Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities
Abstract
1. Introduction
2. HPV Oncogenomics in Cervical Cancer
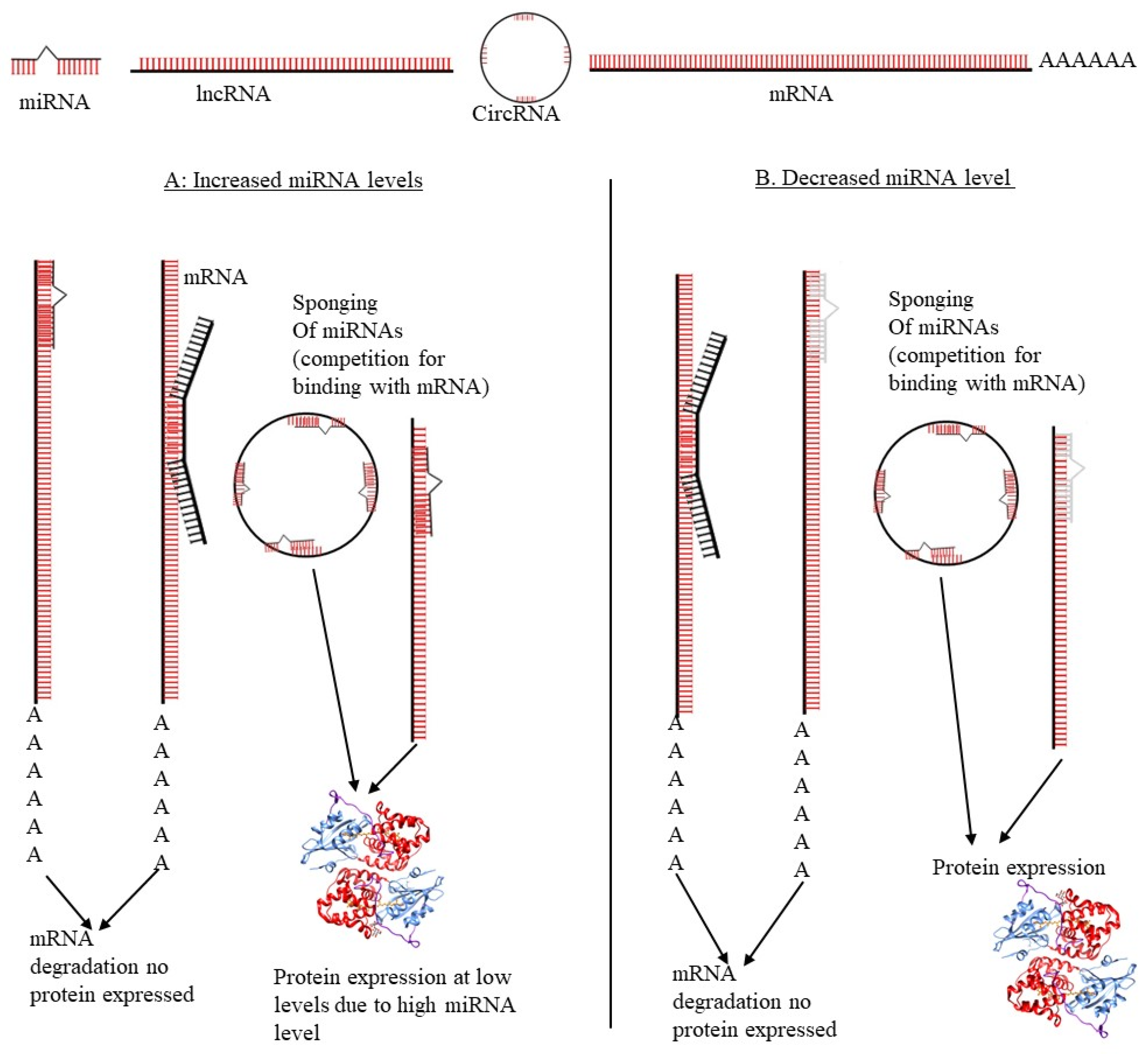
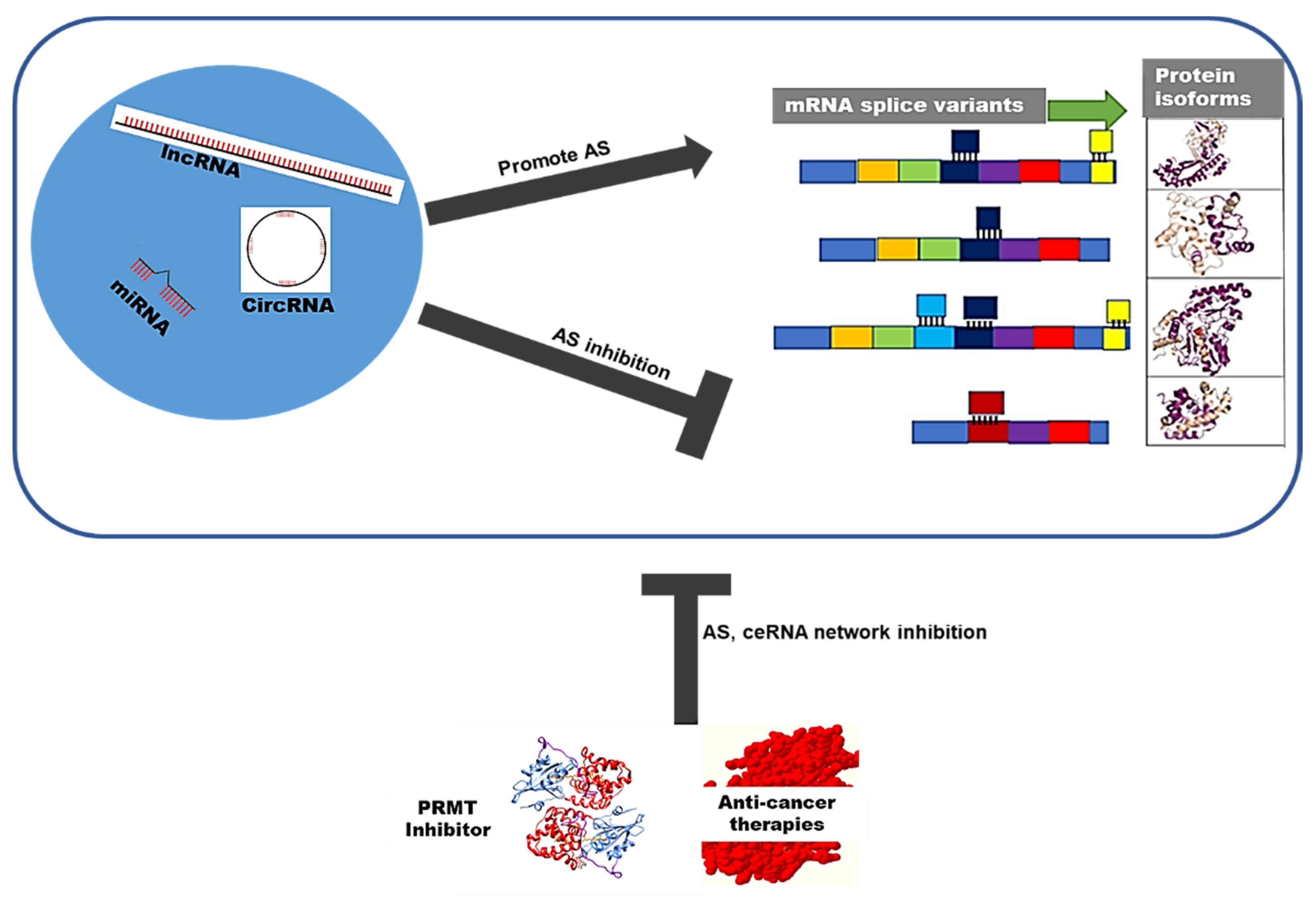
3. ceRNA Networks Regulate Alternative Splicing in Cervical Cancer
3.1. LncRNAs Role in ceRNA Networks Regulation and AS in Cervical Cancer
3.2. miRNAs Role in ceRNA Network Regulation and AS in CC
3.3. circRNAs Role in ceRNA Network Regulation and AS in Cervical Cancer
4. Role of AS and ceRNA Networks in HPV Immune Suppression and Evasion in Cervical Cancer
5. Clinical Significance: ceRNA Networks Regulation and mRNA Splicing Switches in Cervical Cancer
6. Epigenomics of PRMTs and Alternative Splicing
7. PRMT Inhibitor Drugs’ Therapeutic Potential
8. Limitations and Challenges
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.; Yu, J.; Awolude, O.A.; Chuang, L. Cervical cancer worldwide. Curr. Probl. Cancer 2018, 42, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-X.; Zheng, M.-J.; Zhang, W.-C.; Li, X.; Gou, R.; Nie, X.; Liu, Q.; Hao, Y.-Y.; Liu, J.-J.; Lin, B. Systematic profiling of alternative splicing signature reveals prognostic predictor for cervical cancer. J. Transl. Med. 2019, 17, 379. [Google Scholar] [CrossRef]
- Zibako, P.; Hlongwa, M.; Tsikai, N.; Manyame, S.; Ginindza, T.G. Mapping evidence on management of cervical cancer in sub-Saharan Africa: Scoping review protocol. Syst. Rev. 2021, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jedy-Agba, E.; Joko, W.Y.; Liu, B.; Buziba, N.G.; Borok, M.; Korir, A.; Masamba, L.; Manraj, S.S.; Finesse, A.; Wabinga, H. Trends in cervical cancer incidence in sub-Saharan Africa. Br. J. Cancer 2020, 123, 148–154. [Google Scholar] [CrossRef]
- Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today/home (accessed on 20 May 2022).
- Ekwunife, O.I.; O’Mahony, J.F.; Gerber Grote, A.; Mosch, C.; Paeck, T.; Lhachimi, S.K. Challenges in cost-effectiveness analysis modelling of HPV vaccines in low-and middle-income countries: A systematic review and practice recommendations. Pharmacoeconomics 2017, 35, 65–82. [Google Scholar] [CrossRef]
