biosynthesis modules that can be edited to synthesize novel compound [90,91,92,93,94,95,96]. Additional strategies include the combinatorial screening of existing drugs in order to enhance the efficiency of antimicrobial agents through synergistic effects [97]. The discovery of intrinsic antimicrobial resistance components [98,99] might help in the identification of novel drug targets [100,101]. Such sensitizes specifically the loss of the muramyl endopeptidase Spr the Gram-negative bacterium Salmonella typhimurium against vancomycin [102]. Alternatively, upon already underlying multidrug resistance, the identification of novel therapeutic targets can be achieved, for example, by identification of components required for virulence or persistence. Candidate compounds can then be identified by molecular docking, virtual prediction of small molecule binding sites, to the protein’s crystal structure or to its structural model.- More unbiased approaches include binding studies using cell extracts or a compound library screening in vitro, following expression and purification of validated protein targets [103]. Alternatively, already addressed targets can be employed to develop new drugs, through approaches such as modulation of existing antimicrobial agents in case of altered or homologous targets [94]. This is an especially useful approach as multidrug resistant microbes can persist in the environment and do not readily reverse even upon discontinuation of the treatment [104]. For example, N-thiol substituted monocyclic β-lactams covalently inhibits abundant L-D transpeptidase 2 which performs 3,3-diaminopimelic acid crosslinks in peptidoglycan of Mycobacterium tuberculosis thereby being effective against dormant and multidrug resistant isolates [105]. It has also been observed that the frequency of horizontal plasmid transfer increases in the presence of subinhibitory concentrations of antibiotics [106]. Thus, targeting of plasmid maintenance or their conjugation can be novel strategies to lower the frequency of antimicrobial resistance, as exemplified with an IncFIA plasmid [107]. Such strategies, if sufficiently specific for multidrug resistance plasmids, might also be appropriate to remove resistance plasmids or to prevent their transfer from and between the commensal flora.
5. Innovative Strategies to Overcome Antimicrobial Tolerance in Biofilms
Biofilm formation is a major cause of tolerance against antimicrobial treatment [12,14]. Thereby, multiple characteristics of biofilms have been identified which contribute to antimicrobial tolerance. For instance, production of the extracellular matrix serves as physical and chemical barriers [108], induction of reactive oxygen species by antibiotic treatment is less efficient [109], and the metabolism of biofilms is significantly altered and slowed down in chronic infections [110], including the presence of a substantial fraction of metabolically silent persister cells [111]. As such, depletion of the extracellular biofilm matrix by nucleases and/or proteases can enhance the efficiency of antimicrobial treatment [108]. The degradation of the extracellular biofilm matrix by hydrolyzing enzymes is highly efficient. Indeed, production of extracellular matrix components can positively affect P. aeruginosa biofilm formation, and the synthesis of the second messenger cyclic di-GMP, a ubiquitous activator of biofilm formation [112]. Thus, the degradation of the biofilm matrix is not only a simple removal of physical and chemical barriers, but also involves the downregulation of biofilm physiology and metabolism, and hence contributes to the success of removal of the extracellular biofilm matrix as a therapeutic strategy.
A few clinically relevant antimicrobial agents, rifampin and fluoroquinolones, have shown to be effective against Gram-positive and Gram-negative biofilm infections, respectively [113]. Antibiofilm agents, including compounds that even disperse already established biofilms, are widespread in nature and have been developed into effective antibiofilm agents (Table 2; refs. [114,115,116]). For example, sensing of the innate immune agent nitric oxide (NO) at subinhibitory concentration disperses biofilms in a broad range of bacterial species, including human pathogens (Table 2; refs. [117,118]). Biofilm dispersal seems to be affected through downregulation of the second messenger cyclic di-GMP by distinct protein members of conceptually similar signaling pathways. In representative isolates belonging to different species, NO binds to an N-terminal or free-standing signaling domain that subsequently activates a cyclic di-GMP specific phosphodiesterase. With several clinical trials under way (NCT02498535; refs. [119,120]), success in the application of NO as an antibiofilm agent might be based on a combination of antimicrobial, antibiofilm, and host physiological effects [121]. The antimicrobial peptide LL-37 has been shown to possess a potent antibiofilm, rather than antimicrobial activity against various pathogens, such as P. aeruginosa and Escherichia coli (Table 2; refs. [122,123]). A recently developed antimicrobial peptide–vancomycin conjugate combined the antibiofilm with antimicrobial and immunostimulatory effects to reduce bacterial load in an in vivo abscess model [124]. The human hormones brain natriuretic peptide (hBNP) and C-type natriuretic peptide (hCNP) equally efficiently inhibit biofilm formation of P. aeruginosa at concentrations over 1000-fold lower than their antimicrobial concentration (Table 2). On the other hand, a highly differential temperature-dependent effect on biofilm formation of Gram-positive pathogens has been observed with human atrial natriuretic peptide (hANP) and hCNP to inhibit biofilm formation of S. aureus at body temperature. Various established antimicrobial agents, such as macrolides, have been reported to affect biofilm formation at subinhibitory concentrations, although not necessarily through the same mechanism or extent in different microbial species (Table 2; ref. [125]). While macrolides selectively affect the translation of messenger RNA into proteins by interacting with the 23S RNA of ribosomes, and prevent 50S ribosomal subunit assembly [126], transcriptional profiling of the response against subinhibitory concentrations of the semi-synthetic macrolide clarithromycin to prevent ancient rdar biofilm formation of Salmonella typhimurium, a biofilm directed by the transcriptional regulator CsgD via the expression of amyloid curli fimbriae and the exopolysaccharide cellulose, indicated selective upregulation of ribosomal subunit genes with their gene products potentially interacting with clarithromycin. Upregulation of the heat shock stress response with folding and holding chaperons is also indicative of impaired protein homeostasis [127]. Transcriptional analysis further indicated the redirection of microbial metabolism towards an oxygen- and energy-depleted status where energy is derived from L-arginine catabolism and propane-1,2-diol and ethanolamine degradation rather than by oxidative phosphorylation. This defined response might not only be explained by the macrolide clarithromycin to differentially inhibit ribosome assembly or translation, but also indicate an off-target activity of the macrolide antimicrobial agent.

Table 2.
Selected identified anti-biofilm agents and their major physiological effects on the targeted microbes and the host.
The second messenger cyclic di-GMP signaling network is a major determinant of tolerance against antimicrobials and detergents [152,153]. Alternative strategies to combat biofilm formation by mimicking natural situations include direct targeting of the cyclic di-GMP signaling molecules by complexing peptides with high affinity binding sites of protein receptors [154]. Although small molecule compounds can interfere with a variety of regulatory or biosynthetic biofilm pathways, screening identified a hydrazonodiaminopyrazole derivative, (Z)-4-(2-(3-fluorophenyl)hydrazineylidene)-5-imino-4,5-dihydro-1H-pyrazol-3-amine, that activates the breakdown of the cyclic di-GMP biofilm activator by activation of a cyclic di-GMP specific phosphodiesterase, leading to biofilm dispersal [150]. Furthermore, high spider-like biofilms, formed by clinical isolates of the yeast Candida parapsilosis, can be selectively targeted with a benzophenone semicarbazone derivative to elicit a defined transcriptional response (Nadeem, Shafeeq et al., manuscript in preparation). Similar analyses explaining the antibiofilm effect on a molecular level will help in the identification of novel antimicrobial and antibiofilm targets. These strategies combined with transcriptional profiles of alternative antibiofilm and antimicrobial agents can lead to the development of rationalized combinatorial strategies against biofilm infections [155].
