Prevotella: A Key Player in Ruminal Metabolism
Abstract
:1. Introduction
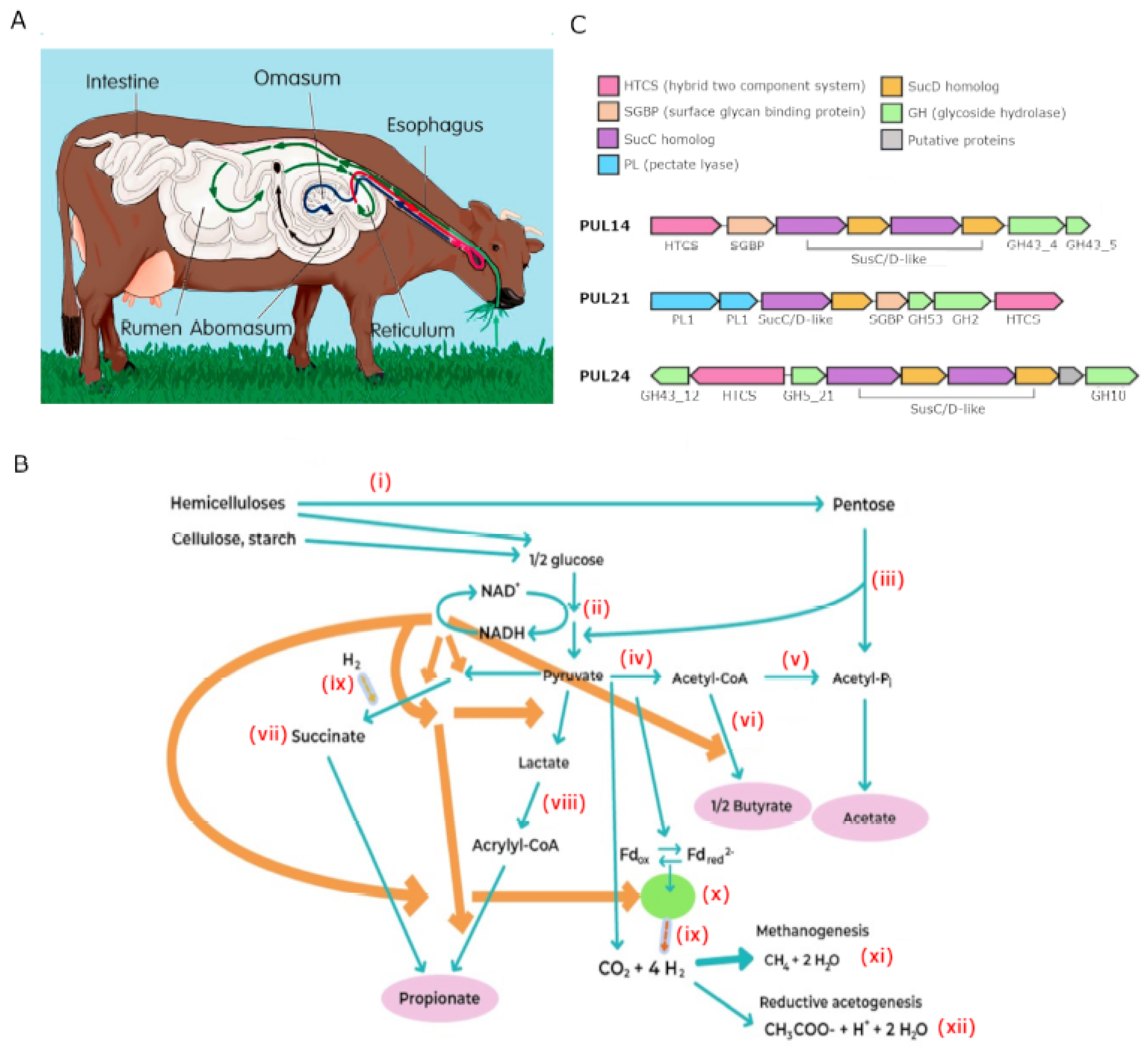
2. Ruminal Carbohydrate Fermentation
3. Role of Prevotella in Lignocellulose Processing
4. Prevotella and Its Association with Reduced Methane Emissions
5. Concluding Remarks
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OTU | Operational taxonomic unit |
| PUL | Polysaccharide utilization loci |
| SCFA | Short-chain fatty acid |
| VFA | Volatile fatty acid |
| H2 | Molecular hydrogen |
| CH4 | Methane |
| NiFe-hydrogenase | A hydrogenase that binds NiFe at its active site |
| FeFe-hydrogenases | A hydrogenase that binds FeFe at its active site |
| Fe-hydrogenases | A hydrogenase that binds Fe at its active site |
| MAG | Metagenome-assembled genome |
| EMP pathway | Embden-Meyerhof-Parnas glycolytic pathway |
| CAZymes | Carbohydrate-active enzymes |
| Methyl-CoM | Methyl coenzyme M |
| MCM | Methylmalonyl CoA mutase |
| CNSL | Cashew nut shell liquid |
| BCM | Bromochloromethane |
| HPF | High content protein and fat |
| KEGG | Kyoto encyclopedia of genes and genomes |
| eggNOG | Clusters of ortholog groups database |
| CAZy | Carbohydrate-active enzymes database |
| NADP-GDH | NADP-specific glutamate dehydrogenase |
| TauE/SafE | Sulfite exporter family protein |
| FeS | Iron-sulfur cluster |
| SufC | ATPase of the ABC superfamily |
| CNSL | Cashew nut shell liquid |
References
- Tett, A.; Pasolli, E.; Masetti, G.; Ercolini, D.; Segata, N. Prevotella Diversity, Niches and Interactions with the Human Host. Nat. Rev. Microbiol. 2021, 19, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E. The Family Prevotellaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 825–827. [Google Scholar]
- Accetto, T.; Avguštin, G. The Diverse and Extensive Plant Polysaccharide Degradative Apparatuses of the Rumen and Hindgut Prevotella Species: A Factor in Their Ubiquity? Syst. Appl. Microbiol. 2019, 42, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-Utilizing Capacity Varies in Prevotella- versus Bacteroides-Dominated Gut Microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.-H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Shibata, K.; Sakamoto, M.; Tomita, S.; Benno, Y. Prevotella copri Sp. Nov. and Prevotella stercorea Sp. Nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2007, 57, 941–946. [Google Scholar] [CrossRef] [Green Version]
- Könönen, E.; Gursoy, U.K. Oral Prevotella Species and Their Connection to Events of Clinical Relevance in Gastrointestinal and Respiratory Tracts. Front. Microbiol. 2021, 12, 798763. [Google Scholar] [CrossRef]
- Richter, H.E.; Carnes, M.U.; Komesu, Y.M.; Lukacz, E.S.; Arya, L.; Bradley, M.; Rogers, R.G.; Sung, V.W.; Siddiqui, N.Y.; Carper, B.; et al. Association between the Urogenital Microbiome and Surgical Treatment Response in Women Undergoing Midurethral Sling Operation for Mixed Urinary Incontinence. Am. J. Obstet. Gynecol. 2022, 226, 93.e1–93.e15. [Google Scholar] [CrossRef]
- Dubourg, G.; Morand, A.; Mekhalif, F.; Godefroy, R.; Corthier, A.; Yacouba, A.; Diakite, A.; Cornu, F.; Cresci, M.; Brahimi, S.; et al. Deciphering the Urinary Microbiota Repertoire by Culturomics Reveals Mostly Anaerobic Bacteria From the Gut. Front. Microbiol. 2020, 11, 513305. [Google Scholar] [CrossRef]
- Thomas-White, K.; Forster, S.C.; Kumar, N.; Van Kuiken, M.; Putonti, C.; Stares, M.D.; Hilt, E.E.; Price, T.K.; Wolfe, A.J.; Lawley, T.D. Culturing of Female Bladder Bacteria Reveals an Interconnected Urogenital Microbiota. Nat. Commun. 2018, 9, 1557. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.-K.; Do, T.-H.; Le, N.-G.; Nguyen, H.-D.; Nguyen, T.-Q.; Le, T.-T.-H.; Truong, N.-H. Understanding the Role of Prevotella Genus in the Digestion of Lignocellulose and Other Substrates in Vietnamese Native Goats’ Rumen by Metagenomic Deep Sequencing. Animals 2021, 11, 3257. [Google Scholar] [CrossRef]
- Dietary, H.-J.; Callaway, R.; Wu, Q.-C.; Wang, W.-K.; Zhang, F.; Li, W.-J.; Wang, Y.-L.; Lv, L.-K.; Yang, H.-J. Dietary Cysteamine Supplementation Remarkably Increased Feed Efficiency and Shifted Rumen Fermentation toward Glucogenic Propionate Production via Enrichment of Prevotella in Feedlot Lambs. Microorganisms 2022, 10, 1105. [Google Scholar]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Emerson, E.L.; Weimer, P.J. Fermentation of Model Hemicelluloses by Prevotella Strains and Butyrivibrio Fibrisolvens in Pure Culture and in Ruminal Enrichment Cultures. Appl. Microbiol. Biotechnol. 2017, 101, 4269–4278. [Google Scholar] [CrossRef]
- Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms 2020, 8, 1584. [Google Scholar] [CrossRef]
- Nagpal, R.; Wang, S.; Solberg Woods, L.C.; Seshie, O.; Chung, S.T.; Shively, C.A.; Register, T.C.; Craft, S.; McClain, D.A.; Yadav, H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-Human Primate, and Human Feces. Front. Microbiol. 2018, 9, 2897. [Google Scholar] [CrossRef] [Green Version]
