Pepsin and Trypsin Treatment Combined with Carvacrol: An Efficient Strategy to Fight Pseudomonas aeruginosa and Enterococcus faecalis Biofilms
Abstract
1. Introduction
2. Materials and Methods
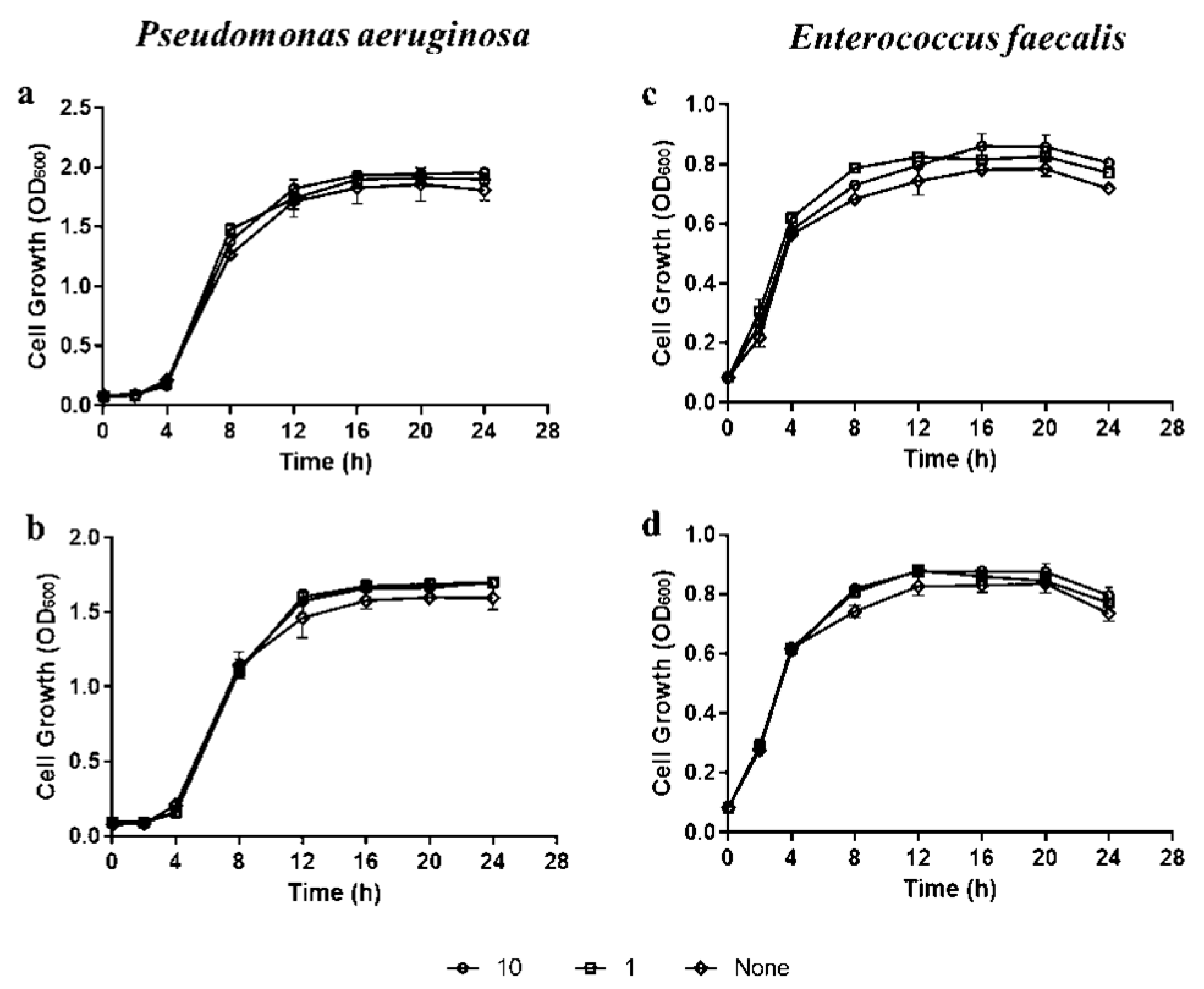
2.1. Preparation of Bacterial Strains, Reagents, and Cell Suspensions
2.2. Bacterial Susceptibility Assay
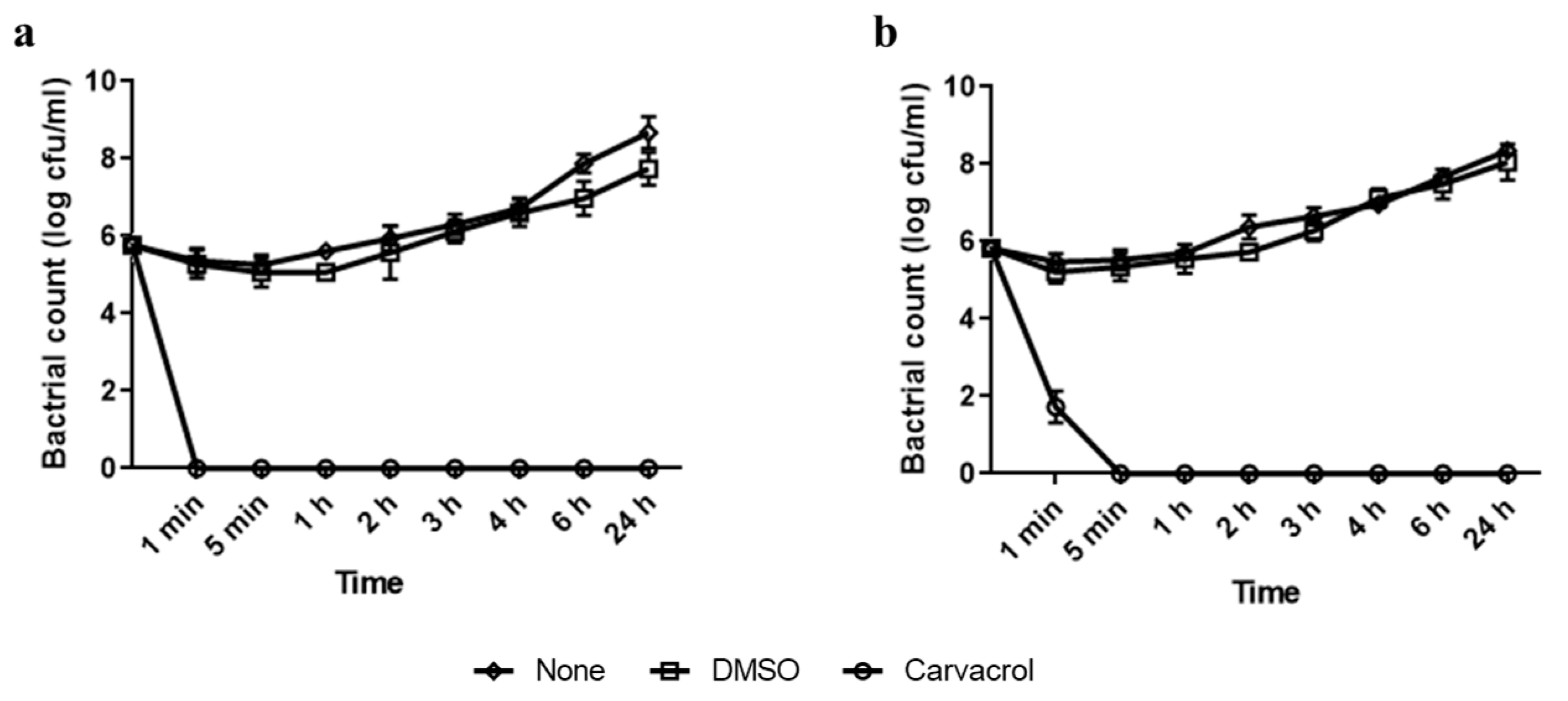
2.3. Time–Kill Assessment
2.4. Assessment of the Enzymatic Effects on Biofilm on Polystyrene Surface
2.5. Combined Treatment for Removal of Biofilms on Stainless Steel
2.5.1. Biofilm Formed on Stainless Steel Surfaces
2.5.2. Biofilm Removal by Single and Combined Treatment
2.5.3. Epifluorescence Microscopy Imaging
2.6. Scanning Electron Microscopy Analysis (SEM)
2.7. Statistical Analysis
3. Results
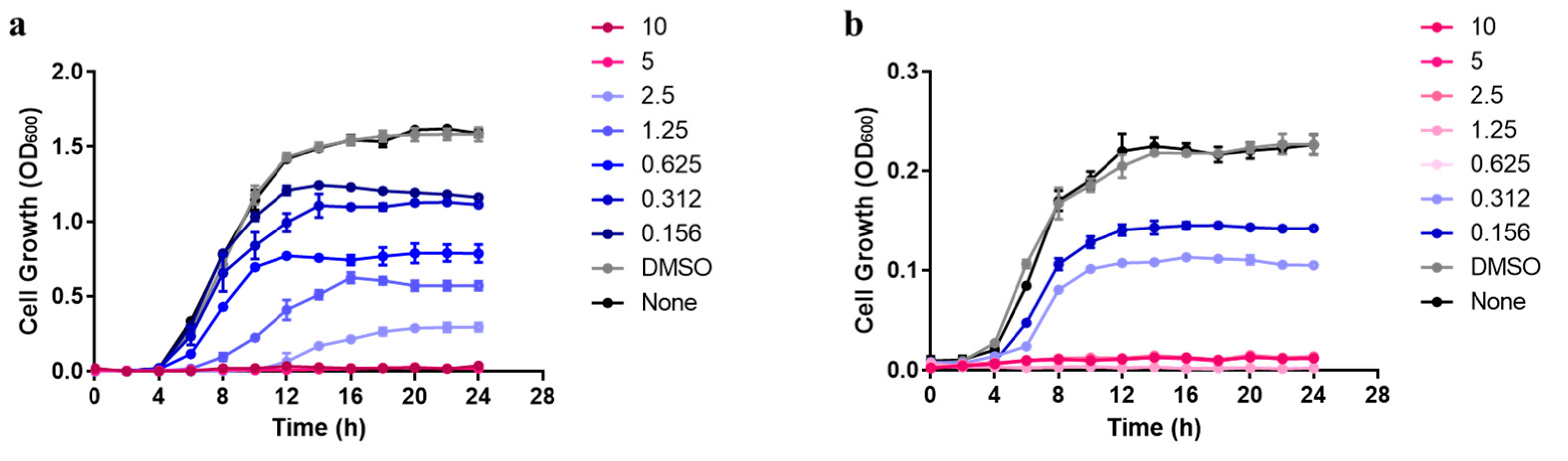
3.1. Antimicrobial Activity of Carvacrol against Planktonic Cells of Pseudomonas aeruginosa and Enterococcus faecalis
3.2. Assessment of the Minimal Dispersive Concentration and Enzyme-Action-Time on Biofilm Developed on Polystyrene Microtiter Plates
3.3. Quantitative Assessment of the Combined Effect of Enzymes and Carvacrol on Biofilm Developed on Stainless Steel
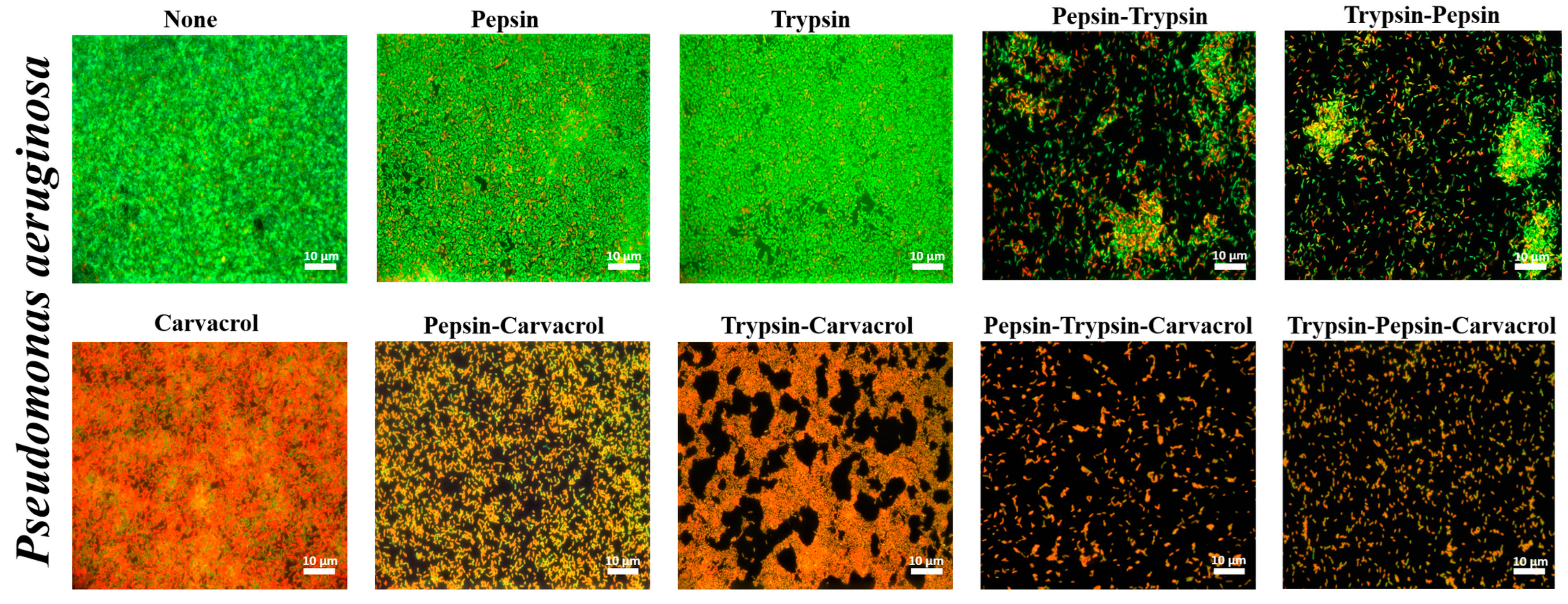
3.4. Qualitative Assessment of the Effect of Enzymes and Carvacrol on Biofilms Viability
3.5. Effect of Enzymes and Carvacrol on the Morphology of Biofilm Cells
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, K.; Lee, K.-M.; Kim, D.; Yoon, S.S. Molecular determinants of the thickened matrix in a dual-species Pseudomonas aeruginosa and Enterococcus faecalis biofilm. Appl. Environ. Microbiol. 2017, 83, e01182-17. [Google Scholar] [CrossRef] [PubMed]