- Marima, R.; Hull, R.; Lolas, G.; Syrigos, K.N.; Kgoebane-Maseko, M.; Kaufmann, A.M.; Dlamini, Z. The catastrophic HPV/HIV dual viral oncogenomics in concert with dysregulated alternative splicing in cervical cancer. Int. J. Mol. Sci. 2021, 22, 10115. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Al-Hendy, A. The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer. Cells 2022, 11, 1149. [Google Scholar] [CrossRef]
- Zhou, C.; Tuong, Z.K.; Frazer, I.H. Papillomavirus immune evasion strategies target the infected cell and the local immune system. Front. Oncol. 2019, 9, 682. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Bzhalava, Z.; Mühr, L.S.A.; Dillner, J. Transcription of human papillomavirus oncogenes in head and neck squamous cell carcinomas. Vaccine 2020, 38, 4066–4070. [Google Scholar] [CrossRef] [PubMed]
- Preti, M.; Rotondo, J.C.; Holzinger, D.; Micheletti, L.; Gallio, N.; McKay-Chopin, S.; Carreira, C.; Privitera, S.S.; Watanabe, R.; Ridder, R. Role of human papillomavirus infection in the etiology of vulvar cancer in Italian women. Infect. Agents Cancer 2020, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- Moscicki, A.-B.; Palefsky, J.M. HPV in men: An update. J. Low. Genit. Tract Dis. 2011, 15, 231. [Google Scholar] [CrossRef] [PubMed]
- Lekoane, K.M.B.; Kuupiel, D.; Mashamba-Thompson, T.P.; Ginindza, T.G. The interplay of HIV and human papillomavirus-related cancers in sub-Saharan Africa: Scoping review. Syst. Rev. 2020, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Singh, M.P.; Rai, B. Human papillomavirus-associated cancers: A growing global problem. Int. J. Appl. Basic Med. Res. 2016, 6, 84. [Google Scholar]
- Monk, B.J.; Tewari, K.S.; Koh, W.-J. Multimodality therapy for locally advanced cervical carcinoma: State of the art and future directions. J. Clin. Oncol. 2007, 25, 2952–2965. [Google Scholar] [CrossRef]
- Gaffney, D.K.; Erickson-Wittmann, B.A.; Jhingran, A.; Mayr, N.A.; Puthawala, A.A.; Moore, D.; Rao, G.G.; Small, W.; Varia, M.A.; Wolfson, A.H. ACR appropriateness criteria® on advanced cervical cancer expert panel on radiation oncology—Gynecology. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 609–614. [Google Scholar] [CrossRef]
- Bradley, K. Cervical Cancer. Available online: NCCN.org (accessed on 17 August 2022).