- Shah, H.N.; Collins, D.M. Prevotella, a New Genus to Include Bacteroides Melaninogenicus and Related Species Formerly Classified in the Genus Bacteroides. Int. J. Syst. Bacteriol. 1990, 40, 205–208. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Duncan, S.H. Bacteroides and Prevotella. In Encyclopedia of Food Microbiology—Reference Work; Lou Tortorello, M., Batt, C.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 203–208. [Google Scholar]
- Rajilić-Stojanović, M.; de Vos, W.M. The First 1000 Cultured Species of the Human Gastrointestinal Microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- Hungate, R.E.; Bryant, M.P.; Mah, R.A. The rumen bacteria and protozoa. Annu. Rev. Microbiol. 1964, 18, 131–166. [Google Scholar] [CrossRef]
- Russell, J.B. Rumen Microbiology and Its Role in Ruminant Nutrition; Department of Microbiology, Cornell University: Ithaca, NY, USA, 2002. [Google Scholar]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The Rumen Microbiome: Balancing Food Security and Environmental Impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Kelly, W.J.; Janssen, P.H.; Attwood, G.T. Rumen Microbial (meta)genomics and Its Application to Ruminant Production. Animal 2013, 7 (Suppl. 1), 184–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hungate, R.E. The Rumen and Its Microbes; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9781483263625. [Google Scholar]
- Pinnell, L.J.; Reyes, A.A.; Wolfe, C.A.; Weinroth, M.D.; Metcalf, J.L.; Delmore, R.J.; Belk, K.E.; Morley, P.S.; Engle, T.E. Bacteroidetes and Firmicutes Drive Differing Microbial Diversity and Community Composition Among Micro-Environments in the Bovine Rumen. Front. Vet. Sci. 2022, 9, 897996. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A Heritable Subset of the Core Rumen Microbiome Dictates Dairy Cow Productivity and Emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svartström, O.; Alneberg, J.; Terrapon, N.; Lombard, V.; de Bruijn, I.; Malmsten, J.; Dalin, A.-M.; El Muller, E.; Shah, P.; Wilmes, P.; et al. Ninety-Nine de Novo Assembled Genomes from the Moose (Alces Alces) Rumen Microbiome Provide New Insights into Microbial Plant Biomass Degradation. ISME J. 2017, 11, 2538–2551. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.; Neves, A.L.A.; Song, Y.; Guan, L.L. The Role of the Gut Microbiome in Cattle Production and Health: Driver or Passenger? Annu. Rev. Anim. Biosci. 2020, 8, 199–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Global Rumen Census Collaborators; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [Green Version]
- Tong, F.; Wang, T.; Gao, N.L.; Liu, Z.; Cui, K.; Duan, Y.; Wu, S.; Luo, Y.; Li, Z.; Yang, C.; et al. The Microbiome of the Buffalo Digestive Tract. Nat. Commun. 2022, 13, 823. [Google Scholar] [CrossRef]
- Xie, F.; Jin, W.; Si, H.; Yuan, Y.; Tao, Y.; Liu, J.; Wang, X.; Yang, C.; Li, Q.; Yan, X.; et al. An Integrated Gene Catalog and over 10,000 Metagenome-Assembled Genomes from the Gastrointestinal Microbiome of Ruminants. Microbiome 2021, 9, 137. [Google Scholar] [CrossRef]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the Bacterial Diversity in the Feces of Cattle Using 16S rDNA Bacterial Tag-Encoded FLX Amplicon Pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Holman, D.B.; Gzyl, K.E. A Meta-Analysis of the Bovine Gastrointestinal Tract Microbiota. FEMS Microbiol. Ecol. 2019, 95, fiz072. [Google Scholar] [CrossRef]
- Furman, O.; Shenhav, L.; Sasson, G.; Kokou, F.; Honig, H.; Jacoby, S.; Hertz, T.; Cordero, O.X.; Halperin, E.; Mizrahi, I. Stochasticity Constrained by Deterministic Effects of Diet and Age Drive Rumen Microbiome Assembly Dynamics. Nat. Commun. 2020, 11, 1904. [Google Scholar] [CrossRef] [Green Version]
- Nathani, N.M.; Kothari, R.K.; Patel, A.K.; Joshi, C.G. Functional Characterization Reveals Novel Putative Coding Sequences in Prevotella Ruminicola Genome Extracted from Rumen Metagenomic Studies. J. Mol. Microbiol. Biotechnol. 2015, 25, 292–299. [Google Scholar] [CrossRef]
- Aguilar-Marin, S.B.; Betancur-Murillo, C.L.; Isaza, G.A.; Mesa, H.; Jovel, J. Lower Methane Emissions Were Associated with Higher Abundance of Ruminal Prevotella in a Cohort of Colombian Buffalos. BMC Microbiol. 2020, 20, 364. [Google Scholar] [CrossRef]
- Singh, K.M.; Reddy, B.; Patel, A.K.; Panchasara, H.; Parmar, N.; Patel, A.B.; Shah, T.M.; Bhatt, V.D.; Joshi, C.G. Metagenomic Analysis of Buffalo Rumen Microbiome: Effect of Roughage Diet on Dormancy and Sporulation Genes. Meta Gene 2014, 2, 252–268. [Google Scholar] [CrossRef]
- Kala, A.; Kamra, D.N.; Kumar, A.; Agarwal, N.; Chaudhary, L.C.; Joshi, C.G. Impact of Levels of Total Digestible Nutrients on Microbiome, Enzyme Profile and Degradation of Feeds in Buffalo Rumen. PLoS ONE 2017, 12, e0172051. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Gu, Q.; Zhou, X.; Xia, Z.; Muhammad, W.I.; Tang, Q.; Liang, M.; Lin, B.; Qin, G. Ruminal Microbiota Composition Associated with Ruminal Fermentation Parameters and Milk Yield in Lactating Buffalo in Guangxi, China-A Preliminary Study. J. Anim. Physiol. Anim. Nutr. 2019, 103, 1374–1379. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, P.; Wang, L.; Zhao, Z.; Chen, Y.; Yang, Y. Bacterial Community Diversity Associated with Different Levels of Dietary Nutrition in the Rumen of Sheep. Appl. Microbiol. Biotechnol. 2017, 101, 3717–3728. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, K.; Zhang, C.; Feng, Y.; Zhang, X.; Wang, X.; Wu, G. Dynamics and Stabilization of the Rumen Microbiome in Yearling Tibetan Sheep. Sci. Rep. 2019, 9, 19620. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Zhang, G.; Liu, Z.; Wu, P.; Yu, Z.; Wang, J. Repeated Inoculation with Fresh Rumen Fluid before or during Weaning Modulates the Microbiota Composition and Co-Occurrence of the Rumen and Colon of Lambs. BMC Microbiol. 2020, 20, 29. [Google Scholar] [CrossRef] [Green Version]