An additional challenge during chronic infections is the presence of so-called small colony variants, comprised of mutated cells with metabolic downregulation, and enhanced antimicrobial resistance, and biofilm formation [156,157,158,159]. Mutations that lead to the emergence of small colony variants can occur in the heme and menaquinone biosynthesis pathways, which leads to impairment of functionality of the respiratory chain, in carbonic anhydrases (that fix inorganic CO2), and in the de novo pyrimidine biosynthesis pathway [160]. Targeting small colony variants might require novel experimental approaches as their metabolism and physiology, including elevated biofilm formation, is substantially different. Metabolic downregulation leading to antimicrobial tolerance is also displayed by some slow growing microorganisms. Such organisms can be resensitized by metabolic stimulation through nutrients [129]. Induction of dormancy can also be prevented by pharmacological interference [129] to stimulate metabolism and respiration and to increase the proton motif force required for the uptake of some classes of antimicrobial agents.
6. Holistic Assessment of Antimicrobial Tolerance
Although effective against microbial cells, several conventionally used antibiotics, such as beta-lactam antibiotics or the aminoglycoside gentamicin, do not penetrate into host cells, and thus fail to reach to intracellular bacteria (Figure 2; ref. [161]). A long-overlooked niche leading to recurrent infections is the intracellular presence of microbes which are evolutionarily not considered to possess an intracellular lifestyle. For example, uropathogenic E. coli survives antimicrobial treatment as an intracellular biofilm in bladder epithelial cells [16], while Staphylococcus aureus invades and survives in phagocytic and non-phagocytic host cells [162]. Antimicrobial agents, such as the fluoroquinolone enrofloxacin, might be poorly effective against intracellular microbes due to insufficient intracellular accumulation. They may also be less effective against microbes which reside in immunologically favorable niches in the host tissues [163,164]. Thus, besides being an effective antimicrobial agent against free-living planktonic and/or biofilm-forming microbes and non-cytotoxic against host cells, accumulation of antimicrobial agents in host cells, their tissue penetration, and immunomodulatory characteristics might add to the success of antimicrobial therapies.
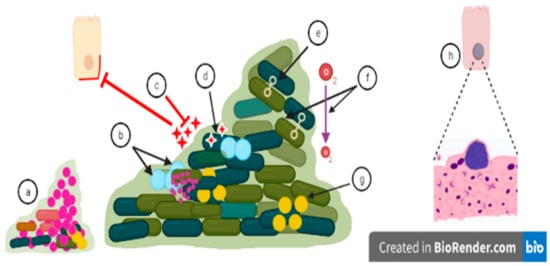
Figure 2.
Biofilm related treatment challenges: (a) Small colony variants are mutants arising during chronic infections due to, for example, a defect in the heme biosynthesis pathway. Small colony variants possess increased antibiotic resistance. (b) Growth rate is not uniform inside biofilm as persister cells are non-growing cells, and antibiotics may be inactive against these cells with low metabolic activity. (c) Antimicrobials fail to penetrate into the surface layer of biofilms or might be inactivated by extracellular matrix components, which decreases the effectiveness of antimicrobials. An increasing dose of the antimicrobial may lead to a cytotoxic effect on host cells [165]. (d) Antibiotics can be degraded or destroyed by enzymes which are secreted into the biofilm matrix. (e) Biofilm formation increases the effectiveness of genes transfer among the microbes constituting the biofilm [166]. Biofilm formation thus facilitates the transfer of antibiotic-resistant genes from resistant microbial strains to susceptible microbial strains. (f) pH and oxygen levels are different in different microenvironments of the biofilm which aid the defensive mechanisms of biofilm and can prevent uptake of antimicrobial agents. (g) Microbial cells communicate through quorum sensing which can trigger increased virulence and biofilm formation by altered gene expression, and accelerating the process of antibiotic tolerance [167]. This mechanism can stimulate biofilm formation, but also disperse biofilms depending on the autoinducer molecule. (h) Intracellular biofilm as, for example, observed in the murine cystitis model. After microbial invasion, these biofilm-like intracellular bacterial communities replicate and persist in host cells, protected from antimicrobial action and can subsequently disperse to other host cells [168].
Antimicrobial agents often have additional biological effects, besides their bactericidal or bacteriostatic effect on the targeted microbes [169]. Such antibiotics can show cross-effectiveness against highly homologous structures present in alternative microbes, fungi, parasites, and helminths and can also exert a substantial toxicity against host cells. This prevents their systemic use for the treatment of infections in humans [170,171,172,173]. Additional adverse effects, such as antibiotic allergy [174] or the selection for alternative pathogens [175], might be experienced upon long-term use. For example, M. abscessus emerged as an opportunistic pathogen in chronic lung infection in immunocompromised individuals [176].
The aminoglycoside antibiotic, nourseothricin, a natural product of the streptothricin class, is composed of a streptolidine, carbamyolated gulosamine, and variable number of β-lysine moieties. This antibiotic causes miscoding by a potentially unique binding mechanism to the ribosome [177,178]. Nourseothricin is effective not only against bacteria, but also against other microorganisms, such as archaea, fungi, protozoa, and microalgae. However, its toxicity against eukaryotic cells prevents its clinical use [179,180,181]. The effect of antimicrobial agents not only on pathogens but on the commensal microflora might increase the risk of viral [182,183] and fungal infections [184]. Besides cytotoxic and cytostatic effects beyond the primarily targeted microbes, antimicrobial agents can have a substantial modulating effect on host metabolism and physiology. Furthermore, antibiotics can affect immune responses, which might substantially alter the outcome of the antimicrobial treatment. For example, some antimicrobial agents, such as the macrolide clarithromycin, accumulate in host cells and provoke an immunomodulatory effect which contributes towards improved conditions [185]. Intriguingly, it has recently been shown that treatment success can be enhanced by stimulation of the immune system, especially in immune compromised host niches [163]. Similar strategies, in combination with strong antibiofilm agents, might also enhance the success of antimicrobial therapies against biofilm infections [107].
7. Discussion and Conclusions
The discovery of antimicrobial agents and their extensive use in the treatment of acute infections has been a major milestone in medical sciences. However, their broad spectrum uses from therapeutics to agriculture and including poultry, cosmetics, and aquaculture have led to the emergence of multidrug resistance in targeted pathogens. As a result, this major therapeutic achievement might be lost with consequences beyond the eradication of acute infections. Not only does immediate effective antimicrobial therapy target acute infections, but also follow-up diseases such as autoimmune diseases, e.g., rheumatic heart disease, can be alleviated or even prevented. Subclinical microbial infections also cause or are associated with lifestyle diseases, such as atherosclerotic cardiovascular disease due to enhanced gut permeability and poor dental health. There are indications that antimicrobial treatment leads to improvement in such situations. Similarly, treatment of chronic infections, often associated with biofilm formation as a major virulence factor, is increasingly challenging as the raise in medical care that requires indwelling devices becomes more frequent. Equally difficult is the treatment of infections in immunocompromised individuals including individuals with diabetes and cancer patients undergoing chemotherapy who are prone to a diversity of microbial infections, more frequently and with more severe outcomes. Antimicrobial agents to treat immunocompromised individuals ideally are bactericidal to kill microbes fast and effectively as the immune system cannot aid in the eradication of the organisms. Tailor-suited immunostimulatory therapies might be able to support eradication and prevent reinfection. An additional challenge is the evolution of tolerant microorganisms manifested by specific mutations to subsequently promote the development of resistance and recurrent infections [8,186] equally as the emergence of reversibly tolerant microorganisms that display enhanced biofilm formation [11]. Thereby, manifested tolerance resembles the phenotype observed with a bacteriostatic antibiotic. Whether either of these two modes of tolerance is increasing among the various pathogens requires further investigation and clinical studies. Thus, multiple strategies need to be deployed in order to develop novel and effective treatment approaches. Antibiofilm approaches might, however, be challenging to develop due to the complexity of biofilm regulation in combination with tolerance. As such, small molecular compounds might have a differential and even opposite effect on biofilm formation of different species of the human microbiome. Among the most promising approaches, molecular genome wide approaches help to identify novel targets of intrinsic antimicrobial resistance. This can aid in identifying species or even strain-specific antimicrobial targets and antibiofilm approaches which can target ubiquitous second messenger pathways mediating biofilm formation, small colony variants, and/or persister cell formation. Genome mining of human and microbial resources, on the other hand, has already provided novel opportunities to discover novel classes of antimicrobial and antibiofilm agents [187,188].