- Khelissa, S.; Abdallah, M.; Jama, C.; Gharsallaoui, A.; Chihib, N.-E. Comparative study of growth temperature impact on the susceptibility of biofilm-detached and planktonic Staphylococcus aureus cells to benzalkonium chloride. Ann. Microbiol. 2019, 69, 291–298. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas Aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas Aeruginosa Lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- An, S.; Murtagh, J.; Twomey, K.B.; Gupta, M.K.; O’Sullivan, T.P.; Ingram, R.; Valvano, M.A.; Tang, J. Modulation of antibiotic sensitivity and biofilm formation in Pseudomonas aeruginosa by interspecies signal analogues. Nat. Commun. 2019, 10, 2334. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef]
- Shridhar, S.; Dhanashree, B. Antibiotic Antibiotic susceptibility pattern and biofilm formation in clinical isolates of Enterococcus spp. Interdiscip. Perspect. Infect. Dis. 2019, 2019, e7854968. [Google Scholar] [CrossRef]
- Al Kassaa, I.; Mechemchani, S.; Zaylaa, M.; Ismail, M.B.; El Omari, K.; Dabboussi, F.; Hamze, M. Characterization of lactobacilli strains isolated from baby’s feces for their potential immunobiotic application. Iran. J. Microbiol. 2019, 11, 379–388. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Mohamed, S.H.; Mohamed, M.S.M.; Khalil, M.S.; Azmy, M.; Mabrouk, M.I. Combination of essential oil and ciprofloxacin to inhibit/eradicate biofilms in multidrug-resistant Klebsiella pneumoniae. J. Appl. Microbiol. 2018, 125, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Lequette, Y.; Boels, G.; Clarisse, M.; Faille, C. Using enzymes to remove biofilms of bacterial isolates sampled in the food-industry. Biofouling 2010, 26, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, A.; Langsrud, S.; Heir, E.; Mikkelsen, M.I.; Møretrø, T. Biofilm matrix composition affects the susceptibility of food associated staphylococci to cleaning and disinfection agents. Front. Microbiol. 2016, 7, 856. [Google Scholar] [CrossRef]
- Lim, E.S.; Koo, O.; Kim, M.; Kim, J.-S. Bio-enzymes for inhibition and elimination of Escherichia coli O157:H7 biofilm and their synergistic effect with sodium hypochlorite. Sci. Rep. 2019, 9, 9920. [Google Scholar] [CrossRef]