- Wallace, R.J.; McKain, N.; Broderick, G.A. Breakdown of Different Peptides by Prevotella (Bacteroides) Ruminicola and Mixed Microorganisms from the Sheep Rumen. Curr. Microbiol. 1993, 26, 333–336. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra Hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [Green Version]
- Wetzels, S.U.; Mann, E.; Metzler-Zebeli, B.U.; Wagner, M.; Klevenhusen, F.; Zebeli, Q.; Schmitz-Esser, S. Pyrosequencing Reveals Shifts in the Bacterial Epimural Community Relative to Dietary Concentrate Amount in Goats. J. Dairy Sci. 2015, 98, 5572–5587. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Chai, J.; Diao, Q.; Huang, W.; Zhuang, Y.; Zhang, N. The Signature Microbiota Drive Rumen Function Shifts in Goat Kids Introduced to Solid Diet Regimes. Microorganisms 2019, 7, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Su, J.; Li, F.; Tian, X.; Liu, Z.; Ding, G.; Bai, J.; Li, Z.; Ma, Z.; Peppelenbosch, M.P. Yak Gut Microbiota: A Systematic Review and Meta-Analysis. Front. Vet. Sci. 2022, 9, 889594. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhou, M.; Ma, T.; Bi, S.; Wang, W.; Zhang, Y.; Huang, X.; Guan, L.L.; Long, R. Survey of Rumen Microbiota of Domestic Grazing Yak during Different Growth Stages Revealed Novel Maturation Patterns of Four Key Microbial Groups and Their Dynamic Interactions. Anim. Microbiome 2020, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Chai, Z.; Zhang, C.; Zhang, Q.; Zhu, Y.; Cao, H.; Zhong, J.; Ji, Q. Comparing the Microbial Community in Four Stomach of Dairy Cattle, Yellow Cattle and Three Yak Herds in Qinghai-Tibetan Plateau. Front. Microbiol. 2019, 10, 1547. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Wang, L.; Ke, S.; Chen, X.; Kenéz, Á.; Xu, W.; Wang, D.; Zhang, F.; Li, Y.; Cui, Z.; et al. Yak Rumen Microbiome Elevates Fiber Degradation Ability and Alters Rumen Fermentation Pattern to Increase Feed Efficiency. Anim. Nutr. 2022, 11, 201–214. [Google Scholar] [CrossRef]
- Li, Z.P.; Liu, H.L.; Li, G.Y.; Bao, K.; Wang, K.Y.; Xu, C.; Yang, Y.F.; Yang, F.H.; Wright, A.-D.G. Molecular Diversity of Rumen Bacterial Communities from Tannin-Rich and Fiber-Rich Forage Fed Domestic Sika Deer (Cervus Nippon) in China. BMC Microbiol. 2013, 13, 151. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Sun, Y.; Shi, Z.; Liu, Z.; Zhao, C.; Lu, T.; Gao, H.; Zhu, F.; Chen, R.; Zhang, J.; et al. Gut Microbiota of Wild and Captive Alpine Musk Deer (Moschus Chrysogaster). Front. Microbiol. 2019, 10, 3156. [Google Scholar] [CrossRef]
- Gruninger, R.J.; Sensen, C.W.; McAllister, T.A.; Forster, R.J. Diversity of Rumen Bacteria in Canadian Cervids. PLoS ONE 2014, 9, e89682. [Google Scholar] [CrossRef]
- Belanche, A.; Patra, A.K.; Morgavi, D.P.; Suen, G.; Newbold, C.J.; Yanez-Ruiz, D.R. Gut Microbiome Modulation in Ruminants: Enhancing Advantages and Minimizing Drawbacks; Frontiers Media SA: Lausanne, Switzerland, 2021; ISBN 9782889664832. [Google Scholar]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial Community Structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.N.; Méndez–García, C.; Geier, R.R.; Iakiviak, M.; Chang, J.; Cann, I.; Mackie, R.I. Metabolic Networks for Nitrogen Utilization in Prevotella Ruminicola 23. Sci. Rep. 2017, 7, 7851. [Google Scholar] [CrossRef]
- Tsuda, T.; Sasaki, Y.; Kawashima, R. Physiological Aspects of Digestion and Metabolism in Ruminants: Proceedings of the Seventh International Symposium on Ruminant Physiology; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780323138611. [Google Scholar]
- Wallace, R.J.; McKain, N.; Broderick, G.A.; Rode, L.M.; Walker, N.D.; Newbold, C.J.; Kopecny, J. Peptidases of the Rumen Bacterium, Prevotella Ruminicola. Anaerobe 1997, 3, 35–42. [Google Scholar] [CrossRef]
- Miyazaki, K.; Martin, J.C.; Marinsek-Logar, R.; Flint, H.J. Degradation and Utilization of Xylans by the Rumen AnaerobePrevotella bryantii(formerlyP. Ruminicolasubsp.brevis) B14. Anaerobe 1997, 3, 373–381. [Google Scholar] [CrossRef]
- Wolin, M.J. The Rumen Fermentation: A Model for Microbial Interactions in Anaerobic Ecosystems. In Advances in Microbial Ecology: Volume 3; Alexander, M., Ed.; Springer: Boston, MA, USA, 1979; pp. 49–77. ISBN 9781461582793. [Google Scholar]
- Hobson, P.N.; Stewart, C.S. The Rumen Microbial Ecosystem; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 9789400914537. [Google Scholar]
- Dodd, D.; Moon, Y.-H.; Swaminathan, K.; Mackie, R.I.; Cann, I.K.O. Transcriptomic Analyses of Xylan Degradation by Prevotella Bryantii and Insights into Energy Acquisition by Xylanolytic Bacteroidetes. J. Biol. Chem. 2010, 285, 30261–30273. [Google Scholar] [CrossRef] [Green Version]