Author Contributions
Conceptualization, U.R. and M.I.C.; Writing—Original Draft Preparation, U.R., H.J., F.N., H.M.B. and M.Z.; Writing—Review and Editing, U.R., H.J., M.I.C., A.-u.-R., F.N., H.M.B. and M.Z.; Visualization, H.M.B.; Supervision, M.I.C., H.J., A.-u.-R. and U.R.; Project Administration, M.I.C., U.R., H.J. and A.-u.-R.; Funding acquisition, M.I.C., U.R., H.J. and A.-u.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding from the Lillan Sagens and Curt Ericssons Research Foundation to UR.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chandra, N.; Kumar, S. Antibiotics Producing Soil Microorganisms. Antibiotics and Antibiotics Resistance Genes in Soils; Hashmi, M., Strezov, V., Varma, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–18. [Google Scholar]
- Jakubczyk, D.; Dussart, F. Selected Fungal Natural Products with Antimicrobial Properties. Molecules 2020, 25, 911. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Melo, M.C.R.; Crescenzi, O.; Notomista, E.; de la Fuente-Nunez, C. Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng. 2022, 6, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Wang, H.H.; Davies, J. Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2007, 362, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef]
- Liu, J.; Gefen, O.; Ronin, I.; Bar-Meir, M.; Balaban, N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020, 367, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Finn, T.J. The evolutionary puzzle of Escherichia coli ST131. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 81, 104265. [Google Scholar] [CrossRef]
- Herrera, C.M.; Henderson, J.C.; Crofts, A.A.; Trent, M.S. Novel coordination of lipopolysaccharide modifications in Vibrio cholerae promotes CAMP resistance. Mol. Microbiol. 2017, 106, 582–596. [Google Scholar] [CrossRef]
- Qu, J.; Cai, Z.; Liu, Y.; Duan, X.; Han, S.; Liu, J.; Zhu, Y.; Jiang, Z.; Zhang, Y.; Zhuo, C.; et al. Persistent Bacterial Coinfection of a COVID-19 Patient Caused by a Genetically Adapted Pseudomonas aeruginosa Chronic Colonizer. Front. Cell. Infect. Microbiol. 2021, 11, 641920. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Magalhaes, C.; Lima, M.; Trieu-Cuot, P.; Ferreira, P. To give or not to give antibiotics is not the only question. Lancet. Infect. Dis. 2021, 21, e191–e201. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Gerdes, K.; Lewis, K.; McKinney, J.D. A problem of persistence: Still more questions than answers? Nat. Rev. Microbiol. 2013, 11, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.G.; Palermo, J.J.; Schilling, J.D.; Roth, R.; Heuser, J.; Hultgren, S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Horn, J.; Stelzner, K.; Rudel, T.; Fraunholz, M. Inside job: Staphylococcus aureus host-pathogen interactions. Int. J. Med. Microbiol. 2018, 308, 607–624. [Google Scholar] [CrossRef]
- Zapotoczna, M.; O’Neill, E.; O’Gara, J.P. Untangling the Diverse and Redundant Mechanisms of Staphylococcus aureus Biofilm Formation. PLoS Pathog. 2016, 12, e1005671. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Bernier, S.P.; Surette, M.G. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013, 4, 20. [Google Scholar] [CrossRef]
- Cepas, V.; Lopez, Y.; Gabasa, Y.; Martins, C.B.; Ferreira, J.D.; Correia, M.J.; Santos, L.M.A.; Oliveira, F.; Ramos, V.; Reis, M.; et al. Inhibition of Bacterial and Fungal Biofilm Formation by 675 Extracts from Microalgae and Cyanobacteria. Antibiotics 2019, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Tasneem, S.; Farooq, S.; Sami, A.; Rahman, A.U.; Choudhary, M.I. Harmaline and its Derivatives Against the Infectious Multi-Drug Resistant Escherichia coli. Med. Chem. 2017, 13, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, F.; Nasrollahzadeh, M.S.; Tajani, A.S.; Soheili, V.; Hadizadeh, F. Bacterial biofilms and their resistance mechanisms: A brief look at treatment with natural agents. Folia Microbiol. 2022, 67, 535–554. [Google Scholar] [CrossRef]
- Yusuf, E.; Bax, H.I.; Verkaik, N.J.; van Westreenen, M. An Update on Eight “New” Antibiotics against Multidrug-Resistant Gram-Negative Bacteria. J. Clin. Med. 2021, 10, 1068. [Google Scholar] [CrossRef]
- Matlock, A.; Garcia, J.A.; Moussavi, K.; Long, B.; Liang, S.Y. Advances in novel antibiotics to treat multidrug-resistant gram-negative bacterial infections. Intern. Emerg. Med. 2021, 16, 2231–2241. [Google Scholar] [CrossRef]
- Giske, C.G.; Turnidge, J.; Canton, R.; Kahlmeter, G. Update from the European Committee on Antimicrobial Susceptibility Testing (EUCAST). J. Clin. Microbiol. 2022, 60, e0027621. [Google Scholar] [CrossRef]
- Ersoy, S.C.; Heithoff, D.M.; Barnes, L.t.; Tripp, G.K.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. EBioMedicine 2017, 20, 173–181. [Google Scholar] [CrossRef]
- Farha, M.A.; French, S.; Stokes, J.M.; Brown, E.D. Bicarbonate Alters Bacterial Susceptibility to Antibiotics by Targeting the Proton Motive Force. ACS Infect. Dis. 2018, 4, 382–390. [Google Scholar] [CrossRef]
- Zak, O.; O’Reilly, T. Animal models in the evaluation of antimicrobial agents. Antimicrob. Agents Chemother. 1991, 35, 1527–1531. [Google Scholar] [CrossRef]
- Rodrigo, M.K.D.; Saiganesh, A.; Hayes, A.J.; Wilson, A.M.; Anstey, J.; Pickering, J.L.; Iwasaki, J.; Hillas, J.; Winslow, S.; Woodman, T.; et al. Host-dependent resistance of Group A Streptococcus to sulfamethoxazole mediated by a horizontally-acquired reduced folate transporter. Nat. Commun. 2022, 13, 6557. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H., Jr.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Rodvold, K.A. Clinical pharmacokinetics of clarithromycin. Clin. Pharmacokinet. 1999, 37, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, J.L.; Bopp, B.A.; Marsh, K.C.; Quigley, S.C.; Johnson, M.J.; Anderson, D.J.; Lamm, J.E.; Tolman, K.G.; Sanders, S.W.; Cavanaugh, J.H.; et al. Metabolism and disposition of clarithromycin in man. Drug Metab. Dispos. Biol. Fate Chem. 1990, 18, 441–446. [Google Scholar]
- Farha, M.A.; MacNair, C.R.; Carfrae, L.A.; El Zahed, S.S.; Ellis, M.J.; Tran, H.R.; McArthur, A.G.; Brown, E.D. Overcoming Acquired and Native Macrolide Resistance with Bicarbonate. ACS Infect. Dis. 2020, 6, 2709–2718. [Google Scholar] [CrossRef] [PubMed]