- Thigpen, T.; Shingleton, H.; Homesley, H.; Lagasse, L.; Blessing, J. Cis-platinum in treatment of advanced or recurrent squamous cell carcinoma of the cervix: A phase II study of the Gynecologic Oncology Group. Cancer 1981, 48, 899–903. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, X.; Lin, C.; Jia, X.; Zhu, H.; Song, J.; Zhang, Y. Noncoding RNAs regulate alternative splicing in Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 1–14. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Faraonio, R.; Buonaguro, L.; Annunziata, C.; Starita, N.; Cerasuolo, A.; Pezzuto, F.; Tornesello, A.L.; Buonaguro, F.M. The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front. Oncol. 2020, 10, 150. [Google Scholar] [CrossRef]
- Ala, U. Competing endogenous RNAs, non-coding RNAs and diseases: An intertwined story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Du, J.; Tang, J.; Tan, B. Integrated analysis of ceRNA regulatory network associated with tumor stage in cervical cancer. Front. Genet. 2021, 12, 618753. [Google Scholar] [CrossRef]
- Gong, J.; Jiang, H.; Shu, C.; Hu, M.-q.; Huang, Y.; Liu, Q.; Li, R.-f.; Wei, Y.-z. Integrated analysis of circular RNA-associated ceRNA network in cervical cancer: Observational study. Medicine 2019, 98, e16922. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Lin, M.; Wu, D.; Zhao, M. LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell Int. 2021, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Huang, H. Long non-coding RNA MIR205HG function as a ceRNA to accelerate tumor growth and progression via sponging miR-122-5p in cervical cancer. Biochem. Biophys. Res. Commun. 2019, 514, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Piersma, S.J. Immunosuppressive tumor microenvironment in cervical cancer patients. Cancer Microenviron. 2011, 4, 361–375. [Google Scholar] [CrossRef]
- Graham, S.V.; Faizo, A.A.A. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2017, 231, 83–95. [Google Scholar] [CrossRef]
- Sharmin, S.; Zohura, F.T.; Islam, M.; Shimonty, A.; Khan, M.; Parveen, R.; Sharmin, F.; Ahsan, C.R.; Islam, A.B.M.M.; Yasmin, M. Mutational profiles of marker genes of cervical carcinoma in Bangladeshi patients. BMC Cancer 2021, 21, 289. [Google Scholar] [CrossRef]
- Molina-Pineda, A.; López-Cardona, M.G.; Limón-Toledo, L.P.; Cantón-Romero, J.C.; Martínez-Silva, M.G.; Ramos-Sánchez, H.V.; Flores-Miramontes, M.G.; de la Mata-González, P.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. High frequency of HPV genotypes 59, 66, 52, 51, 39 and 56 in women from Western Mexico. BMC Infect. Dis. 2020, 20, 889. [Google Scholar] [CrossRef]
- (WHO), W.H.O. Human Papilloma. Available online: https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccines-quality/human-papillomavirus (accessed on 17 August 2022).
- Steinbach, A.; Riemer, A.B. Immune evasion mechanisms of human papillomavirus: An update. Int. J. Cancer 2018, 142, 224–229. [Google Scholar] [CrossRef]
- Paz-Zulueta, M.; Álvarez-Paredes, L.; Rodríguez Díaz, J.C.; Parás-Bravo, P.; Andrada Becerra, M.; Rodríguez Ingelmo, J.M.; Ruiz García, M.M.; Portilla, J.; Santibañez, M. Prevalence of high-risk HPV genotypes, categorised by their quadrivalent and nine-valent HPV vaccination coverage, and the genotype association with high-grade lesions. BMC Cancer 2018, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, J.-Y.; Choi, S.; Kim, D.S.; Oh, Y.L. Carcinogenic risk of human papillomavirus (HPV) genotypes and potential effects of HPV vaccines in Korea. Sci. Rep. 2019, 9, 12556. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qi, Y.; Cui, X.; Huo, Q.; Zhu, L.; Zhang, A.; Tan, M.; Hong, Q.; Yang, Y.; Zhang, H.; et al. Characteristic of HPV Integration in the Genome and Transcriptome of Cervical Cancer Tissues. BioMed Res. Int. 2018, 2018, 6242173. [Google Scholar] [CrossRef] [PubMed]
- Olmedo-Nieva, L.; Muñoz-Bello, J.O.; Contreras-Paredes, A.; Lizano, M. The Role of E6 Spliced Isoforms (E6*) in Human Papillomavirus-Induced Carcinogenesis. Viruses 2018, 10, 45. [Google Scholar] [CrossRef]
- Wang, X.; Huang, X.; Zhang, Y. Involvement of human papillomaviruses in cervical cancer. Front. Microbiol. 2018, 9, 2896. [Google Scholar] [CrossRef]
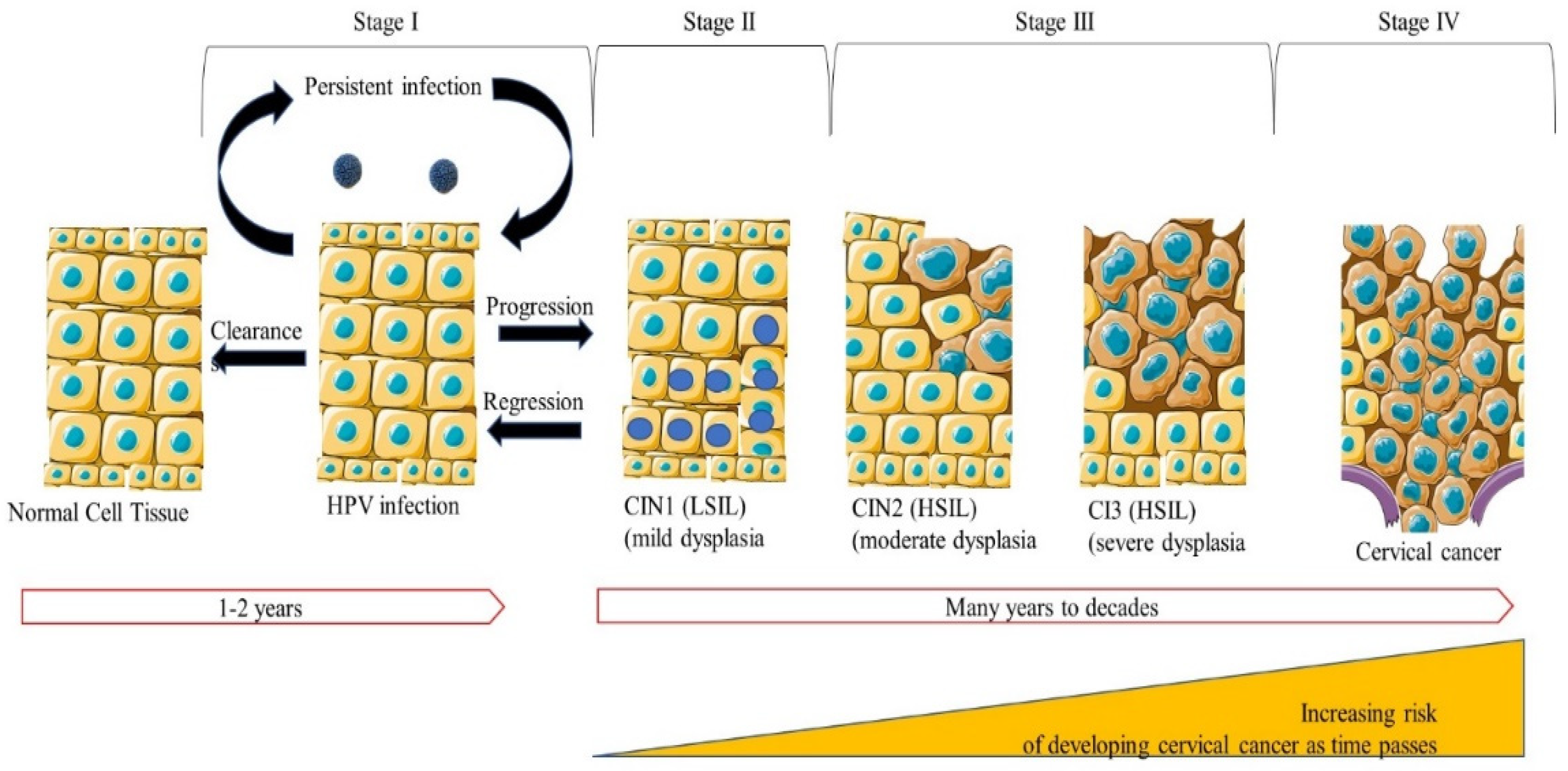
- Tainio, K.; Athanasiou, A.; Tikkinen, K.A.; Aaltonen, R.; Cárdenas, J.; Glazer-Livson, S.; Jakobsson, M.; Joronen, K.; Kiviharju, M.; Louvanto, K.; et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: Systematic review and meta-analysis. BMJ 2018, 360, k499. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, C.-x. Spontaneous regression of cervical intraepithelial neoplasia 2: A meta-analysis. Gynecol. Obstet. Investig. 2019, 84, 562–567. [Google Scholar] [CrossRef]