- Kabel, M.A.; Yeoman, C.J.; Han, Y.; Dodd, D.; Abbas, C.A.; de Bont, J.A.M.; Morrison, M.; Cann, I.K.O.; Mackie, R.I. Biochemical Characterization and Relative Expression Levels of Multiple Carbohydrate Esterases of the Xylanolytic Rumen Bacterium Prevotella Ruminicola 23 Grown on an Ester-Enriched Substrate. Appl. Environ. Microbiol. 2011, 77, 5671–5681. [Google Scholar] [CrossRef] [Green Version]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella Copri Isolates. Cell Host Microbe 2019, 26, 680–690.e5. [Google Scholar] [CrossRef]
- Grondin, J.M.; Tamura, K.; Déjean, G.; Abbott, D.W.; Brumer, H. Polysaccharide Utilization Loci: Fueling Microbial Communities. J. Bacteriol. 2017, 199, e00860-16. [Google Scholar] [CrossRef] [Green Version]
- Gálvez, E.J.C.; Iljazovic, A.; Amend, L.; Lesker, T.R.; Renault, T.; Thiemann, S.; Hao, L.; Roy, U.; Gronow, A.; Charpentier, E.; et al. Distinct Polysaccharide Utilization Determines Interspecies Competition between Intestinal Prevotella spp. Cell Host Microbe 2020, 28, 838–852.e6. [Google Scholar] [CrossRef]
- Strobel, H.J. Vitamin B12-Dependent Propionate Production by the Ruminal Bacterium Prevotella Ruminicola 23. Appl. Environ. Microbiol. 1992, 58, 2331–2333. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Huang, S.; Huang, J.; Peng, P.; Liu, Y.; Han, B.; Sun, D. Identification of the Potential Role of the Rumen Microbiome in Milk Protein and Fat Synthesis in Dairy Cows Using Metagenomic Sequencing. Animals 2021, 11, 1247. [Google Scholar] [CrossRef]
- Takahashi, N.; Yamada, T. Pathways for Amino Acid Metabolism by Prevotella Intermedia and Prevotella Nigrescens. Oral Microbiol. Immunol. 2000, 15, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Griswold, K.E.; Mackie, R.I. Degradation of Protein and Utilization of the Hydrolytic Products by a Predominant Ruminal Bacterium, Prevotella Ruminicola B14. J. Dairy Sci. 1997, 80, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.D.; Prather, M.J.; Søvde, O.A.; Myhre, G. Future Methane, Hydroxyl, and Their Uncertainties: Key Climate and Emission Parameters for Future Predictions. Atmos. Chem. Phys. 2013, 13, 285–302. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.A.; Johnson, D.E. Methane Emissions from Cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Mayne, C.S.; Gordon, F.G.; Porter, M.G.; Agnew, R.E.; Patterson, D.C.; Ferris, C.P.; Kilpatrick, D.J. Mitigation of Enteric Methane Emissions through Improving Efficiency of Energy Utilization and Productivity in Lactating Dairy Cows. J. Dairy Sci. 2010, 93, 2630–2638. [Google Scholar] [CrossRef] [Green Version]
- Ungerfeld, E.M. Metabolic Hydrogen Flows in Rumen Fermentation: Principles and Possibilities of Interventions. Front. Microbiol. 2020, 11, 589. [Google Scholar] [CrossRef] [Green Version]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse Hydrogen Production and Consumption Pathways Influence Methane Production in Ruminants. ISME J. 2019, 13, 2617–2632. [Google Scholar] [CrossRef]
- Lan, W.; Yang, C. Ruminal Methane Production: Associated Microorganisms and the Potential of Applying Hydrogen-Utilizing Bacteria for Mitigation. Sci. Total Environ. 2019, 654, 1270–1283. [Google Scholar] [CrossRef]
- Baldwin, R.L.; Allison, M.J. Rumen Metabolism. J. Anim. Sci. 1983, 57 (Suppl. 2), 461–477. [Google Scholar]
- Sutton, J.D. Carbohydrate Digestion and Glucose Supply in the Gut of the Ruminant. Proc. Nutr. Soc. 1971, 30, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Andrieu, S.; Wilde, D. Gut Efficiency; the Key Ingredient in Ruminant Production: Elevating Animal Performance and Health; Wageningen Academic Publishers: Wageningen, The Netherlands, 2008; ISBN 9789086866403. [Google Scholar]
- Fettke, J.; Fernie, A.R. Intracellular and Cell-to-Apoplast Compartmentation of Carbohydrate Metabolism. Trends Plant Sci. 2015, 20, 490–497. [Google Scholar] [CrossRef]
- Leegood, R.C.; Sharkey, T.D.; Von Caemmerer, S.; von Caemmerer, S. Photosynthesis: Physiology and Metabolism; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2000; ISBN 9780792361435. [Google Scholar]
- Umphrey, J.E.; Staples, C.R. General Anatomy of the Ruminant Digestive System; University of Florida, Cooperative Extension Service, Institute of Foods and Agriculture Sciences (EDIS): Gainesville, FL, USA, 1992. [Google Scholar]
- Russell, J.B.; Hespell, R.B. Microbial Rumen Fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]
- Murphy, M.R. Nutrition, Digestion and Absorption: Fermentation in the Rumen. In Encyclopedia of Dairy Sciences (Third Edition); McSweeney, P.L.H., McNamara, J.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 93–97. [Google Scholar]
- McSweeney, P.L.H.; McNamara, J.P. (Eds.) Encyclopedia of Dairy Sciences—Reference Work; Elsevier: San Diego, CA, USA, 2022; ISBN 9780128187678. [Google Scholar]
- Li, J.; Gálvez, E.J.C.; Amend, L.; Almási, É.; Iljazovic, A.; Lesker, T.R.; Bielecka, A.A.; Schorr, E.-M.; Strowig, T. A Versatile Genetic Toolbox for Prevotella Copri Enables Studying Polysaccharide Utilization Systems. EMBO J. 2021, 40, e108287. [Google Scholar] [CrossRef]