- Kim, L.H.; Kim, S.-J.; Kim, C.-M.; Shin, M.S.; Kook, S.; Kim, I.S. Effects of enzymatic treatment on the reduction of extracellular polymeric substances (EPS) from biofouled membranes. Desalination Water Treat. 2013, 51, 6355–6361. [Google Scholar] [CrossRef]
- Zhou, J.-W.; Luo, H.-Z.; Jiang, H.; Jian, T.-K.; Chen, Z.-Q.; Jia, A.-Q. Hordenine: A novel quorum sensing inhibitor and antibiofilm agent against Pseudomonas aeruginosa. J. Agric. Food Chem. 2018, 66, 1620–1628. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Xing, T.; Wu, N.; Xu, X.; Zhou, G. Removal of Salmonella biofilm formed under meat processing environment by surfactant in combination with bio-enzyme. LWT-Food Sci. Technol. 2016, 66, 298–304. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Puga, C.H.; Orgaz, B.; Cabo, M.L. Quantifying the combined effects of pronase and benzalkonium chloride in removing late-stage Listeria monocytogenes–Escherichia coli dual-species biofilms. Biofouling 2017, 33, 690–702. [Google Scholar] [CrossRef]
- Baidamshina, D.R.; Koroleva, V.A.; Olshannikova, S.S.; Trizna, E.Y.; Bogachev, M.I.; Artyukhov, V.G.; Holyavka, M.G.; Kayumov, A.R. Biochemical properties and anti-biofilm activity of chitosan-immobilized papain. Mar. Drugs 2021, 19, 197. [Google Scholar] [CrossRef]
- Jiang, Y.; Geng, M.; Bai, L. Targeting biofilms therapy: Current research strategies and development hurdles. Microorganisms 2020, 8, 1222. [Google Scholar] [CrossRef]
- Isenberg, H.D. Clinical Microbiology Procedures Handbook; ASM Press: Almere, The Netherlands, 2004; ISBN 978-1-55581-290-4. [Google Scholar]