- Hinnu, M.; Putrins, M.; Kogermann, K.; Bumann, D.; Tenson, T. Making Antimicrobial Susceptibility Testing More Physiologically Relevant with Bicarbonate? Antimicrob. Agents Chemother. 2022, 66, e0241221. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Huante, M.; Martinez, H.; Bustamante, V.H.; Puente, J.L.; Sanchez, J. Bicarbonate enhances the in vitro antibiotic activity of kanamycin in Escherichia coli. Lett. Appl. Microbiol. 2015, 60, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Jaikumpun, P.; Ruksakiet, K.; Stercz, B.; Pallinger, E.; Steward, M.; Lohinai, Z.; Dobay, O.; Zsembery, A. Antibacterial Effects of Bicarbonate in Media Modified to Mimic Cystic Fibrosis Sputum. Int. J. Mol. Sci. 2020, 21, 8614. [Google Scholar] [CrossRef]
- Thulin, E.; Thulin, M.; Andersson, D.I. Reversion of High-level Mecillinam Resistance to Susceptibility in Escherichia coli During Growth in Urine. EBioMedicine 2017, 23, 111–118. [Google Scholar] [CrossRef]
- Kubicek-Sutherland, J.Z.; Heithoff, D.M.; Ersoy, S.C.; Shimp, W.R.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Host-dependent Induction of Transient Antibiotic Resistance: A Prelude to Treatment Failure. EBioMedicine 2015, 2, 1169–1178. [Google Scholar] [CrossRef]
- Hoiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Hola, V.; Imbert, C.; Kirketerp-Moller, K.; et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015, 21 (Suppl. S1), S1–S25. [Google Scholar] [CrossRef] [PubMed]
- Harrington, N.E.; Sweeney, E.; Harrison, F. Building a better biofilm—Formation of in vivo-like biofilm structures by Pseudomonas aeruginosa in a porcine model of cystic fibrosis lung infection. Biofilm 2020, 2, 100024. [Google Scholar] [CrossRef]
- Sriramulu, D.D.; Lünsdorf, H.; Lam, J.S.; Römling, U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005, 54, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Cornforth, D.M.; Diggle, F.L.; Melvin, J.A.; Bomberger, J.M.; Whiteley, J. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 2020, 11, e03042-19. [Google Scholar] [CrossRef]
- Anderson, M.J.; Scholz, M.T.; Parks, P.J.; Peterson, M.L. Ex vivo porcine vaginal mucosal model of infection for determining effectiveness and toxicity of antiseptics. J. Appl. Microbiol. 2013, 115, 679–688. [Google Scholar] [CrossRef]
- Brooks, T.; Keevil, C.W. A simple artificial urine for the growth of urinary pathogens. Lett. Appl. Microbiol. 1997, 24, 203–206. [Google Scholar] [CrossRef]
- Polzin, S.; Huber, C.; Eylert, E.; Elsenhans, I.; Eisenreich, W.; Schmidt, H. Growth media simulating ileal and colonic environments affect the intracellular proteome and carbon fluxes of enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Appl. Environ. Microbiol. 2013, 79, 3703–3715. [Google Scholar] [CrossRef]
- Crawford, R.W.; Reeve, K.E.; Gunn, J.S. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 2010, 192, 2981–2990. [Google Scholar] [CrossRef]
- Hahn, M.M.; Gonzalez, J.F.; Hitt, R.; Tucker, L.; Gunn, J.S. The Abundance and Organization of Salmonella Extracellular Polymeric Substances in Gallbladder-Mimicking Environments and In Vivo. Infect. Immun. 2021, 89, e0031021. [Google Scholar] [CrossRef]
- Pouget, C.; Dunyach-Remy, C.; Bernardi, T.; Provot, C.; Tasse, J.; Sotto, A.; Lavigne, J.P. A Relevant Wound-Like in vitro Media to Study Bacterial Cooperation and Biofilm in Chronic Wounds. Front. Microbiol. 2022, 13, 705479. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Smith, E.; Wolcott, R.; Dowd, S.E. Propagation of anaerobic bacteria within an aerobic multi-species chronic wound biofilm model. J. Wound Care 2009, 18, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Dowd, S.E.; Smith, E.; Rhoads, D.D.; Wolcott, R.D. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008, 16, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Knott, S.; Curry, D.; Zhao, N.; Metgud, P.; Dastgheyb, S.S.; Purtill, C.; Harwood, M.; Chen, A.F.; Schaer, T.P.; Otto, M.; et al. Staphylococcus aureus Floating Biofilm Formation and Phenotype in Synovial Fluid Depends on Albumin, Fibrinogen, and Hyaluronic Acid. Front. Microbiol. 2021, 12, 655873. [Google Scholar] [CrossRef]
- Coombes, B.K.; Brown, N.F.; Valdez, Y.; Brumell, J.H.; Finlay, B.B. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 2004, 279, 49804–49815. [Google Scholar] [CrossRef]
- Mondal, M.; Nag, D.; Koley, H.; Saha, D.R.; Chatterjee, N.S. The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS ONE 2014, 9, e103119. [Google Scholar] [CrossRef]
- Barth, A.L.; Pitt, T.L. Microbial Pathogens Associated With Cystic Fibrosis: Special Focus on Pseudomonas aeruginosa. Braz. J. Infect. Dis. Off. Publ. Braz. Soc. Infect. Dis. 1998, 2, 43–61. [Google Scholar]
- Lozano, C.; Lopez, M.; Rojo-Bezares, B.; Saenz, Y. Antimicrobial Susceptibility Testing in Pseudomonas aeruginosa Biofilms: One Step Closer to a Standardized Method. Antibiotics 2020, 9, 880. [Google Scholar] [CrossRef]
- Kirchner, S.; Fothergill, J.L.; Wright, E.A.; James, C.E.; Mowat, E.; Winstanley, C. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J. Vis. Exp. JoVE 2012, e3857. [Google Scholar] [CrossRef]
- Neve, R.L.; Carrillo, B.D.; Phelan, V.V. Impact of Artificial Sputum Medium Formulation on Pseudomonas aeruginosa Secondary Metabolite Production. J. Bacteriol. 2021, 203, e0025021. [Google Scholar] [CrossRef]
- Lam, J.; Chan, R.; Lam, K.; Costerton, J.W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 1980, 28, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.R.; Ray, A.; Hodson, M.E.; Pitt, T.L. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax 2000, 55, 795–797. [Google Scholar] [CrossRef]
- Ruksakiet, K.; Stercz, B.; Toth, G.; Jaikumpun, P.; Grof, I.; Tengolics, R.; Lohinai, Z.M.; Horvath, P.; Deli, M.A.; Steward, M.C.; et al. Bicarbonate Evokes Reciprocal Changes in Intracellular Cyclic di-GMP and Cyclic AMP Levels in Pseudomonas Aeruginosa. Biology 2021, 10, 519. [Google Scholar] [CrossRef]
- Vieira, J.; Gallagher, T.; Sui, H.Y.; Jesudasen, S.; Whiteson, K.; O’Toole, G.A.; Hanselmann, K.; Lai, P.S. Design and Development of a Model to Study the Effect of Supplemental Oxygen on the Cystic Fibrosis Airway Microbiome. J. Vis. Exp. JoVE 2021, 174, e62888. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Brown, S.A.; Whiteley, M. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 4449–4455. [Google Scholar] [CrossRef] [PubMed]
- Eichner, A.; Gunther, N.; Arnold, M.; Schobert, M.; Heesemann, J.; Hogardt, M. Marker genes for the metabolic adaptation of Pseudomonas aeruginosa to the hypoxic cystic fibrosis lung environment. Int. J. Med. Microbiol. 2014, 304, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, E.; Schaumann, A.; Schmitz-Afonso, I.; Afonso, C.; De, E.; Loutelier-Bourhis, C.; Alexandre, S. Membrane phospholipid composition of Pseudomonas aeruginosa grown in a cystic fibrosis mucus-mimicking medium. Biochim. Biophys. Acta. Biomembr. 2021, 1863, 183482. [Google Scholar] [CrossRef]