- Puniya, A.K.; Singh, R.; Kamra, D.N. Rumen Microbiology: From Evolution to Revolution; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 9788132224013. [Google Scholar]
- Maekawa, M.; Beauchemin, K.A.; Christensen, D.A. Chewing Activity, Saliva Production, and Ruminal pH of Primiparous and Multiparous Lactating Dairy Cows. J. Dairy Sci. 2002, 85, 1176–1182. [Google Scholar] [CrossRef]
- Moran, J. Tropical Dairy Farming: Feeding Management for Small Holder Dairy Farmers in Humid Tropics; Csiro Publishing: Clayton, Australia, 2005. [Google Scholar]
- Leng, R.A. The Rumen—A Fermentation Vat or a Series of Organized Structured Microbial Consortia: Implications for the Mitigation of Enteric Methane Production by Feed Additives. Livestock Res. Rural Dev. 2011, 23, 258. [Google Scholar]
- Nakamura, S.; Nakashio, S.; Yamakawa, K.; Tanabe, N.; Nishida, S. Carbohydrate Fermentation by Clostridium Difficile. Microbiol. Immunol. 1982, 26, 107–111. [Google Scholar] [CrossRef]
- Annison, E.F.; Lewis, D. Metabolism in the Rumen; John Wiley & Sons: Hoboken, NJ, USA, 1959. [Google Scholar]
- Hamar, D.; Borchers, R. Glycolytic Pathway in Rumen Microorganisms. J. Anim. Sci. 1967, 26, 654–657. [Google Scholar] [CrossRef]
- Romano, A.H.; Conway, T. Evolution of Carbohydrate Metabolic Pathways. Res. Microbiol. 1996, 147, 448–455. [Google Scholar] [CrossRef]
- Ibrahim, R.; Varin, L.; De Luca, V.; Romeo, J. Evolution of Metabolic Pathways; Elsevier: Amsterdam, The Netherlands, 2000; ISBN 9780080531328. [Google Scholar]
- Ungerfeld, E.M.; James Newbold, C. Engineering Rumen Metabolic Pathways: Where We Are, and Where Are We Heading; Frontiers Media SA: Lausanne, Switzerland, 2018; ISBN 9782889454266. [Google Scholar]
- Thurston, B.; Dawson, K.A.; Strobel, H.J. Pentose Utilization by the Ruminal Bacterium Ruminococcus Albus. Appl. Environ. Microbiol. 1994, 60, 1087–1092. [Google Scholar] [CrossRef] [Green Version]
- McAllister, T.A.; Newbold, C.J. Redirecting Rumen Fermentation to Reduce Methanogenesis. Aust. J. Exp. Agric. 2008, 48, 7–13. [Google Scholar] [CrossRef]
- Baldwin, R.L., 6th; Connor, E.E. Rumen Function and Development. Vet. Clin. N. Am. Food Anim. Pract. 2017, 33, 427–439. [Google Scholar] [CrossRef]
- Karekar, S.; Stefanini, R.; Ahring, B. Homo-Acetogens: Their Metabolism and Competitive Relationship with Hydrogenotrophic Methanogens. Microorganisms 2022, 10, 397. [Google Scholar] [CrossRef]
- Harrison, J.J.; Turner, R.J.; Marques, L.L.R.; Ceri, H. Biofilms: A New Understanding of These Microbial Communities Is Driving a Revolution That May Transform the Science of Microbiology. Am. Sci. 2005, 93, 508–515. [Google Scholar] [CrossRef]
- Tapio, I.; Snelling, T.J.; Strozzi, F.; Wallace, R.J. The Ruminal Microbiome Associated with Methane Emissions from Ruminant Livestock. J. Anim. Sci. Biotechnol. 2017, 8, 7. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Álvaro, M.; Auffret, M.D.; Stewart, R.D.; Dewhurst, R.J.; Duthie, C.-A.; Rooke, J.A.; Wallace, R.J.; Shih, B.; Freeman, T.C.; Watson, M.; et al. Identification of Complex Rumen Microbiome Interaction Within Diverse Functional Niches as Mechanisms Affecting the Variation of Methane Emissions in Bovine. Front. Microbiol. 2020, 11, 659. [Google Scholar] [CrossRef] [PubMed]
- Sejian, V.; Gaughan, J.; Baumgard, L.; Prasad, C. (Eds.) Climate Change Impact on Livestock: Adaptation and Mitigation; Springer: New Delhi, India, 2015; ISBN 9788132222651. [Google Scholar]
- Loor, J.J.; Elolimy, A.A.; McCann, J.C. Dietary Impacts on Rumen Microbiota in Beef and Dairy Production. Anim. Front. 2016, 6, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific Microbiome-Dependent Mechanisms Underlie the Energy Harvest Efficiency of Ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moe, P.W. Energy Metabolism of Dairy Cattle. J. Dairy Sci. 1981, 64, 1120–1139. [Google Scholar] [CrossRef] [PubMed]
- McBride, B.W.; Kelly, J.M. Energy Cost of Absorption and Metabolism in the Ruminant Gastrointestinal Tract and Liver: A Review. J. Anim. Sci. 1990, 68, 2997–3010. [Google Scholar] [CrossRef]
- Oltjen, J.W.; Kebreab, E.; Lapierre, H. Energy and Protein Metabolism and Nutrition in Sustainable Animal Production; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9789086867813. [Google Scholar]
- Pryce, J.E.; Arias, J.; Bowman, P.J.; Davis, S.R.; Macdonald, K.A.; Waghorn, G.C.; Wales, W.J.; Williams, Y.J.; Spelman, R.J.; Hayes, B.J. Accuracy of Genomic Predictions of Residual Feed Intake and 250-Day Body Weight in Growing Heifers Using 625,000 Single Nucleotide Polymorphism Markers. J. Dairy Sci. 2012, 95, 2108–2119. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.S.; Mujibi, F.D.; Sherman, E.L. Molecular Basis for Residual Feed Intake in Beef Cattle. J. Anim. Sci. 2009, 87, E41–E47. [Google Scholar] [CrossRef] [Green Version]
- Rubino, F.; Carberry, C.; Waters, S.M.; Kenny, D.; McCabe, M.S.; Creevey, C.J. Divergent Functional Isoforms Drive Niche Specialisation for Nutrient Acquisition and Use in Rumen Microbiome. ISME J. 2017, 11, 932–944. [Google Scholar] [CrossRef] [Green Version]