- Abdallah, M.; Khelissa, O.; Ibrahim, A.; Benoliel, C.; Heliot, L.; Dhulster, P.; Chihib, N.-E. Impact of growth temperature and surface type on the resistance of Pseudomonas aeruginosa and Staphylococcus aureus biofilms to disinfectants. Int. J. Food Microbiol. 2015, 214, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Toté, K.; Horemans, T.; Berghe, D.V.; Maes, L.; Cos, P. Inhibitory effect of biocides on the viable masses and matrices of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2010, 76, 3135–3142. [Google Scholar] [CrossRef] [PubMed]
- Meireles, A.; Borges, A.; Giaouris, E.; Simões, M. The current knowledge on the application of anti-biofilm enzymes in the food industry. Food Res. Int. 2016, 86, 140–146. [Google Scholar] [CrossRef]
- Taglialegna, A.; Matilla-Cuenca, L.; Dorado-Morales, P.; Navarro, S.; Ventura, S.; Garnett, J.A.; Lasa, I.; Valle, J. The biofilm-associated surface protein Esp of Enterococcus faecalis forms amyloid-like fibers. NPJ Biofilms Microbiomes 2020, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Molobela, I.P. Proteolytic and Amylolytic Enzymes for Bacterial Biofilm Control. Ph.D. Thesis, University of Pretoria, Hatfield, South Africa, 2010. [Google Scholar]
- Brindle, E.R.; Miller, D.A.; Stewart, P.S. Hydrodynamic deformation and removal of Staphylococcus epidermidis biofilms treated with urea, chlorhexidine, iron chloride, or DispersinB. Biotechnol. Bioeng. 2011, 108, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Banar, M.; Emaneini, M.; Satarzadeh, M.; Abdellahi, N.; Beigverdi, R.; van Leeuwen, W.B.; Jabalameli, F. Evaluation of Mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS ONE 2016, 11, e0164622. [Google Scholar] [CrossRef]
- Chaignon, P.; Sadovskaya, I.; Ragunah, C.; Ramasubbu, N.; Kaplan, J.B.; Jabbouri, S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 2007, 75, 125–132. [Google Scholar] [CrossRef]
- Patterson, J.L.; Girerd, P.H.; Karjane, N.W.; Jefferson, K.K. Effect of biofilm phenotype on resistance of Gardnerella vaginalis to hydrogen peroxide and lactic acid. Am. J. Obstet. Gynecol. 2007, 197, 170.e1–170.e7. [Google Scholar] [CrossRef]