- Kirchhoff, L.; Weisner, A.K.; Schrepffer, M.; Hain, A.; Scharmann, U.; Buer, J.; Rath, P.M.; Steinmann, J. Phenotypical Characteristics of the Black Yeast Exophiala dermatitidis Are Affected by Pseudomonas aeruginosa in an Artificial Sputum Medium Mimicking Cystic Fibrosis-Like Conditions. Front. Microbiol. 2020, 11, 471. [Google Scholar] [CrossRef]
- Hunt-Serracin, A.C.; Parks, B.J.; Boll, J.; Boutte, C.C. Mycobacterium abscessus Cells Have Altered Antibiotic Tolerance and Surface Glycolipids in Artificial Cystic Fibrosis Sputum Medium. Antimicrob. Agents Chemother. 2019, 63, e02488-18. [Google Scholar] [CrossRef]
- Willsey, G.G.; Eckstrom, K.; LaBauve, A.E.; Hinkel, L.A.; Schutz, K.; Meagher, R.J.; LiPuma, J.J.; Wargo, M.J. Stenotrophomonas maltophilia Differential Gene Expression in Synthetic Cystic Fibrosis Sputum Reveals Shared and Cystic Fibrosis Strain-Specific Responses to the Sputum Environment. J. Bacteriol. 2019, 201, e00074-19. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, Y.; Zimmermann, M.; Lee, Y.; Lim, H.W.; Leong Tan, A.S.; Choi, I.; Ko, Y.; Lee, S.; Seo, J.J.; et al. Pharmacological perturbation of thiamine metabolism sensitizes Pseudomonas aeruginosa to multiple antibacterial agents. Cell Chem. Biol. 2022, 29, 1317–1324.e5. [Google Scholar] [CrossRef] [PubMed]
- Maisetta, G.; Grassi, L.; Esin, S.; Kaya, E.; Morelli, A.; Puppi, D.; Piras, M.; Chiellini, F.; Pifferi, M.; Batoni, G. Targeting Pseudomonas aeruginosa in the Sputum of Primary Ciliary Dyskinesia Patients with a Combinatorial Strategy Having Antibacterial and Anti-Virulence Potential. Int. J. Mol. Sci. 2019, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Stokniene, J.; Varache, M.; Rye, P.D.; Hill, K.E.; Thomas, D.W.; Ferguson, E.L. Alginate oligosaccharides enhance diffusion and activity of colistin in a mucin-rich environment. Sci. Rep. 2022, 12, 4986. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.J.; Records, A.R.; Orr, M.W.; Linden, S.B.; Lee, V.T. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 2014, 82, 2048–2058. [Google Scholar] [CrossRef]
- Müsken, A.; Bielaszewska, M.; Greune, L.; Schweppe, C.H.; Muthing, J.; Schmidt, H.; Schmidt, M.A.; Karch, H.; Zhang, W. Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM to human intestinal epithelial cells. Appl. Environ. Microbiol. 2008, 74, 1087–1093. [Google Scholar] [CrossRef]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef]
- Gil-Gil, T.; Martinez, J.L. Fosfomycin resistance evolutionary pathways of Stenotrophomonas maltophilia in different growing conditions. Int. J. Mol. Sci. 2022, 23, 1132. [Google Scholar] [CrossRef]
- Pestrak, M.J.; Gupta, T.T.; Dusane, D.H.; Guzior, D.V.; Staats, A.; Harro, J.; Horswill, A.R.; Stoodley, P. Investigation of synovial fluid induced Staphylococcus aureus aggregate development and its impact on surface attachment and biofilm formation. PLoS ONE 2020, 15, e0231791. [Google Scholar]
- Dastgheyb, S.S.; Villaruz, A.E.; Le, K.Y.; Tan, V.Y.; Duong, A.C.; Chatterjee, S.S.; Cheung, G.Y.; Joo, H.S.; Hickok, N.J.; Otto, M. Role of Phenol-Soluble Modulins in Formation of Staphylococcus aureus Biofilms in Synovial Fluid. Infect. Immun. 2015, 83, 2966–2975. [Google Scholar] [CrossRef]
- Staats, A.; Burback, P.W.; Schwieters, A.; Li, D.; Sullivan, A.; Horswill, A.R.; Stoodley, P. Rapid Aggregation of Staphylococcus aureus in Synovial Fluid Is Influenced by Synovial Fluid Concentration, Viscosity, and Fluid Dynamics, with Evidence of Polymer Bridging. mBio 2022, 13, e0023622. [Google Scholar] [CrossRef] [PubMed]
- Yelloji Rao, M.K.; Somasundaran, P.; Schilling, K.M.; Carson, B.; Ananthapadmanabhan, K.P. Bacterial adhesion onto apatite minerals—Electrokinetic aspects. Colloids Surf. A Physicochem. Eng. Asp. 1993, 79, 293–300. [Google Scholar]
- Sweeney, E.; Lovering, A.M.; Bowker, K.E.; MacGowan, A.P.; Nelson, S.M. An in vitro biofilm model of Staphylococcus aureus infection of bone. Lett. Appl. Microbiol. 2019, 68, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Perni, S.; Prokopovich, P. Micropatterning with conical features can control bacterial adhesion on silicone. Soft Matter 2013, 9, 1844–1851. [Google Scholar] [CrossRef]
- Al-Fouzan, A.F.; Al-Mejrad, L.A.; Albarrag, A.M. Adherence of Candida to complete denture surfaces in vitro: A comparison of conventional and CAD/CAM complete dentures. J. Adv. Prosthodont. 2017, 9, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, L.; Leigh, R.; Cavaliere, R.; Osvath, S.R.; Nolan, L.M.; Smyth, D.; Verhoeven, K.; Chole, R.A.; Whitchurch, C.B. Device Design Modifications Informed by In Vitro Testing of Bacterial Attachment Reduce Infection Rates of Cochlear Implants in Clinical Practice. Microorganisms 2021, 9, 1809. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, C.; Christensen, B.B.; Johansen, T.; Toftgaard Nielsen, A.; Andersen, J.B.; Givskov, M.; Molin, S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 1999, 65, 4108–4117. [Google Scholar] [CrossRef]
- Abbas, Z.; Siddiqui, B.S.; Shahzad, S.; Sattar, S.; Begum, S.; Batool, A.; Choudhary, M.I. Lawsozaheer, a new chromone produced by an endophytic fungus Paecilomyces variotii isolated from Lawsonia Alba Lam. inhibits the growth of Staphylococcus aureus. Nat. Prod. Res. 2021, 35, 4448–4453. [Google Scholar]
- Wahab, A.T.; Ilyas, Q.; Farooq, S.; Javaid, S.; Ahmed, S.; Rahman, A.U.; Choudhary, M.I. In-vitro and in-vivo anticandidal activity of Trachyspermum ammi (L.) sprague seeds ethanolic extract and thymol-containing hexanes fraction. Nat. Prod. Res. 2021, 35, 4833–4838. [Google Scholar] [CrossRef]
- Hobson, C.; Chan, A.N.; Wright, G.D. The Antibiotic Resistome: A Guide for the Discovery of Natural Products as Antimicrobial Agents. Chem. Rev. 2021, 121, 3464–3494. [Google Scholar] [CrossRef]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet. Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 181, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Fodouop Chegaing, S.P.; Kengni, A.D.M.; Siddiqui, M.; Fowa, A.B.; Gatsing, D.; Choudhary, M.I. Fungal transformation of norandrostenedione with Cunninghamella blakesleeana and anti-bacterial activity of the transformed products. Steroids 2020, 162, 108679. [Google Scholar] [CrossRef] [PubMed]
