Abstract
Plastic polymer waste management is an increasingly prevalent issue. In this paper, Rhodococcus genomes were explored to predict new plastic-degrading enzymes based on recently discovered biodegrading enzymes for diverse plastic polymers. Bioinformatics prediction analyses were conducted using 124 gene products deriving from diverse microorganisms retrieved from databases, literature data, omic-approaches, and functional analyses. The whole results showed the plastic-degrading potential of Rhodococcus genus. Among the species with high plastic-degrading potential, R. erythropolis, R. equi, R. opacus, R. qingshengii, R. fascians, and R. rhodochrous appeared to be the most promising for possible plastic removal. A high number of genetic determinants related to polyester biodegradation were obtained from different Rhodococcus species. However, score calculation demonstrated that Rhodococcus species (especially R. pyridinivorans, R. qingshengii, and R. hoagii) likely possess PE-degrading enzymes. The results identified diverse oxidative systems, including multicopper oxidases, alkane monooxygenases, cytochrome P450 hydroxylases, para-nitrobenzylesterase, and carboxylesterase, and they could be promising reference sequences for the biodegradation of plastics with C−C backbone, plastics with heteroatoms in the main chain, and polyesters, respectively. Notably, the results of this study could be further exploited for biotechnological applications in biodegradative processes using diverse Rhodococcus strains and through catalytic reactions.
1. Introduction
Within Actinobacteria phylum (Corynebacteriaceae order), Rhodococcus represents one of the most extraordinary genera in terms of catabolic versatility, since the members of this genus deploy an extensive asset of genes encoding enzymes enabling the biotransformation and/or biodegradation of a wide array of organic compounds, contaminants, pharmaceuticals, and wastes [1,2,3,4,5,6,7,8,9]. The remarkable metabolic capabilities of Rhodococcus species together with their strong persistence under difficult conditions are due to their peculiar cell features including the cell envelope (containing mycolic acids, and peptidoglycan), hydrophobicity, a specific phospholipid profile, and large and plastic genomes with a multiplicity of genes for catabolic and anabolic processes, and stress response [10,11,12,13,14]. In line with this aspect, rhodococci can inhabit different environments from groundwater to water systems, soils, diverse extreme and harsh environmental niches, and contaminated ecosystems; they have been also isolated from plants (R. fascians), insects (R. rhodnii), diseased and healthy animals (e.g., R. equi), and humans (e.g., R. equi, R. rhodochrous, and R. erythropolis) [15,16]. However, until now, Rhodococcus has received relatively little attention for plastic biodegrading capabilities of diverse biodegradable and non-biodegradable polymers and the corresponding genetic determinants [17,18,19].
Synthetic plastic constitutes a large group of compounds with profitable features, e.g., strength, flexibility, extreme durability, low weight, and easy and low-cost production, that are crucial in everyday life, which led to a global plastic production of almost 360 million tonnes in 2018 that is still growing year by year [20]. Synthetic plastics are commonly grouped into two main categories, based on their chemical structure: (i) the most recalcitrant carbon−carbon (C−C) backbone plastics, including low and high molecular density polyethylene (PE), and polypropylene (PP); (ii) the most prone to biodegradation, heteroatomic backbone plastics, including polyethylene terephthalate (PET) and polyurethane (PU) [21,22].
As a consequence of plastic utilization, emerging issues in disposal management are rising and are becoming an extreme priority. Up to now, environmental plastic pollution has been considered merely a facade disturbance, but recent research revealed potential risks of plastic impacts on biota in diverse ecosystems and on human health [23]. The actual plastic waste crisis, predicting around 12 thousand million metric tons of plastic waste accumulating in landfills and the natural environment by 2050 [24], seeks new technologies for recycling post-consumer plastics for waste valorization and/or the optimization of the biodegradation of discarded plastic polymers. Biocatalytic degradation and/or biotransformation by enzymes is an efficient and sustainable alternative for synthetic plastic (usually considered non-biodegradable) depolymerization to support and complement plastic recycling with mechanical, chemical, or chemical-physical treatments [25].
On the other hand, all polyester-based plastics are considered theoretically biodegradable, since the esterification process for their production is chemically reversible, even by biocatalysis [26]. This category includes, among others, (i) aliphatic polyesters, such as polyhydroxyalkanoates (PHAs), poly(propiolactone) (PPL), poly(ε-caprolactone) (PCL), poly(L-lactic acid) (PLA), and poly(ethylene succinate) (PES); (ii) co-polyesters containing aliphatic and aromatic components, such as poly(butylene succinate) (PBS), poly(butylene succinate)-co-(butylene adipate) (PBSA), poly(butylene adipate-co-terephthalate) (PBAT), poly(butylene succinate- co-terephthalate) (PBST), and poly(butylene succinate/terephthalate/isophthalate)-co-(lactate) (PBSTIL) [27].