- Diederichs, S.; Bartsch, L.; Berkmann, J.C.; Fröse, K.; Heitmann, J.; Hoppe, C.; Iggena, D.; Jazmati, D.; Karschnia, P.; Linsenmeier, M. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016, 8, 442–457. [Google Scholar] [CrossRef]
- Chang, Z.; Huang, R.; Fu, W.; Li, J.; Ji, G.; Huang, J.; Shi, W.; Yin, H.; Wang, W.; Meng, T.; et al. The Construction and Analysis of ceRNA Network and Patterns of Immune Infiltration in Colon Adenocarcinoma Metastasis. Front. Cell Dev. Biol. 2020, 8, 688. [Google Scholar] [CrossRef]
- Hong, H.; Zhu, H.; Zhao, S.; Wang, K.; Zhang, N.; Tian, Y.; Li, Y.; Wang, Y.; Lv, X.; Wei, T. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019, 10, 950. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, J.; Yang, Z.; Chen, P.; Wang, D.-b. CircRNA_400029 promotes the aggressive behaviors of cervical cancer by regulation of miR-1285-3p/TLN1 axis. J. Cancer 2022, 13, 541. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Wang, W.; Guo, X. Overexpression of circular RNA hsa_circ_0001038 promotes cervical cancer cell progression by acting as a ceRNA for miR-337-3p to regulate cyclin-M3 and metastasis-associated in colon cancer 1 expression. Gene 2020, 733, 144273. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, L.; Li, Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR-218/HOXA1 pathway. Mol. Med. Rep. 2019, 19, 3714–3722. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.-M.; Dai, F.-F.; Zhou, Q.; Cheng, Y.-X. Hsa_circ_0000301 facilitates the progression of cervical cancer by targeting miR-1228-3p/IRF4 Axis. BMC Cancer 2021, 21, 583. [Google Scholar] [CrossRef]
- Shang, C.; Li, Y.; He, T.; Liao, Y.; Du, Q.; Wang, P.; Qiao, J.; Guo, H. The prognostic miR-532-5p-correlated ceRNA-mediated lipid droplet accumulation drives nodal metastasis of cervical cancer. J. Adv. Res. 2022, 37, 169–184. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, D.; Ye, L.; Wang, K.; Huang, L.; Mei, S.; Wu, J.; Chen, S.; Lai, X.; Zheng, L.; et al. Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 2019, 19, 45. [Google Scholar] [CrossRef]
- Zheng, P.; Yin, Z.; Wu, Y.; Xu, Y.; Luo, Y.; Zhang, T.-C. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun. Signal. 2018, 16, 5. [Google Scholar] [CrossRef]
- Liu, M.; Jia, J.; Wang, X.; Liu, Y.; Wang, C.; Fan, R. Long non-coding RNA HOTAIR promotes cervical cancer progression through regulating BCL2 via targeting miR-143-3p. Cancer Biol. Ther. 2018, 19, 391–399. [Google Scholar] [CrossRef]
- Sun, J.; Chu, H.; Ji, J.; Huo, G.; Song, Q.; Zhang, X. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int. J. Oncol. 2016, 49, 943–952. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Zhou, M.; Lv, H.; Qin, X.; Zhou, J.; Mao, X.; Li, X.; Xu, Y.; Liu, Y.; Xing, H. Involvement of NEAT1/miR-133a axis in promoting cervical cancer progression via targeting SOX4. J. Cell. Physiol. 2019, 234, 18985–18993. [Google Scholar] [CrossRef]
- Hu, Y.; Ma, Y.; Liu, J.; Cai, Y.; Zhang, M.; Fang, X. LINC01128 expedites cervical cancer progression by regulating miR-383-5p/SFN axis. BMC Cancer 2019, 19, 1157. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Song, L.; Zeng, S.; Zhang, L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016, 37, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Wang, L.; Pan, X.; Yang, C. lncRNA OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit cell apoptosis through a mechanism involving miR-143-3p in cervical cancer. Braz. J. Med. Biol. Res. 2020, 53, e8883. [Google Scholar] [CrossRef]
- Chang, S.; Sun, L.; Feng, G. SP1-mediated long noncoding RNA POU3F3 accelerates the cervical cancer through miR-127-5p/FOXD1. Biomed. Pharmacother. 2019, 117, 109133. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, W.; Zhang, C.; Liu, X.; Lv, J.; Li, X.; Zhao, L.; Li, W.; Li, J.; Ren, Y. Long non-coding RNA RP11-552M11. 4 favors tumorigenesis and development of cervical cancer via modulating miR-3941/ATF1 signaling. Int. J. Biol. Macromol. 2019, 130, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hong, J.; Wijayakulathilaka, W.S.M.A. Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p. Cell Cycle 2019, 18, 3313–3324. [Google Scholar] [CrossRef]
- Guo, H.; Yang, S.; Li, S.; Yan, M.; Li, L.; Zhang, H. LncRNA SNHG20 promotes cell proliferation and invasion via miR-140-5p-ADAM10 axis in cervical cancer. Biomed. Pharmacother. 2018, 102, 749–757. [Google Scholar] [CrossRef]
- Clarke, T.L.; Mostoslavsky, R. DNA repair as a shared hallmark in cancer and ageing. Mol. Oncol. 2022. [Google Scholar] [CrossRef]
- Marima, R.; Francies, F.Z.; Hull, R.; Molefi, T.; Oyomno, M.; Khanyile, R.; Mbatha, S.; Mabongo, M.; Owen Bates, D.; Dlamini, Z. MicroRNA and Alternative mRNA Splicing Events in Cancer Drug Response/Resistance: Potent Therapeutic Targets. Biomedicines 2021, 9, 1818. [Google Scholar] [CrossRef]