- Ghazal, F.; Farooq, S.; Wahab, A.T.; Maharjan, R.; Zafar, H.; Siddiqui, H.; Shafi, S.; Choudhary, M.I. Identification of quinoline derivatives as growth inhibitors of MDR pathogen Kleb. Pneumoniae. Future Microbiol. 2022, 17, 843–859. [Google Scholar] [CrossRef] [PubMed]
- Thong, W.L.; Zhang, Y.; Zhuo, Y.; Robins, K.J.; Fyans, J.K.; Herbert, A.J.; Law, B.J.C.; Micklefield, J. Gene editing enables rapid engineering of complex antibiotic assembly lines. Nat. Commun. 2021, 12, 6872. [Google Scholar] [CrossRef]
- Brem, J.; Panduwawala, T.; Hansen, J.U.; Hewitt, J.; Liepins, E.; Donets, P.; Espina, L.; Farley, A.J.M.; Shubin, K.; Campillos, G.G.; et al. Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors. Nat. Chem. 2022, 14, 15–24. [Google Scholar] [CrossRef]
- Cantrell, J.M.; Chung, C.H.; Chandrasekaran, S. Machine learning to design antimicrobial combination therapies: Promises and pitfalls. Drug Discov. Today 2022, 27, 1639–1651. [Google Scholar] [CrossRef]
- Duval, M.; Dar, D.; Carvalho, F.; Rocha, E.P.C.; Sorek, R.; Cossart, P. HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 13359–13364. [Google Scholar] [CrossRef]
- Marr, A.K.; Overhage, J.; Bains, M.; Hancock, R.E.W. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 2007, 153, 474–482. [Google Scholar] [CrossRef]
- Moller, T.S.; Rau, M.H.; Bonde, C.S.; Sommer, M.O.; Guardabassi, L.; Olsen, J.E. Adaptive responses to cefotaxime treatment in ESBL-producing Escherichia coli and the possible use of significantly regulated pathways as novel secondary targets. J. Antimicrob. Chemother. 2016, 71, 2449–2459. [Google Scholar] [CrossRef]
- Brochmann, P.R.; Hesketh, A.; Jana, B.; Brodersen, G.H.; Guardabassi, L. Transcriptome analysis of extended-spectrum beta-lactamase-producing Escherichia coli and methicillin-resistant Staphylococcus aureus exposed to cefotaxime. Sci. Rep. 2018, 8, 16076. [Google Scholar] [CrossRef]
- Vesto, K.; Huseby, D.L.; Snygg, I.; Wang, H.; Hughes, D.; Rhen, M. Muramyl Endopeptidase Spr Contributes to Intrinsic Vancomycin Resistance in Salmonella enterica Serovar Typhimurium. Front. Microbiol. 2018, 9, 2941. [Google Scholar] [CrossRef] [PubMed]
- Zayed, M.F.; Ibrahim, S.R.M.; Habib, E.E.; Hassan, M.H.; Ahmed, S.; Rateb, H.S. Design, Synthesis, Antimicrobial and Anti-biofilm Evaluation, and Molecular Docking of Newly Substituted Fluoroquinazolinones. Med. Chem. 2019, 15, 659–675. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1689. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.; Pessatti, T.B.; Steiner, E.M.; Cirillo, M.; Caso, C.; Bisognin, F.; Landreh, M.; Monte, P.D.; Giacomini, D.; Schnell, R. N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium Tuberc. Cell Chem. Biol. 2021, 28, 1321–1332.e5. [Google Scholar] [CrossRef]
- Moller, T.S.B.; Liu, G.; Boysen, A.; Thomsen, L.E.; Luthje, F.L.; Mortensen, S.; Moller-Jensen, J.; Olsen, J.E. Treatment with Cefotaxime Affects Expression of Conjugation Associated Proteins and Conjugation Transfer Frequency of an IncI1 Plasmid in Escherichia coli. Front. Microbiol. 2017, 8, 2365. [Google Scholar] [CrossRef]
- Zulauf, K.E.; Kirby, J.E. Discovery of small-molecule inhibitors of multidrug-resistance plasmid maintenance using a high-throughput screening approach. Proc. Natl. Acad. Sci. USA 2020, 117, 29839–29850. [Google Scholar] [CrossRef]
- Redman, W.K.; Welch, G.S.; Williams, A.C.; Damron, A.J.; Northcut, W.O.; Rumbaugh, K.P. Efficacy and safety of biofilm dispersal by glycoside hydrolases in wounds. Biofilm 2021, 3, 100061. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Katharios-Lanwermeyer, S.; O’Toole, G.A. Biofilm Maintenance as an Active Process: Evidence that Biofilms Work Hard to Stay Put. J. Bacteriol. 2022, 204, e0058721. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Lewis, K. Persister cells in biofilm associated infections. In Biofilm-Based Healthcare-Associated Infections. Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2015; Volume 831, pp. 1–9. [Google Scholar]
- Irie, Y.; Borlee, B.R.; O’Connor, J.R.; Hill, P.J.; Harwood, C.S.; Wozniak, D.J.; Parsek, M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 2012, 109, 20632–20636. [Google Scholar] [CrossRef]
- Zimmerli, W.; Sendi, P. Orthopaedic biofilm infections. Apmis 2017, 125, 353–364. [Google Scholar] [CrossRef]
- Rendueles, O.; Kaplan, J.B.; Ghigo, J.M. Antibiofilm polysaccharides. Environ. Microbiol. 2013, 15, 334–346. [Google Scholar] [CrossRef]
- Hughes, G.; Webber, M.A. Novel approaches to the treatment of bacterial biofilm infections. Br. J. Pharm. 2017, 174, 2237–2246. [Google Scholar] [CrossRef]
- Lancaster, J.R., Jr. Nitric oxide: A brief overview of chemical and physical properties relevant to therapeutic applications. Future Sci. OA 2015, 1, FSO59. [Google Scholar] [CrossRef]
- Rinaldo, S.; Giardina, G.; Mantoni, F.; Paone, A.; Cutruzzola, F. Beyond nitrogen metabolism: Nitric oxide, cyclic-di-GMP and bacterial biofilms. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Goldbart, A.; Gatt, D.; Golan Tripto, I. Non-nuberculous mycobacteria infection treated with intermittently inhaled high-dose nitric oxide. BMJ Case Rep. 2021, 14, e243979. [Google Scholar] [CrossRef]
- Bentur, L.; Gur, M.; Ashkenazi, M.; Livnat-Levanon, G.; Mizrahi, M.; Tal, A.; Ghaffari, A.; Geffen, Y.; Aviram, M.; Efrati, O. Pilot study to test inhaled nitric oxide in cystic fibrosis patients with refractory Mycobacterium abscessus lung infection. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2020, 19, 225–231. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E. Nitric oxide signaling in health and disease. Cell 2022, 185, 2853–2878. [Google Scholar] [CrossRef]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef]
- Kai-Larsen, Y.; Lüthje, P.; Chromek, M.; Peters, V.; Wang, X.; Holm, A.; Kadas, L.; Hedlund, K.O.; Johansson, J.; Chapman, M.R.; et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010, 6, e1001010. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Alford, M.; Akhoundsadegh, N.; Drayton, M.; Straus, S.K.; Hancock, R.E.W. Multifunctional Antibiotic-Host Defense Peptide Conjugate Kills Bacteria, Eradicates Biofilms, and Modulates the Innate Immune Response. J. Med. Chem. 2021, 64, 16854–16863. [Google Scholar] [CrossRef] [PubMed]
- Parra-Ruiz, J.; Vidaillac, C.; Rybak, M.J. Macrolides and staphylococcal biofilms. Rev. Esp. Quimioter. Publ. Of. Soc. Esp. Quimioter. 2012, 25, 10–16. [Google Scholar]