- Sasson, G.; Kruger Ben-Shabat, S.; Seroussi, E.; Doron-Faigenboim, A.; Shterzer, N.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Halperin, E.; Mizrahi, I. Heritable Bovine Rumen Bacteria Are Phylogenetically Related and Correlated with the Cow’s Capacity to Harvest Energy from Its Feed. MBio 2017, 8, e00703-17. [Google Scholar] [CrossRef]
- Sasson, G.; Moraïs, S.; Kokou, F.; Plate, K.; Trautwein-Schult, A.; Jami, E.; Bayer, E.A.; Becher, D.; Mizrahi, I. Metaproteome Plasticity Sheds Light on the Ecology of the Rumen Microbiome and Its Connection to Host Traits. ISME J. 2022, 16, 2610–2621. [Google Scholar] [CrossRef]
- Roehe, R.; Dewhurst, R.J.; Duthie, C.-A.; Rooke, J.A.; McKain, N.; Ross, D.W.; Hyslop, J.J.; Waterhouse, A.; Freeman, T.C.; Watson, M.; et al. Bovine Host Genetic Variation Influences Rumen Microbial Methane Production with Best Selection Criterion for Low Methane Emitting and Efficiently Feed Converting Hosts Based on Metagenomic Gene Abundance. PLoS Genet. 2016, 12, e1005846. [Google Scholar] [CrossRef]
- Foley, M.H.; Cockburn, D.W.; Koropatkin, N.M. The Sus Operon: A Model System for Starch Uptake by the Human Gut Bacteroidetes. Cell. Mol. Life Sci. 2016, 73, 2603–2617. [Google Scholar] [CrossRef] [Green Version]
- Terrapon, N.; Lombard, V.; Drula, É.; Lapébie, P.; Al-Masaudi, S.; Gilbert, H.J.; Henrissat, B. PULDB: The Expanded Database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018, 46, D677–D683. [Google Scholar] [CrossRef]
- Ausland, C.; Zheng, J.; Yi, H.; Yang, B.; Li, T.; Feng, X.; Zheng, B.; Yin, Y. dbCAN-PUL: A Database of Experimentally Characterized CAZyme Gene Clusters and Their Substrates. Nucleic Acids Res. 2021, 49, D523–D528. [Google Scholar] [CrossRef]
- Terrapon, N.; Lombard, V.; Gilbert, H.J.; Henrissat, B. Automatic Prediction of Polysaccharide Utilization Loci in Bacteroidetes Species. Bioinformatics 2015, 31, 647–655. [Google Scholar] [CrossRef] [Green Version]
- Stewart, R.D.; Auffret, M.D.; Roehe, R.; Watson, M. Open Prediction of Polysaccharide Utilisation Loci (PUL) in 5414 Public Bacteroidetes Genomes Using PULpy. bioRxiv 2018, 421024. [Google Scholar] [CrossRef] [Green Version]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Accetto, T.; Avguštin, G. Polysaccharide Utilization Locus and CAZYme Genome Repertoires Reveal Diverse Ecological Adaptation of Prevotella Species. Syst. Appl. Microbiol. 2015, 38, 453–461. [Google Scholar] [CrossRef] [PubMed]
- McGregor, N.; Morar, M.; Fenger, T.H.; Stogios, P.; Lenfant, N.; Yin, V.; Xu, X.; Evdokimova, E.; Cui, H.; Henrissat, B.; et al. Structure-Function Analysis of a Mixed-Linkage β-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella Bryantii B(1)4. J. Biol. Chem. 2016, 291, 1175–1197. [Google Scholar] [CrossRef] [PubMed]
- Rosewarne, C.P.; Pope, P.B.; Cheung, J.L.; Morrison, M. Analysis of the Bovine Rumen Microbiome Reveals a Diversity of Sus-like Polysaccharide Utilization Loci from the Bacterial Phylum Bacteroidetes. J. Ind. Microbiol. Biotechnol. 2014, 41, 601–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naas, A.E.; Mackenzie, A.K.; Mravec, J.; Schückel, J.; Willats, W.G.T.; Eijsink, V.G.H.; Pope, P.B. Do Rumen Bacteroidetes Utilize an Alternative Mechanism for Cellulose Degradation? MBio 2014, 5, e01401–e01414. [Google Scholar] [CrossRef] [Green Version]
- Glendinning, L.; Genç, B.; Wallace, R.J.; Watson, M. Metagenomic Analysis of the Cow, Sheep, Reindeer and Red Deer Rumen. Sci. Rep. 2021, 11, 1990. [Google Scholar] [CrossRef]
- Ferrer, M.; Golyshina, O.V.; Chernikova, T.N.; Khachane, A.N.; Reyes-Duarte, D.; Santos, V.; Martins Dos, A.P.; Strompl, C.; Elborough, K.; Jarvis, G.; et al. Novel Hydrolase Diversity Retrieved from a Metagenome Library of Bovine Rumen Microflora. Environ. Microbiol. 2005, 7, 1996–2010. [Google Scholar] [CrossRef]
- Mackenzie, A.K.; Naas, A.E.; Kracun, S.K.; Schückel, J.; Fangel, J.U.; Agger, J.W.; Willats, W.G.T.; Eijsink, V.G.H.; Pope, P.B. A Polysaccharide Utilization Locus from an Uncultured Bacteroidetes Phylotype Suggests Ecological Adaptation and Substrate Versatility. Appl. Environ. Microbiol. 2015, 81, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Pope, P.B.; Mackenzie, A.K.; Gregor, I.; Smith, W.; Sundset, M.A.; McHardy, A.C.; Morrison, M.; Eijsink, V.G.H. Metagenomics of the Svalbard Reindeer Rumen Microbiome Reveals Abundance of Polysaccharide Utilization Loci. PLoS ONE 2012, 7, e38571. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Bayer, E.A. Plant Cell Wall Breakdown by Anaerobic Microorganisms from the Mammalian Digestive Tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef]
- Conte, G.; Dimauro, C.; Daghio, M.; Serra, A.; Mannelli, F.; McAmmond, B.M.; Van Hamme, J.D.; Buccioni, A.; Viti, C.; Mantino, A.; et al. Exploring the Relationship between Bacterial Genera and Lipid Metabolism in Bovine Rumen. Animal 2022, 16, 100520. [Google Scholar] [CrossRef]
- Hu, R.; Zou, H.; Wang, H.; Wang, Z.; Wang, X.; Ma, J.; Shah, A.M.; Peng, Q.; Xue, B.; Wang, L.; et al. Dietary Energy Levels Affect Rumen Bacterial Populations That Influence the Intramuscular Fat Fatty Acids of Fattening Yaks (Bos Grunniens). Animals 2020, 10, 1474. [Google Scholar] [CrossRef]