- Magee, P.; Shi, L.; Garofalo, M. Role of microRNAs in chemoresistance. Ann. Transl. Med. 2015, 3, 332. [Google Scholar]
- Jiang, Y.; Li, Y.; Fang, S.; Jiang, B.; Qin, C.; Xie, P.; Zhou, G.; Li, G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol. Lett. 2014, 7, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhong, Y.; Zhang, Y.; Yang, L.; Wu, P.; Hou, X.; Xiong, F.; Li, X.; Zhang, S.; Gong, Z. Long non-coding RNAs are involved in alternative splicing and promote cancer progression. Br. J. Cancer 2022, 126, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Noncoding RNA 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Pruszko, M.; Milano, E.; Forcato, M.; Donzelli, S.; Ganci, F.; Di Agostino, S.; De Panfilis, S.; Fazi, F.; Bates, D.O.; Bicciato, S.; et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017, 18, 1331–1351. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Akimitsu, N. Long non-coding RNAs in cancer progression. Front. Genet. 2012, 3, 219. [Google Scholar] [CrossRef]
- Dong, M.; Dong, Z.; Zhu, X.; Zhang, Y.; Song, L. Long non-coding RNA MIR205HG regulates KRT17 and tumor processes in cervical cancer via interaction with SRSF1. Exp. Mol. Pathol. 2019, 111, 104322. [Google Scholar] [CrossRef]
- Guo, L.; Yang, G.; Kang, Y.; Li, S.; Duan, R.; Shen, L.; Jiang, W.; Qian, B.; Yin, Z.; Liang, T. Construction and analysis of a ceRNA network reveals potential prognostic markers in colorectal cancer. Front. Genet. 2020, 11, 418. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, L.J. The role of miRNAs in the invasion and metastasis of cervical cancer. Biosci. Rep. 2019, 39, BSR20181377. [Google Scholar] [CrossRef]
- Mole, S.; Faizo, A.A.A.; Hernandez-Lopez, H.; Griffiths, M.; Stevenson, A.; Roberts, S.; Graham, S.V. Human papillomavirus type 16 infection activates the host serine arginine protein kinase 1 (SRPK1)—Splicing factor axis. J. Gen. Virol. 2020, 101, 523–532. [Google Scholar] [CrossRef]
- Zhang, Q.; Lv, R.; Guo, W.; Li, X. microRNA-802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine-rich splicing factor 9. J. Cell. Biochem. 2019, 120, 10370–10379. [Google Scholar] [CrossRef]
- Wu, P.; Li, C.; Ye, D.M.; Yu, K.; Li, Y.; Tang, H.; Xu, G.; Yi, S.; Zhang, Z. Circular RNA circEPSTI1 accelerates cervical cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging (Albany NY) 2021, 13, 4663–4673. [Google Scholar] [CrossRef] [PubMed]
- Song, T.-F.; Xu, A.-L.; Chen, X.-H.; Gao, J.-Y.; Gao, F.; Kong, X.-C. Circular RNA circRNA_101996 promoted cervical cancer development by regulating miR-1236-3p/TRIM37 axis. Kaohsiung J. Med. Sci. 2021, 37, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, B.; Li, F.; Yu, K.; Bai, Y. The mechanism and detection of alternative splicing events in circular RNAs. PeerJ 2020, 8, e10032. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Sinha, T.; Shyamal, S.; Panda, A.C. Emerging Role of Circular RNA-Protein Interactions. Noncoding RNA 2021, 7, 48. [Google Scholar] [CrossRef]
- Chen, Z.; Gan, J.; Wei, Z.; Zhang, M.; Du, Y.; Xu, C.; Zhao, H. The Emerging Role of PRMT6 in Cancer. Front. Oncol. 2022, 12, 841381. [Google Scholar] [CrossRef]
- Dong, S.-H.; Wang, X.; Tian, S.-C.; Ma, N.-Q.; Zhang, X.-Y.; Liu, Y.; Zhang, B.-L.; Wu, Y.-J. Arginine methyltransferase inhibitor 1 exhibits antitumor effects against cervical cancer in vitro and in vivo. Die Pharm.-Int. J. Pharm. Sci. 2018, 73, 269–273. [Google Scholar]
- Papatsirou, M.; Diamantopoulos, M.A.; Katsaraki, K.; Kletsas, D.; Kontos, C.K.; Scorilas, A. Identification of Novel Circular RNAs of the Human Protein Arginine Methyltransferase 1 (PRMT1) Gene, Expressed in Breast Cancer Cells. Genes 2022, 13, 1133. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Feng, Z.; Xu, X.; Zhang, J.; Yu, A.; Zhu, L.; Xiao, J.; Du, J.; Chen, M. Circular RNA circPRMT5 is upregulated in breast cancer and is required for cellproliferation and migration. Turk. J. Med. Sci. 2022, 52, 303–312. [Google Scholar] [CrossRef]
- Jin, J.; Martin, M.; Hartley, A.-V.; Lu, T. PRMTs and miRNAs: Functional cooperation in cancer and beyond. Cell Cycle 2019, 18, 1676–1686. [Google Scholar] [CrossRef]
- Qian, K.; Pietilä, T.; Rönty, M.; Michon, F.; Frilander, M.J.; Ritari, J.; Tarkkanen, J.; Paulín, L.; Auvinen, P.; Auvinen, E. Identification and validation of human papillomavirus encoded microRNAs. PLoS ONE 2013, 8, e70202. [Google Scholar] [CrossRef]