- Gaynor, M.; Mankin, A.S. Macrolide antibiotics: Binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 2003, 3, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.; Jahan, H.; Shafeeq, S.; Nimtz, M.; Jänsch, L.; Römling, U.; Choudhary, M.I. Clarithromycin Exerts an Antibiofilm Effect against Salmonella enterica Serovar Typhimurium rdar Biofilm Formation and Transforms the Physiology towards an Apparent Oxygen-Depleted Energy and Carbon Metabolism. Infect. Immun. 2020, 88, e00510-20. [Google Scholar] [CrossRef] [PubMed]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef]
- Meylan, S.; Andrews, I.W.; Collins, J.J. Targeting Antibiotic Tolerance, Pathogen by Pathogen. Cell 2018, 172, 1228–1238. [Google Scholar] [CrossRef]
- Krishna, S.; Bustamante, L.; Haynes, R.K.; Staines, H.M. Artemisinins: Their growing importance in medicine. Trends Pharm. Sci. 2008, 29, 520–527. [Google Scholar] [CrossRef]
- Rohatgi, A.; Gupta, P. Natural and synthetic plant compounds as anti-biofilm agents against Escherichia coli O157:H7 biofilm. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2021, 95, 105055. [Google Scholar] [CrossRef]
- Moskaug, J.O.; Carlsen, H.; Myhrstad, M.; Blomhoff, R. Molecular imaging of the biological effects of quercetin and quercetin-rich foods. Mech. Ageing Dev. 2004, 125, 315–324. [Google Scholar] [CrossRef]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Li, M. Developing natural products as potential anti-biofilm agents. Chin. Med. 2019, 14, 11. [Google Scholar] [CrossRef]
- Barraud, N.; Hassett, D.J.; Hwang, S.H.; Rice, S.A.; Kjelleberg, S.; Webb, J.S. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7344–7353. [Google Scholar] [CrossRef]
- Hassett, D.J.; Cuppoletti, J.; Trapnell, B.; Lymar, S.V.; Rowe, J.J.; Yoon, S.S.; Hilliard, G.M.; Parvatiyar, K.; Kamani, M.C.; Wozniak, D.J.; et al. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: Rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 2002, 54, 1425–1443. [Google Scholar] [CrossRef]
- Louis, M.; Clamens, T.; Tahrioui, A.; Desriac, F.; Rodrigues, S.; Rosay, T.; Harmer, N.; Diaz, S.; Barreau, M.; Racine, P.J.; et al. Pseudomonas aeruginosa Biofilm Dispersion by the Human Atrial Natriuretic Peptide. Adv. Sci. 2022, 9, e2103262. [Google Scholar] [CrossRef]
- Song, W.; Wang, H.; Wu, Q. Atrial natriuretic peptide in cardiovascular biology and disease (NPPA). Gene 2015, 569, 1–6. [Google Scholar] [CrossRef]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C. Inhibition and dispersal of Agrobacterium tumefaciens biofilms by a small diffusible Pseudomonas aeruginosa exoproduct(s). Arch. Microbiol. 2012, 194, 391–403. [Google Scholar] [CrossRef]
- Jennings, J.A.; Courtney, H.S.; Haggard, W.O. Cis-2-decenoic acid inhibits S. aureus growth and biofilm in vitro: A pilot study. Clin. Orthop. Relat. Res. 2012, 470, 2663–2670. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, C.; Yang, Y.; Weng, L.; Wang, L. Blocking of Candida albicans biofilm formation by cis-2-dodecenoic acid and trans-2-dodecenoic acid. J. Med. Microbiol. 2011, 60, 1643–1650. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, Y.; Li, J.; Wang, X.; He, S.; Yan, X.; Shi, Y.; Zhang, W.; Ding, L. Marine natural products and their synthetic analogs as promising antibiofilm agents for antibiotics discovery and development. Eur. J. Med. Chem. 2022, 239, 114513. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Scocchi, M.; Pomponio, S.; Guida, F.; Di Primio, A.; Fiscarelli, E.; Gennaro, R.; Di Bonaventura, G. Antibacterial and anti-biofilm effects of cathelicidin peptides against pathogens isolated from cystic fibrosis patients. Peptides 2011, 32, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- De Le Fuente-Nunez, C.; Cardoso, M.H.; de Souza Candida, E.; Franco, O.L.; Hancock, R.E. Synthetic antibiofilm peptides. Biochim. Biophys. Acta (BBA)—Biomembr. 2016, 1856, 1061–1069. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef]
- Tao, R.; Tong, Z.; Lin, Y.; Xue, Y.; Wang, W.; Kuang, R.; Wang, P.; Tian, Y.; Ni, L. Antimicrobial and antibiofilm activity of pleurocidin against cariogenic microorganisms. Peptides 2011, 32, 1748–1754. [Google Scholar] [CrossRef]
- Qian, C.D.; Wu, X.C.; Teng, Y.; Zhao, W.P.; Li, O.; Fang, S.G.; Huang, Z.H.; Gao, H.C. Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2012, 56, 1458–1465. [Google Scholar] [CrossRef]
- Park, J.S.; Choi, H.Y.; Kim, W.G. The Nitrite Transporter Facilitates Biofilm Formation via Suppression of Nitrite Reductase and Is a New Antibiofilm Target in Pseudomonas Aeruginosa. MBio 2020, 11, e00878-20. [Google Scholar] [CrossRef]
- Andersen, J.B.; Hultqvist, L.D.; Jansen, C.U.; Jakobsen, T.H.; Nilsson, M.; Rybtke, M.; Uhd, J.; Fritz, B.G.; Seifert, R.; Berthelsen, J.; et al. Identification of small molecules that interfere with c-di-GMP signaling and induce dispersal of Pseudomonas aeruginosa biofilms. NPJ Biofilms Microbiomes 2021, 7, 59. [Google Scholar] [CrossRef]
- Souza, E.S.P.; Ferreira, M.A.; de Moraes, L.F.R.; de Barros, E.; Preza, S.L.E.; Cardoso, M.H.; Franco, O.L.; Migliolo, L. Synthetic peptides bioinspired in temporin-PTa with antibacterial and antibiofilm activity. Chem. Biol. Drug Des. 2022, 100, 51–63. [Google Scholar] [CrossRef]
- Poudyal, B.; Sauer, K. The PA3177 Gene Encodes an Active Diguanylate Cyclase That Contributes to Biofilm Antimicrobial Tolerance but Not Biofilm Formation by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01049-18. [Google Scholar] [CrossRef]
- Kim, H.K.; Harshey, R.M. A Diguanylate Cyclase Acts as a Cell Division Inhibitor in a Two-Step Response to Reductive and Envelope Stresses. MBio 2016, 7, e00822-16. [Google Scholar] [CrossRef]
- Hee, C.S.; Habazettl, J.; Schmutz, C.; Schirmer, T.; Jenal, U.; Grzesiek, S. Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP. Proc. Natl. Acad. Sci. USA 2020, 117, 17211–17220. [Google Scholar] [CrossRef]
- Zheng, E.J.; Andrews, I.W.; Grote, A.T.; Manson, A.L.; Alcantar, M.A.; Earl, A.M.; Collins, J.J. Modulating the evolutionary trajectory of tolerance using antibiotics with different metabolic dependencies. Nat. Commun. 2022, 13, 2525. [Google Scholar] [CrossRef]
- Proctor, R.A.; Kahl, B.; von Eiff, C.; Vaudaux, P.E.; Lew, D.P.; Peters, G. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 1998, 27 (Suppl. S1), S68–S74. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef]
- Haussler, S.; Rohde, M.; Steinmetz, I. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental melioidosis. Med. Microbiol. Immunol. 1999, 188, 91–97. [Google Scholar]