- Niazi, S.A.; Clark, D.; Do, T.; Gilbert, S.C.; Foschi, F.; Mannocci, F.; Beighton, D. The effectiveness of enzymic irrigation in removing a nutrient-stressed endodontic multispecies biofilm. Int. Endod. 2014, 47, 756–768. [Google Scholar] [CrossRef]
- Marcato-Romain, C.E.; Pechaud, Y.; Paul, E.; Girbal-Neuhauser, E.; Dossat-Létisse, V. Removal of microbial multi-species biofilms from the paper industry by enzymatic treatments. Biofouling 2012, 28, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, J.; Ding, W.; Lin, J.; Tian, R.; Lu, L.; Liu, X.; Shen, X.; Qian, P.-Y. Extracellular matrix-associated proteins form an integral and dynamic system during Pseudomonas aeruginosa biofilm development. Front. Cell Infect. Microbiol. 2015, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, M.; Greenberg, E.P. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 4449–4456. [Google Scholar] [CrossRef]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842. [Google Scholar] [CrossRef]
- Schooling, S.R.; Beveridge, T.J. Membrane vesicles: An overlooked component of the matrices of biofilms. J. Bacteriol. 2006, 188, 5945–5957. [Google Scholar] [CrossRef]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, Z.; Yin, B.; Wu, H.; Tang, S.; Wu, L.; Su, Y.N.; Lin, Y.; Liu, X.Q.; Pang, B.; et al. A complex of trypsin and chymotrypsin effectively inhibited growth of pathogenic bacteria inducing cow mastitis and showed synergistic antibacterial activity with antibiotics. Livest. Sci. 2016, 188, 25–36. [Google Scholar] [CrossRef]
- Esteban, J.; García-Coca, M. Mycobacterium biofilms. Front. Microbiol. 2017, 8, 2651. [Google Scholar] [CrossRef]
- Mauriello, E.; Ferrari, G.; Donsì, F. Effect of formulation on properties, stability, carvacrol release and antimicrobial activity of carvacrol emulsions. Colloids Surf. B Biointerfaces 2021, 197, 111424. [Google Scholar] [CrossRef]