- Gao, Y.-L.; Zhang, M.-Y.; Xu, B.; Han, L.-J.; Lan, S.-F.; Chen, J.; Dong, Y.-J.; Cao, L.-L. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget 2017, 8, 86625. [Google Scholar] [CrossRef] [PubMed]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Farzanehpour, M.; Faghihloo, E.; Salimi, V.; Jalilvand, S.; Akhavan, S.; Muhammadnejad, A.; Razavi, A.N.E.; Kakavandi, E.; Azad, T.M. Comparison of Snail1, ZEB1, E-Cadherin Expression Levels in HPV-Induced Cervical Cancer. Iran. J. Public Health 2020, 49, 2179. [Google Scholar] [CrossRef] [PubMed]
- Boudreault, S.; Roy, P.; Lemay, G.; Bisaillon, M. Viral modulation of cellular RNA alternative splicing: A new key player in virus-host interactions? Wiley Interdiscip. Rev. RNA 2019, 10, e1543. [Google Scholar] [CrossRef]
- El Marabti, E.; Younis, I. The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Qiu, Z.; Kang, Y.; Liu, J.; Ning, S.; Yin, Y.; Pang, D.; Xu, S. Noncoding RNAs: The shot callers in tumor immune escape. Signal Transduct. Target. Ther. 2020, 5, 1–24. [Google Scholar]
- Zheng, J.; Cao, B.; Zhang, X.; Niu, Z.; Tong, J. Immune-related four-lncRNA signature for patients with cervical cancer. BioMed Res. Int. 2020, 2020, 3641231. [Google Scholar] [CrossRef]
- Su, K.; Zhao, Q.; Bian, A.; Wang, C.; Cai, Y.; Zhang, Y. A novel positive feedback regulation between long noncoding RNA UICC and IL-6/STAT3 signaling promotes cervical cancer progression. Am. J. Cancer Res. 2018, 8, 1176. [Google Scholar]
- Liu, Q.; Liu, S.; Wang, D. Overexpression of microRNA-21 decreased the sensitivity of advanced cervical cancer to chemoradiotherapy through SMAD7. Anti-Cancer Drugs 2020, 31, 272–281. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, Z.; Zheng, P.; Zhao, W.; Han, N. MicroRNA-1285-5p influences the proliferation and metastasis of non-small-cell lung carcinoma cells via downregulating CDH1 and Smad4. Tumour Biol. 2017, 39, 1010428317705513. [Google Scholar] [CrossRef]
- Feng, Y.; Ma, J.; Fan, H.; Liu, M.; Zhu, Y.; Li, Y.; Tang, H. TNF-α-induced lncRNA LOC105374902 promotes the malignant behavior of cervical cancer cells by acting as a sponge of miR-1285-3p. Biochem. Biophys. Res. Commun. 2019, 513, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Bukur, J.; Jasinski, S.; Seliger, B. The role of classical and non-classical HLA class I antigens in human tumors. Semin. Cancer Biol. 2012, 22, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yang, J.; Yu, J.; Liu, X.; Yu, C.; Hu, J.; Shi, H.; Ma, X. Long non-coding RNAs: Emerging roles in the immunosuppressive tumor microenvironment. Front. Oncol. 2020, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, J.-B.; Deng, J.; Zou, D.-Z.; Wu, J.-J.; Cao, Y.-H.; Yin, J.; Ma, Y.-S.; Da, F.; Li, W. The role of ceRNA-mediated diagnosis and therapy in hepatocellular carcinoma. Hereditas 2021, 158, 1–14. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Liu, X.-h.; Sun, M.; Nie, F.-q.; Ge, Y.-b.; Zhang, E.-b.; Yin, D.-d.; Kong, R.; Xia, R.; Lu, K.-h.; Li, J.-h. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef]
- Li, F.; Yang, Q.; He, A.T.; Yang, B.B. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin. Cancer Biol. 2021, 75, 49–61. [Google Scholar] [CrossRef]
- Ouyang, D.; Yang, P.; Cai, J.; Sun, S.; Wang, Z. Comprehensive analysis of prognostic alternative splicing signature in cervical cancer. Cancer Cell Int. 2020, 20, 221. [Google Scholar] [CrossRef]
- He, J.; Huang, B.; Zhang, K.; Liu, M.; Xu, T. Long non-coding RNA in cervical cancer: From biology to therapeutic opportunity. Biomed. Pharmacother. 2020, 127, 110209. [Google Scholar] [CrossRef]
- Zhang, X.; Mao, L.; Li, L.; He, Z.; Wang, N.; Song, Y. Long noncoding RNA GIHCG functions as an oncogene and serves as a serum diagnostic biomarker for cervical cancer. J. Cancer 2019, 10, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, G.; Zhang, L.; Cong, J.; Hou, J.; Liu, C. LncRNA PVT1 promotes the growth of HPV positive and negative cervical squamous cell carcinoma by inhibiting TGF-β1. Cancer Cell Int. 2018, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Liu, K.; Yang, M.; Gao, X. ceRNAs in Cancer: Mechanism and Functions in a Comprehensive Regulatory Network. J. Oncol. 2021, 2021, 4279039. [Google Scholar] [CrossRef]
- Shi, D.; Zhang, C.; Liu, X. Long noncoding RNAs in cervical cancer. J. Cancer Res. Ther. 2018, 14, 745. [Google Scholar] [PubMed]
- Zhang, Y.; Wu, D.; Wang, D. Long non-coding RNA ARAP1-AS1 promotes tumorigenesis and metastasis through facilitating proto-oncogene c-Myc translation via dissociating PSF/PTB dimer in cervical cancer. Cancer Med. 2020, 9, 1855–1866. [Google Scholar] [CrossRef]
- Liu, J.; Wang, D.; Long, Z.; Liu, J.; Li, W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell. Physiol. Biochem. 2018, 48, 173–184. [Google Scholar] [CrossRef]
- Chen, S.; Yang, X.; Yu, C.; Zhou, W.; Xia, Q.; Liu, Y.; Chen, Q.; Chen, X.; Lv, Y.; Lin, Y. The potential of circRNA as a novel diagnostic biomarker in cervical cancer. J. Oncol. 2021, 2021, 5529486. [Google Scholar] [CrossRef]