Diverse plastic-degrading enzymes have been discovered from microbial sources, but in-depth investigations are still necessary for biotechnological industrial applications [28]. Despite the lack of enzymes able to oxidize highly stable C−C bonds of polyolefins hampering their biodegradation, few of them were retrieved from transcriptomic studies and/or functional analysis, including hydroxylases/monooxygenases, oxidases, laccases, and peroxidases [17,29,30,31]. Among the few plastic-degrading enzymes closest to achieving feasible biotechnological industrial processes, PET hydrolases are the most prominent/valuable, although it is still unknown how frequently they appear in different bacterial species. The majority of this enzyme category comprises cutinases, showing a broad substrate specificity, lipases, possessing a lower activity that is generally specific for long-chain substrates, and esterases, which usually hydrolyze esters with a shorter aliphatic chain in comparison to lipases [32].
Different PU-degrading enzymes were also reported (e.g., cutinases, esterases, lipases, laccases, peroxidases, proteases, and ureases) [25,33,34], but most of them showed ester-linked PU activity and were unlikely to show ether-linked PU activity [35].
In general, a single specific enzyme is not able to catalyze the biodegradation of a certain plastic group, but enzyme categories are recognized to be able to perform a step of polymer biotransformation, and the overall biodegradation can occur only due to the involvement of a multiplicity of enzymatic arrays and their subsequent activity [21,28].
Since Rhodococcus members play an important role in biotechnological applications [7], and only a few rhodococci have been revealed to possess plastic-degrading abilities and in a few cases their genetic determinants have been investigated [17,18,19], the present study investigates for the first time 669 Rhodococcus genomes mining predictive plastic-degrading enzymes. Comparative bioinformatics analyses were performed using 124 gene products related to plastic degradation of diverse polymers from different bacterial genera; they were retrieved from databases, literature data, omic-approaches, and functional analyses. The level of similarity of these enzymes from other organisms against Rhodococcus genomes in connection with the analysis of clustering contributes to evaluate the evolutional relationships and/or the putative horizontal gene transfer of new Rhodococcus identified sequences. In addition, score calculation provides a weight level to evaluate the real potential role of the different Rhodococcus species and different genes involved in the metabolism of diverse polymers.
2. Material and Methods
2.1. Input Data
Predictive analyses were performed on 669 Rhodococcus genomes from NCBI (http://www.ncbi.nlm.nih.gov, accessed on 27 May 2022) and analyzed by blast search.
Rhodococcus genomes were screened for 124 gene products related to plastic degradation of diverse polymers from different microorganisms retrieved from PlasticDB database [36], literature data, omic studies, and functional analyses in order to investigate the plastic-degrading potential of Rhodococcus genus (Table S1). The considered gene products were reported to be involved in the biodegradation and/or biotransformation of at least one considered plastic polymer. Among the diverse polymers, synthetic (non-biodegradable) or biodegradable plastics taken into account were organized into three major groups: plastics with C−C backbone comprising PE, plastics with heteroatoms in the main chain, including PET and PU, and polyesters (both aliphatic and co-polyesters containing aliphatic and aromatic components), such as polyhydroxybutyrate (PHB), PES, PPL, PCL, PLA, (poly(3-hydroxyvalerate) (PHV), poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), poly(hydroxyphenyl-valerate) (PHPV), PBS, PBSA, PBAT, and Ecoflex.
The unique input gene product sequences were also aligned with Clustal Omega program using the multiple sequence alignment (MSA) tool with default parameters (Neighbor-Joining method, the Gonnet transition matrix, gap opening penalty of 6 bits, maintain gaps with an extension of 1 bit, used bed-like clustering during subsequent iterations, and zero number of combined iterations) [37] in order to establish a clusterization framework of gene products used as input against Rhodococcus genomes. MSA was used as input of cluster analysis inferred using the maximum likelihood (ML) method by MEGA (version 10.2) software [38], with the following settings: JTT substitution matrix and gamma distribution of mutation rates with gamma optimized to 2. As a test of inferred phylogeny, 50 bootstrap replicates were used.
2.2. Comparative Analyses for Plastic-Degrading Gene Products in Rhodococcus Species
A list of 124 gene products known to be involved in the degradation of plastic polymers from different microorganisms was used as a reference for blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 27 May 2022) against the Rhodococcus dataset (Taxid 1827) of genomes. The outcomes of the analyses were defined as HIT sequences, i.e., all the output sequences presenting a degree of similarity with respect to the query sequence. The threshold is 5% and it is expressed as an e-value: if the statistical significance of the match (query against the random model) is greater than 5%, the match is not reported. The hit sequences of all results have been collected and annotated according to the following features in a single table: Gene ID, Plastic Group, Plastic, Plastic Enzyme and Rhodococcus Species, and Blast output (Score, e-value, and Identity).
Statistical analysis has been performed in order to count the number of normalized unique genes identified in Rhodococcus genomes grouped by different criteria (by plastic type, Rhodococcus species, enzymes, and others), as reported in the resulting plots. In addition, the score calculation has been obtained as a sum of Blast Score over all the groups of selected genes identified in Rhodococcus genomes and it indicates the plastic degradation potential of the diverse Rhodococcus species; manual curation was performed using the BLASTx tool of NCBI pipeline [39], Clustal Omega [37], Protein Data Bank [40], UniProt [41], and KEGG [42].