- Atasoglu, C.; Valdés, C.; Walker, N.D.; Newbold, C.J.; Wallace, R.J. De Novo Synthesis of Amino Acids by the Ruminal Bacteria Prevotella Bryantii B14, Selenomonas Ruminantium HD4, and Streptococcus Bovis ES1. Appl. Environ. Microbiol. 1998, 64, 2836–2843. [Google Scholar] [CrossRef]
- Franco-Lopez, J.; Duplessis, M.; Bui, A.; Reymond, C.; Poisson, W.; Blais, L.; Chong, J.; Gervais, R.; Rico, D.E.; Cue, R.I.; et al. Correlations between the Composition of the Bovine Microbiota and Vitamin B12 Abundance. mSystems 2020, 5, e00107-20. [Google Scholar] [CrossRef] [Green Version]
- Hertzberg, M.; Siddons, A.; Schreuder, H. Role of Greenhouse Gases in Climate Change. Energy Environ. 2017, 28, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Hoegh-Guldberg, O.; Poloczanska, E.S.; Skirving, W.; Dove, S. Coral Reef Ecosystems under Climate Change and Ocean Acidification. Front. Mar. Sci. 2017, 4, 158. [Google Scholar] [CrossRef] [Green Version]
- Cornwall, C.E.; Comeau, S.; Kornder, N.A.; Perry, C.T.; van Hooidonk, R.; DeCarlo, T.M.; Pratchett, M.S.; Anderson, K.D.; Browne, N.; Carpenter, R.; et al. Global Declines in Coral Reef Calcium Carbonate Production under Ocean Acidification and Warming. Proc. Natl. Acad. Sci. USA 2021, 118, e2015265118. [Google Scholar] [CrossRef]
- Buccioni, A.; Cappucci, A.; Mele, M. Methane Emission from Enteric Fermentation: Methanogenesis and Fermentation. In Climate Change Impact on Livestock: Adaptation and Mitigation; Sejian, V., Gaughan, J., Baumgard, L., Prasad, C., Eds.; Springer: New Delhi, India, 2015; pp. 171–186. ISBN 9788132222651. [Google Scholar]
- Bhatta, R.; Enishi, O. Measurement of Methane Production from Ruminants. Asian-Australas. J. Anim. Sci. 2007, 20, 1305–1318. [Google Scholar] [CrossRef]
- Moss, A.R.; Jouany, J.-P.; Newbold, J. Methane Production by Ruminants: Its Contribution to Global Warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef] [Green Version]
- Seid, A. Review on Methane Emission from Dairy Farms and Its Impact on Global Warming. Austin J. Vet. Sci. Anim. Husb. 2019, 6, 1052–1056. [Google Scholar] [CrossRef]
- Lyu, Z.; Shao, N.; Akinyemi, T.; Whitman, W.B. Methanogenesis. Curr. Biol. 2018, 28, R727–R732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appuhamy, J.A.D.R.N.; France, J.; Kebreab, E. Models for Predicting Enteric Methane Emissions from Dairy Cows in North America, Europe, and Australia and New Zealand. Glob. Chang. Biol. 2016, 22, 3039–3056. [Google Scholar] [CrossRef] [PubMed]
- Neill, A.R.; Grime, D.W.; Dawson, R.M. Conversion of Choline Methyl Groups through Trimethylamine into Methane in the Rumen. Biochem. J 1978, 170, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.; Schwab, C.; Jensen, B.B.; Engberg, R.M.; Spang, A.; Canibe, N.; Højberg, O.; Milinovich, G.; Fragner, L.; Schleper, C.; et al. Methylotrophic Methanogenic Thermoplasmata Implicated in Reduced Methane Emissions from Bovine Rumen. Nat. Commun. 2013, 4, 1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, A.; Sharma, A. Use of Proteomics and Transcriptomics to Identify Proteins for Cold Adaptation in Microbes. In Survival Strategies in Cold-Adapted Microorganisms; Springer: Singapore, 2022; pp. 285–319. [Google Scholar]
- Liu, Y.; Whitman, W.B. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- Denman, S.E.; Tomkins, N.W.; McSweeney, C.S. Quantitation and Diversity Analysis of Ruminal Methanogenic Populations in Response to the Antimethanogenic Compound Bromochloromethane. FEMS Microbiol. Ecol. 2007, 62, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Goel, G.; Makkar, H.P.S.; Becker, K. Inhibition of Methanogens by Bromochloromethane: Effects on Microbial Communities and Rumen Fermentation Using Batch and Continuous Fermentations. Br. J. Nutr. 2009, 101, 1484–1492. [Google Scholar] [CrossRef] [Green Version]
- Denman, S.E.; Martinez Fernandez, G.; Shinkai, T.; Mitsumori, M.; McSweeney, C.S. Metagenomic Analysis of the Rumen Microbial Community Following Inhibition of Methane Formation by a Halogenated Methane Analog. Front. Microbiol. 2015, 6, 1087. [Google Scholar] [CrossRef] [Green Version]
- Mitsumori, M.; Shinkai, T.; Takenaka, A.; Enishi, O.; Higuchi, K.; Kobayashi, Y.; Nonaka, I.; Asanuma, N.; Denman, S.E.; McSweeney, C.S. Responses in Digestion, Rumen Fermentation and Microbial Populations to Inhibition of Methane Formation by a Halogenated Methane Analogue. Br. J. Nutr. 2012, 108, 482–491. [Google Scholar] [CrossRef]
- Purushe, J.; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, K.E. Comparative Genome Analysis of Prevotella Ruminicola and Prevotella Bryantii: Insights into Their Environmental Niche. Microb. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Takahashi-Iñiguez, T.; García-Hernandez, E.; Arreguín-Espinosa, R.; Flores, M.E. Role of Vitamin B12 on Methylmalonyl-CoA Mutase Activity. J. Zhejiang Univ. Sci. B 2012, 13, 423–437. [Google Scholar] [CrossRef] [Green Version]
- Daugherty, M.S.; Galyean, M.L.; Hallford, D.M.; Hageman, J.H. Vitamin B12 and monensin effects on performance, liver and serum vitamin B12 concentrations and activity of propionate metabolizing hepatic enzymes in feedlot lambs. J. Anim. Feed Sci. 1986, 62, 452–463. [Google Scholar] [CrossRef]