- Wu, M.; Han, Y.; Gong, X.; Wan, K.; Liu, Y.; Zhou, Y.; Zhang, L.; Tang, G.; Fang, H.; Chen, B. Novel Insight of CircRNAs in Cervical Cancer: Potential Biomarkers and Therapeutic Target. Front. Med. 2022, 9, 759928. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Zhang, J.; Zheng, X.; Li, F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem. Biophys. Res. Commun. 2018, 501, 428–433. [Google Scholar] [CrossRef]
- Jiang, Y.; Hu, Z.; Zuo, Z.; Li, Y.; Pu, F.; Wang, B.; Tang, Y.; Guo, Y.; Tao, H. Identification of Circulating MicroRNAs as a Promising Diagnostic Biomarker for Cervical Intraepithelial Neoplasia and Early Cancer: A Meta-Analysis. BioMed Res. Int. 2020, 2020, 4947381. [Google Scholar] [CrossRef]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.E.; Zhang, C.; Xiang, Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol. 2015, 3, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- He, R.-q.; Zhou, X.-g.; Yi, Q.-y.; Deng, C.-w.; Gao, J.-m.; Chen, G.; Wang, Q.-y. Prognostic signature of alternative splicing events in bladder urothelial carcinoma based on spliceseq data from 317 cases. Cell. Physiol. Biochem. 2018, 48, 1355–1368. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Li, Q.Q.; Liang, L.; Qiu, L.L.; Wei, H.W.; Huang, B.Y.; Gang, C.; He, R.Q.; Huang, Z.G.; Hou, W.; et al. Prognostic alternative splicing signature in cervical squamous cell carcinoma. IET Syst. Biol. 2020, 14, 314–322. [Google Scholar] [CrossRef]
- Baldwin, R.M.; Morettin, A.; Côté, J. Role of PRMTs in cancer: Could minor isoforms be leaving a mark? World J. Biol. Chem. 2014, 5, 115. [Google Scholar]
- Goulet, I.; Gauvin, G.; Boisvenue, S.; Cote, J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 2007, 282, 33009–33021. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, Y.; Chen, M.; Li, S.; Bai, J.; Zhang, Y.; Sun, Y.; Wang, G.; Xu, H.; Wang, Z. PRMT5 disruption drives antitumor immunity in cervical cancer by reprogramming T cell-mediated response and regulating PD-L1 expression. Theranostics 2021, 11, 9162. [Google Scholar] [CrossRef]
- Xiao, W.; Chen, X.; Liu, L.; Shu, Y.; Zhang, M.; Zhong, Y. Role of protein arginine methyltransferase 5 in human cancers. Biomed. Pharm. 2019, 114, 108790. [Google Scholar] [CrossRef]
- Hwang, J.W.; Cho, Y.; Bae, G.-U.; Kim, S.-N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef]
- Vhuiyan, M.I.; Pak, M.L.; Park, M.A.; Thomas, D.; Lakowski, T.M.; Chalfant, C.E.; Frankel, A. PRMT2 interacts with splicing factors and regulates the alternative splicing of BCL-X. J. Biochem. 2017, 162, 17–25. [Google Scholar] [CrossRef]
- Maron, M.I.; Casill, A.D.; Gupta, V.; Roth, J.S.; Sidoli, S.; Query, C.C.; Gamble, M.J.; Shechter, D. Type I and II PRMTs inversely regulate post-transcriptional intron detention through Sm and CHTOP methylation. eLife 2022, 11, e72867. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.-H.; Fan, X.-J.; Hu, Y.; Tian, X.-X.; Guo, M.; Mao, M.-W.; Fang, Z.-Y.; Wu, P.; Gao, S.-X.; Peng, C. A systematic survey of PRMT interactomes reveals the key roles of arginine methylation in the global control of RNA splicing and translation. Sci. Bull. 2021, 66, 1342–1357. [Google Scholar] [CrossRef]
- Li, W.-j.; He, Y.-h.; Yang, J.-j.; Hu, G.-s.; Lin, Y.-a.; Ran, T.; Peng, B.-l.; Xie, B.-l.; Huang, M.-f.; Gao, X.; et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 2021, 12, 1946. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Avasarala, S.; Van Scoyk, M.; Karuppusamy Rathinam, M.K.; Zerayesus, S.; Zhao, X.; Zhang, W.; Pergande, M.R.; Borgia, J.A.; DeGregori, J.; Port, J.D.; et al. PRMT1 Is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-small Cell Lung Cancer. J. Biol. Chem. 2015, 290, 13479–13489. [Google Scholar] [CrossRef]
- Dowhan, D.H.; Harrison, M.J.; Eriksson, N.A.; Bailey, P.; Pearen, M.A.; Fuller, P.J.; Funder, J.W.; Simpson, E.R.; Leedman, P.J.; Tilley, W.D. Protein arginine methyltransferase 6-dependent gene expression and splicing: Association with breast cancer outcomes. Endocr.-Relat. Cancer 2012, 19, 509–526. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine methylation: The coming of age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Ding, Z.; Guo, L.; Deng, Z.; Li, P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann. Hepatol. 2020, 19, 269–279. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, H.; Wang, F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene 2019, 720, 144099. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Li, D.; Guo, X.; Li, P.; Li, X.; Tong, S.; Tong, J.; Kuang, L.; Liang, D. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4120–4130. [Google Scholar] [CrossRef]