- Tang, Q.; Precit, M.R.; Thomason, M.K.; Blanc, S.F.; Ahmed-Qadri, F.; McFarland, A.P.; Wolter, D.J.; Hoffman, L.R.; Woodward, J.J. Thymidine starvation promotes c-di-AMP-dependent inflammation during pathogenic bacterial infection. Cell Host Microbe 2022, 30, 961–974.e6. [Google Scholar] [CrossRef]
- Kahl, B.C.; Becker, K.; Loffler, B. Clinical Significance and Pathogenesis of Staphylococcal Small Colony Variants in Persistent Infections. Clin. Microbiol. Rev. 2016, 29, 401–427. [Google Scholar] [CrossRef]
- Vaudaux, P.; Waldvogel, F.A. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob. Agents Chemother. 1979, 16, 743–749. [Google Scholar] [CrossRef][Green Version]
- Fraunholz, M.; Sinha, B. Intracellular Staphylococcus aureus: Live-in and let die. Front. Cell. Infect. Microbiol. 2012, 2, 43. [Google Scholar] [CrossRef]
- Li, J.; Claudi, B.; Fanous, J.; Chicherova, N.; Cianfanelli, F.R.; Campbell, R.A.A.; Bumann, D. Tissue compartmentalization enables Salmonella persistence during chemotherapy. Proc. Natl. Acad. Sci. USA 2021, 118, e2113951118. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, N.F.; Kendall, S.; Good, L. Targeting the hard to reach: Challenges and novel strategies in the treatment of intracellular bacterial infections. Br. J. Pharm. 2017, 174, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon 2019, 5, e02192. [Google Scholar] [CrossRef]
- Del Pozo, J.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.A.; Hooton, T.M.; Stamm, W.E.; Humphrey, P.A.; Hultgren, S.J. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007, 4, e329. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial biofilm eradication agents: A current review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef]
- Kricker, J.A.; Page, C.P.; Gardarsson, F.R.; Baldursson, O.; Gudjonsson, T.; Parnham, M.J. Nonantimicrobial Actions of Macrolides: Overview and Perspectives for Future Development. Pharm. Rev. 2021, 73, 233–262. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, J.Y.; Fu, H.; Harvey, C.; Ravindran, S.; Roush, W.R.; Boothroyd, J.C.; Khosla, C. Chemistry and biology of macrolide antiparasitic agents. J. Med. Chem. 2011, 54, 2792–2804. [Google Scholar] [CrossRef]
- Karpinski, T.M. Marine Macrolides with Antibacterial and/or Antifungal Activity. Mar. Drugs 2019, 17, 241. [Google Scholar] [CrossRef]
- Dzhafarov, M.K.; Vasilevich, F.I. Ecological, physiological and biochemical adaptation in helminth: Trends in evolution of antihelminthic chemical agents. Adv. Pharmacol. Pharm. 2014, 2, 30–45. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit. Care 2006, 10, R27. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, K.G.; Peter, J.G.; Trubiano, J.A.; Phillips, E.J. Antibiotic allergy. Lancet 2019, 393, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Wetzstein, N.; Diricks, M.; Kohl, T.A.; Wichelhaus, T.A.; Andres, S.; Paulowski, L.; Schwarz, C.; Lewin, A.; Kehrmann, J.; Kahl, B.C.; et al. Molecular Epidemiology of Mycobacterium abscessus Isolates Recovered from German Cystic Fibrosis Patients. Microbiol. Spectr. 2022, 10, e0171422. [Google Scholar] [CrossRef] [PubMed]
- Bronson, R.A.; Gupta, C.; Manson, A.L.; Nguyen, J.A.; Bahadirli-Talbott, A.; Parrish, N.M.; Earl, A.M.; Cohen, K.A. Global phylogenomic analyses of Mycobacterium abscessus provide context for non cystic fibrosis infections and the evolution of antibiotic resistance. Nat. Commun. 2021, 12, 5145. [Google Scholar] [CrossRef] [PubMed]
- Haupt, I.; Hubener, R.; Thrum, H. Streptothricin F, an inhibitor of protein synthesis with miscoding activity. J. Antibiot. 1978, 31, 1137–1142. [Google Scholar] [CrossRef]
- Smith, K.P.; Kang, Y.-S.; Green, A.B.; Dowgiallo, M.G.; Miller, B.C.; Chiaraviglio, L.; Truelson, K.A.; Zulauf, K.E.; Rodriguez, S.; Manetsch, R.; et al. Profiling the in vitro and in vivo activity of streptothricin-F against carbapenem-resistant Enterobacterales: A historic scaffold with a novel mechanism of action. Biorxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Reuss, O.; Vik, A.; Kolter, R.; Morschhauser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef]
- Farley, K.R.; Metcalf, W.W. The streptothricin acetyltransferase (sat) gene as a positive selectable marker for methanogenic archaea. FEMS Microbiol. Lett. 2019, 366, fnz216. [Google Scholar] [CrossRef]
- Joshi, P.B.; Webb, J.R.; Davies, J.E.; McMaster, W.R. The gene encoding streptothricin acetyltransferase (sat) as a selectable marker for Leishmania expression vectors. Gene 1995, 156, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Bradley, K.C.; Finsterbusch, K.; Schnepf, D.; Crotta, S.; Llorian, M.; Davidson, S.; Fuchs, S.Y.; Staeheli, P.; Wack, A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019, 28, 245–256.e4. [Google Scholar] [CrossRef]
- Erttmann, S.F.; Swacha, P.; Aung, K.M.; Brindefalk, B.; Jiang, H.; Hartlova, A.; Uhlin, B.E.; Wai, S.N.; Gekara, N.O. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity 2022, 55, 847–861.e10. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.A.; Desai, J.V.; Ricotta, E.E.; Swamydas, M.; Deming, C.; Conlan, S.; Quinones, M.; Matei-Rascu, V.; Sherif, L.; Lecky, D.; et al. Long-term antibiotic exposure promotes mortality after systemic fungal infection by driving lymphocyte dysfunction and systemic escape of commensal bacteria. Cell Host Microbe 2022, 30, 1020–1033.e6. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Chalmers, J.D. The immunomodulatory effects of macrolide antibiotics in respiratory disease. Pulm. Pharmacol. Ther. 2021, 71, 102095. [Google Scholar] [CrossRef] [PubMed]
- Lazarovits, G.; Gefen, O.; Cahanian, N.; Adler, K.; Fluss, R.; Levin-Reisman, I.; Ronin, I.; Motro, Y.; Moran-Gilad, J.; Balaban, N.Q.; et al. Prevalence of Antibiotic Tolerance and Risk for Reinfection Among, E. coli Bloodstream Isolates: A Prospective Cohort Study. Clin. Infect. Dis. 2022, 75, 1706–1713. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Latendorf, T.; Bartels, J.; Becker, A.; Tholey, A.; Schroder, J.M. Hornerin contains a Linked Series of Ribosome-Targeting Peptide Antibiotics. Sci. Rep. 2018, 8, 16158. [Google Scholar] [CrossRef] [PubMed]
- Lesouhaitier, O.; Clamens, T.; Rosay, T.; Desriac, F.; Louis, M.; Rodrigues, S.; Gannesen, A.; Plakunov, V.K.; Bouffartigues, E.; Tahrioui, A.; et al. Host Peptidic Hormones Affecting Bacterial Biofilm Formation and Virulence. J. Innate Immun. 2019, 11, 227–241. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).