- Fang, S.; Zhou, Q.; Hu, Y.; Liu, F.; Mei, J.; Xie, J. Antimicrobial carvacrol incorporated in flaxseed gum-sodium alginate active films to improve the quality attributes of chinese sea bass (Lateolabrax maculatus) during cold storage. Molecules 2019, 24, 3292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gan, R.-Y.; Ge, Y.-Y.; Yang, Q.-Q.; Ge, J.; Li, H.-B.; Corke, H. Research progress on the antibacterial mechanisms of carvacrol: A mini review. Bioact. Compd. Health Dis. 2018, 1, 71–81. [Google Scholar] [CrossRef]
- Liu, Q.; Qiao, K.; Zhang, S. Potential of a small molecule carvacrol in management of vegetable diseases. Molecules 2019, 24, 1932. [Google Scholar] [CrossRef] [PubMed]
- Siroli, L.; Braschi, G.; de Jong, A.; Kok, J.; Patrignani, F.; Lanciotti, R. Transcriptomic approach and membrane fatty acid analysis to study the response mechanisms of Escherichia coli to thyme essential oil, carvacrol, 2-(E)-hexanal and citral exposure. J. Appl. Microbiol. 2018, 125, 1308–1320. [Google Scholar] [CrossRef] [PubMed]
- Ghafari, O.; Sharifi, A.; Ahmadi, A.; Nayeri Fasaei, B. Antibacterial and anti-PmrA activity of plant essential oils against fluoroquinolone-resistant Streptococcus pneumoniae clinical isolates. Lett. Appl. Microbiol. 2018, 67, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.B.; Heckler, C.; Tondo, E.C.; Daroit, D.J.; da Silva Malheiros, P. Antimicrobial activity of free and liposome-encapsulated thymol and carvacrol against Salmonella and Staphylococcus aureus adhered to stainless steel. Int. J. Food Microbiol. 2017, 252, 18–23. [Google Scholar] [CrossRef]
- Trevisan, D.A.C.; da Silva, A.F.; Negri, M.; de Abreu Filho, B.A.; Machinski Junior, M.; Patussi, E.V.; Campanerut-Sá, P.A.Z.; Mikcha, J.M.G.; Trevisan, D.A.C.; da Silva, A.F.; et al. Antibacterial and antibiofilm activity of carvacrol against Salmonella enterica serotype Typhimurium. Braz. J. Pharm. Sci. 2018, 54, e17229. [Google Scholar] [CrossRef]