- Pang, J.; Ye, L.; Zhao, D.; Zhao, D.; Chen, Q. Circular RNA PRMT5 confers cisplatin-resistance via miR-4458/REV3L axis in non-small-cell lung cancer. Cell Biol. Int. 2020, 44, 2416–2426. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Sun, X.; Zhang, S.; Chen, Z.; Yu, J. Circ_0039960 regulates growth and Warburg effect of breast cancer cells via modulating miR-1178/PRMT7 axis. Mol. Cell. Probes 2022, 64, 101829. [Google Scholar] [CrossRef] [PubMed]
- Samuel, S.F.; Barry, A.; Greenman, J.; Beltran-Alvarez, P. Arginine methylation: The promise of a ’silver bullet’ for brain tumours? Amino Acids 2021, 53, 489–506. [Google Scholar] [CrossRef]
- Bonday, Z.Q.; Cortez, G.S.; Grogan, M.J.; Antonysamy, S.; Weichert, K.; Bocchinfuso, W.P.; Li, F.; Kennedy, S.; Li, B.; Mader, M.M. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 2018, 9, 612–617. [Google Scholar] [CrossRef]
- Fedoriw, A.; Rajapurkar, S.R.; O’Brien, S.; Gerhart, S.V.; Mitchell, L.H.; Adams, N.D.; Rioux, N.; Lingaraj, T.; Ribich, S.A.; Pappalardi, M.B. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell 2019, 36, 100–114. [Google Scholar] [CrossRef]
- Yang, Z.; Xiao, T.; Li, Z.; Zhang, J.; Chen, S. Novel Chemicals Derived from Tadalafil Exhibit PRMT5 Inhibition and Promising Activities against Breast Cancer. Int. J. Mol. Sci. 2022, 23, 4806. [Google Scholar] [CrossRef]
- AbuHammad, S.; Cullinane, C.; Martin, C.; Bacolas, Z.; Ward, T.; Chen, H.; Slater, A.; Ardley, K.; Kirby, L.; Chan, K.T. Regulation of PRMT5–MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 17990–18000. [Google Scholar] [CrossRef]

| ceRNA | Regulatory Axis | Role in Cervical Cancer | Ref |
|---|---|---|---|
| circRNA 400029 | miR-1285-3p/TLN1 | Aggressive behaviors of cervical cancer | [44] |
| circCLK3 | MiR-320a/Fox M1 | Cervical cancer progression | [43] |
| hsa_circ_0001038 | miR-337-3p/cyclin-M3 | Promotes cell growth, migration, and invasion | [45] |
| hsa_circRNA_101996 | miR-8075/TPX2 | Promotes cell growth and invasion | [45] |
| hsa_circ_0023404 | miR-136/TFCP2 | Cervical cancer development and progression | [25] |
| circ-EIF4G2 | miR-218/HOXA1 | Modulates malignant biological behaviors | [46] |
| Hsa_circ_0000301 | miR-1228-3p/IRF4 | Cancer progression | [47] |
| miR-532-5p | LINC01410/FASN | Tumour metastasis | [48] |
| LncRNA XIST | miR-200a/Fus | Cancer progression | [43] |
| LncRNA XIST | miR-140-5p/ORC1 | Cell proliferation and increased expression of Bcl-2 | [49] |
| LncRNA HOTAIR | miR-206/MKL1 | Migration and invasion | [50] |
| LncRNA HOTAIR | miR-143-3p/BCL2 | Inhibit tumor suppression | [51] |
| LncRNA HOTAIR | miR-148a/human leucocyteantigen-G (HLA-G) | Proliferation, migration, and invasion of cervical cancer cells | [52] |
| LncRNA NEAT1 | miR-133a/Sox4 | Cell proliferation, migration, and invasion | [53] |
| LncRNA LINC01128 | miR-383-5p/SFN | Inhibits apoptosisproliferation, migration, and invasion of cervical cancer cells | [54] |
| LncRNA MALAT1 | miR-124/RBG2 | Proliferation, migration, and invasion | [55] |
| lncRNA OIP5-AS1 | miR-143-3p/ROCK1 | Inhibit apoptosis and promotes cell proliferation | [56] |
| LncRNA RNA POU3F3 | miR-127-5p/FOXD1 | Promoted the proliferation and invasion | [57] |
| LncRNA RP11-552M11.4 | miR-3941/ATF1 | Cell proliferation | [58] |
| SNHG4 | miR-148a-3p/c-Met | Improve cell viability and inhibit apoptosis | [59] |
| SNHG12 | miR-125b/STAT3 | Proliferation and invasion of cervical cancer | [59] |
| lncRNA SU1P2 | let-7a/IGF1R, let-7a/N-myc, and let-7a/EphA4 | Promotes tumorigenesis | [55] |
| LncRNA SNHG20 | miR-140-5p-ADAM10 | Promote cervical cancer cells proliferation and invasion | [60] |
| ZNF667-AS1 | microRNA-93-3p/PEG3 | Decreases tumor invasion and metastasis | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basera, A.; Hull, R.; Demetriou, D.; Bates, D.O.; Kaufmann, A.M.; Dlamini, Z.; Marima, R. Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities. Microorganisms 2022, 10, 1852. https://doi.org/10.3390/microorganisms10091852
Basera A, Hull R, Demetriou D, Bates DO, Kaufmann AM, Dlamini Z, Marima R. Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities. Microorganisms. 2022; 10(9):1852. https://doi.org/10.3390/microorganisms10091852
Chicago/Turabian StyleBasera, Afra, Rodney Hull, Demetra Demetriou, David Owen Bates, Andreas Martin Kaufmann, Zodwa Dlamini, and Rahaba Marima. 2022. "Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities" Microorganisms 10, no. 9: 1852. https://doi.org/10.3390/microorganisms10091852
APA StyleBasera, A., Hull, R., Demetriou, D., Bates, D. O., Kaufmann, A. M., Dlamini, Z., & Marima, R. (2022). Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities. Microorganisms, 10(9), 1852. https://doi.org/10.3390/microorganisms10091852