- Islam, M.; Kim, S.-H.; Ramos, S.C.; Mamuad, L.L.; Son, A.-R.; Yu, Z.; Lee, S.-S.; Cho, Y.-I.; Lee, S.-S. Holstein and Jersey Steers Differ in Rumen Microbiota and Enteric Methane Emissions Even Fed the Same Total Mixed Ration. Front. Microbiol. 2021, 12, 601061. [Google Scholar] [CrossRef]
- Tamori, K.; Matsunaga, B.; Boonsaen, P.; Khongpradit, A.; Sawanon, S.; Nagashima, K.; Koike, S.; Kobayashi, Y. Feeding Cashew Nut Shell Liquid Decreases Methane Production from Feces by Altering Fecal Bacterial and Archaeal Communities in Thai Local Ruminants. Anim. Sci. J. 2021, 92, e13569. [Google Scholar] [CrossRef]
- Su, C.; Shinkai, T.; Miyazawa, N.; Mitsumori, M.; Enishi, O.; Nagashima, K.; Koike, S.; Kobayashi, Y. Microbial Community Structure of the Bovine Rumen as Affected by Feeding Cashew Nut Shell Liquid, a Methane-inhibiting and Propionate-enhancing Agent. Anim. Sci. J. 2021, 92, e13503. [Google Scholar] [CrossRef]
- Watanabe, Y.; Suzuki, R.; Koike, S.; Nagashima, K.; Mochizuki, M.; Forster, R.J.; Kobayashi, Y. In Vitro Evaluation of Cashew Nut Shell Liquid as a Methane-Inhibiting and Propionate-Enhancing Agent for Ruminants. J. Dairy Sci. 2010, 93, 5258–5267. [Google Scholar] [CrossRef] [Green Version]
- Narabe, C.; Kamiyama, S.; Saito, M.; Boonsaen, P.; Khongpradit, A.; Sawanon, S.; Suzuki, Y.; Koike, S.; Kobayashi, Y. Cashew Nut Shell Liquid Potentially Mitigates Methane Emission from the Feces of Thai Native Ruminant Livestock by Modifying Fecal Microbiota. Anim. Sci. J. 2021, 92, e13614. [Google Scholar] [CrossRef]
- Shinkai, T.; Enishi, O.; Mitsumori, M.; Higuchi, K.; Kobayashi, Y.; Takenaka, A.; Nagashima, K.; Mochizuki, M.; Kobayashi, Y. Mitigation of Methane Production from Cattle by Feeding Cashew Nut Shell Liquid. J. Dairy Sci. 2012, 95, 5308–5316. [Google Scholar] [CrossRef] [Green Version]
- Veneman, J.B.; Muetzel, S.; Hart, K.J.; Faulkner, C.L.; Moorby, J.M.; Perdok, H.B.; Newbold, C.J. Does Dietary Mitigation of Enteric Methane Production Affect Rumen Function and Animal Productivity in Dairy Cows? PLoS ONE 2015, 10, e0140282. [Google Scholar] [CrossRef]
- Dixit, S.; Kumar, S.; Sharma, R.; Banakar, P.S.; Deb, R.; Tyagi, A.K. Rumen Microbial Diversity, Enteric Methane Emission and Nutrient Utilization of Crossbred Karan-Fries Cattle (Bos Taurus) and Murrah Buffalo (Bubalus Bubalis) Consuming Varied Roughage Concentrate Ratio. Anim. Biotechnol. 2022, 1–19. [Google Scholar] [CrossRef]
- Ross, E.M.; Moate, P.J.; Marett, L.; Cocks, B.G.; Hayes, B.J. Investigating the Effect of Two Methane-Mitigating Diets on the Rumen Microbiome Using Massively Parallel Sequencing. J. Dairy Sci. 2013, 96, 6030–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weimer, P.J.; Cox, M.S.; de Paula, T.V.; Lin, M.; Hall, M.B.; Suen, G. Transient Changes in Milk Production Efficiency and Bacterial Community Composition Resulting from near-Total Exchange of Ruminal Contents between High- and Low-Efficiency Holstein Cows. J. Dairy Sci. 2017, 100, 7165–7182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weimer, P.J.; Stevenson, D.M.; Mantovani, H.C.; Man, S.L.C. Host Specificity of the Ruminal Bacterial Community in the Dairy Cow Following near-Total Exchange of Ruminal Contents. J. Dairy Sci. 2010, 93, 5902–5912. [Google Scholar] [CrossRef] [PubMed]
- Lassen, J.; Løvendahl, P. Heritability Estimates for Enteric Methane Emissions from Holstein Cattle Measured Using Noninvasive Methods. J. Dairy Sci. 2016, 99, 1959–1967. [Google Scholar] [CrossRef] [Green Version]
- Donoghue, K.A.; Bird-Gardiner, T.; Arthur, P.F.; Herd, R.M.; Hegarty, R.F. Genetic and Phenotypic Variance and Covariance Components for Methane Emission and Postweaning Traits in Angus Cattle. J. Anim. Sci. 2016, 94, 1438–1445. [Google Scholar] [CrossRef]
- Zhu, Z.; Difford, G.F.; Noel, S.J.; Lassen, J.; Løvendahl, P.; Højberg, O. Stability Assessment of the Rumen Bacterial and Archaeal Communities in Dairy Cows Within a Single Lactation and Its Association With Host Phenotype. Front. Microbiol. 2021, 12, 636223. [Google Scholar] [CrossRef]
- Zhang, Q.; Difford, G.; Sahana, G.; Løvendahl, P.; Lassen, J.; Lund, M.S.; Guldbrandtsen, B.; Janss, L. Bayesian Modeling Reveals Host Genetics Associated with Rumen Microbiota Jointly Influence Methane Emission in Dairy Cows. ISME J. 2020, 14, 2019–2033. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2023, 11, 1. https://doi.org/10.3390/microorganisms11010001
Betancur-Murillo CL, Aguilar-Marín SB, Jovel J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms. 2023; 11(1):1. https://doi.org/10.3390/microorganisms11010001
Chicago/Turabian StyleBetancur-Murillo, Claudia Lorena, Sandra Bibiana Aguilar-Marín, and Juan Jovel. 2023. "Prevotella: A Key Player in Ruminal Metabolism" Microorganisms 11, no. 1: 1. https://doi.org/10.3390/microorganisms11010001
APA StyleBetancur-Murillo, C. L., Aguilar-Marín, S. B., & Jovel, J. (2023). Prevotella: A Key Player in Ruminal Metabolism. Microorganisms, 11(1), 1. https://doi.org/10.3390/microorganisms11010001