- Tapia-Rodriguez, M.R.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Martins, C.M.; Ayala-Zavala, J.F. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on stainless steel surfaces. Food Control 2017, 75, 255–261. [Google Scholar] [CrossRef]
- dos Santos Rodrigues, J.B.; de Carvalho, R.J.; de Souza, N.T.; de Sousa Oliveira, K.; Franco, O.L.; Schaffner, D.; de Souza, E.L.; Magnani, M. Effects of oregano essential oil and carvacrol on biofilms of Staphylococcus aureus from food-contact surfaces. Food Control 2017, 73, 1237–1246. [Google Scholar] [CrossRef]
- Kalia, M.; Yadav, V.K.; Singh, P.K.; Sharma, D.; Pandey, H.; Narvi, S.S.; Agarwal, V. Effect of cinnamon oil on quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas aeruginosa. PLoS ONE 2015, 10, e0135495. [Google Scholar] [CrossRef]
- de Oliveira Negreiros, M.; Pawlowski, Â.; Zini, C.A.; Soares, G.L.G.; de Souza Motta, A.; Frazzon, A.P.G. Antimicrobial and antibiofilm activity of Baccharis psiadioides essential oil against antibiotic-resistant Enterococcus faecalis strains. Pharm. Biol. 2016, 54, 3272–3279. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Fulaz, S.; Vitale, S.; Quinn, L.; Casey, E. Nanoparticle–biofilm interactions: The role of the eps matrix. Trends Microbiol. 2019, 27, 915–926. [Google Scholar] [CrossRef]
- Pinto, R.M.; Soares, F.A.; Reis, S.; Nunes, C.; Van Dijck, P. Innovative strategies toward the disassembly of the eps matrix in bacterial biofilms. Front. Microbiol. 2020, 11, 952. [Google Scholar] [CrossRef]
- Cui, H.; Ma, C.; Lin, L. Synergetic antibacterial efficacy of cold nitrogen plasma and clove oil against Escherichia coli O157:H7 biofilms on lettuce. Food Control 2016, 66, 8–16. [Google Scholar] [CrossRef]
- Saggu, S.K.; Jha, G.; Mishra, P.C. Enzymatic degradation of biofilm by metalloprotease from Microbacterium sp. SKS10. Front. Bioeng. Biotechnol. 2019, 7, 192. [Google Scholar] [CrossRef]
- Cordeiro, A.L.; Werner, C. Enzymes for antifouling strategies. J. Adhes. Sci. Technol. 2011, 25, 2317–2344. [Google Scholar] [CrossRef]
- Dorman, H.J.D.; Deans, S.G. Antimicrobial agents from plants: Antibacterial activity of plant volatile oils. J. Appl. Microbiol. 2000, 88, 308–316. [Google Scholar] [CrossRef]
- Liu, C.; Guo, D.-A.; Liu, L. Quality transitivity and traceability system of herbal medicine products based on quality markers. Phytomedicine 2018, 44, 247–257. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mechmechani, S.; Gharsallaoui, A.; Karam, L.; EL Omari, K.; Fadel, A.; Hamze, M.; Chihib, N.-E. Pepsin and Trypsin Treatment Combined with Carvacrol: An Efficient Strategy to Fight Pseudomonas aeruginosa and Enterococcus faecalis Biofilms. Microorganisms 2023, 11, 143. https://doi.org/10.3390/microorganisms11010143
Mechmechani S, Gharsallaoui A, Karam L, EL Omari K, Fadel A, Hamze M, Chihib N-E. Pepsin and Trypsin Treatment Combined with Carvacrol: An Efficient Strategy to Fight Pseudomonas aeruginosa and Enterococcus faecalis Biofilms. Microorganisms. 2023; 11(1):143. https://doi.org/10.3390/microorganisms11010143
Chicago/Turabian StyleMechmechani, Samah, Adem Gharsallaoui, Layal Karam, Khaled EL Omari, Alexandre Fadel, Monzer Hamze, and Nour-Eddine Chihib. 2023. "Pepsin and Trypsin Treatment Combined with Carvacrol: An Efficient Strategy to Fight Pseudomonas aeruginosa and Enterococcus faecalis Biofilms" Microorganisms 11, no. 1: 143. https://doi.org/10.3390/microorganisms11010143
APA StyleMechmechani, S., Gharsallaoui, A., Karam, L., EL Omari, K., Fadel, A., Hamze, M., & Chihib, N.-E. (2023). Pepsin and Trypsin Treatment Combined with Carvacrol: An Efficient Strategy to Fight Pseudomonas aeruginosa and Enterococcus faecalis Biofilms. Microorganisms, 11(1), 143. https://doi.org/10.3390/microorganisms11010143