2.3. Rhodococcus HIT Sequence Tree Clusterization Analysis
On the basis of the bioinformatics analyses for the search for plastic-degrading gene products, the identified Rhodococcus genetic determinants were aligned by BLASTp tool [39] to determine sequence homology and retrieve the corresponding sequences. Reference proteins were selected among 124 gene products used as input data selected for the plastic-degrading ability from different microorganisms.
The sequences identified in Rhodococcus genomes belonging to multicopper oxidases, alkane 1-monooxygenases, and cytochrome P450 hydroxylases putatively involved in C−C backbone plastics (PE) degradation, and gene products putatively involved in plastics with heteroatoms in the main chain (PET and PU) degradation were aligned on Clustal Omega program using the MSA tool with default parameters [37], using the corresponding reference sequences from other microorganisms. For multiple sequence alignment with more than 4000 sequences, MAFFT software v. 7 with default parameters has been used [43]. NCBI Tree Viewer 1.19.2 was used to generate the corresponding tree clusterization images.
In order to generate subtree clusterization, MSA was used as input of cluster analysis inferred using the maximum likelihood (ML) method by MEGA (v. 10.2) software [38], with the same setting used for the unique input gene product sequences clusterization. The resulting groups allowed to define different trees, showing clades with putative functions identified by the UniProt database [41].
3. Results
3.1. Overview of Rhodococcus Genus Potential for Plastic Degradation
In order to explore the genetic determinants related to plastic degradation of Rhodococcus genomes, diverse gene products deriving from other microorganisms were retrieved from literature data, genomic and transcriptomic studies, and the PlasticDB database [36]. In total, 124 input gene products (Table S1) were listed for specific polymer degradation capabilities, including the degradation of plastics with C−C backbones, plastics with heteroatoms in the main chain, and polyesters. Among them, 75 sequences were unique and belong to Gram-negative bacteria, Gram-positive bacteria, and fungal strains. Since only a few studies investigate the genetic determinants of Rhodococcus strains for plastic degradation, other microorganisms were considered to investigate in-depth the plastic biodegrading potential of Rhodococcus genus. Within these microorganisms, 27 bacterial genera and 5 fungal genera belong, respectively, to four different bacterial phyla (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) and two fungal phyla (Aspergillus and Cryptococcus). In order to establish a clusterization framework of gene products used as input against Rhodococcus genomes, 75 unique input gene product sequences were considered. The generated tree reveals that these amino acid sequences for polymer degradation capabilities are divided into fifteen clades (Figure 1): cutinase I (clade I), polyester-hydrolase/PET-hydrolase (clade II), depolymerase (clade III), oxygenase (clade IV), multicopper oxidase (clade V), cytochrome P450 hydroxylase (clade VI), PLA-depolymerase (clade VII), esterase (clade VIII), PU esterase (clade IX), membrane transporters (clade X), carboxylesterase (clade XI), PHB-depolymerase I (clade XII), polyurethanase (clade XIII), cutinase II (clade XIV), and PHB-depolymerase II (clade XV).
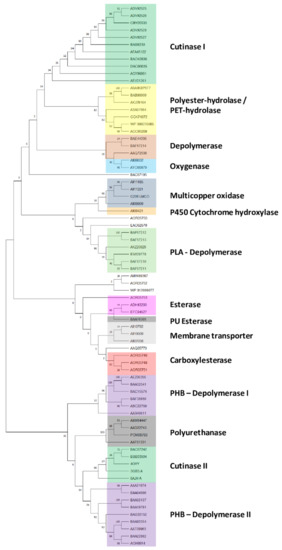
Figure 1.
Clusterization tree of unique input gene products belonging to different microorganisms (retrieved from PlasticDB database, literature data, omic studies, and functional analyses for the capability of degrading at least one type of plastic) divided into fifteen clades: cutinase I (clade I), polyester-hydrolase/PET-hydrolase (clade II), depolymerase (clade III), oxygenase (clade IV), multicopper oxidase (clade V), cytochrome P450 hydroxylase (clade VI), PLA-depolymerase (clade VII), esterase (clade VIII), PU esterase (clade IX), membrane transporters (clade X), carboxylesterase (clade XI), PHB-depolymerase I (clade XII), polyurethanase (clade XIII), cutinase II (clade XIV), and PHB-depolymerase II (clade XV).
These considered genes deriving from different microorganisms were reported for their involvement in the degradation of the following polymers, classified into three main categories: (i) C−C backbone plastics, including PE; (ii) heteroatomic backbone plastics including PET and PU; (iii) polyesters including PHB, PES, PPL, PCL, PLA, and co-polyesters containing aliphatic and aromatic components such as PHV, PHBV, PHPV, PBS, PBSA, PBAT, and Ecoflex.
For every polymer type, at least one plastic-degrading gene (retrieved from the PlasticDB database, literature data, omic studies, and functional analyses) was used for the comparative analysis against Rhodococcus genomes, and, in case of more than one gene from different microorganisms described for their ability to biodegrade a specific plastic polymer, multiple genes were selected to broaden the spectrum of phyla used in the overall comparison.
After aligning all 124-input degrading-genes against 669 Rhodococcus genomes, the total number of identified sequences was almost 50,000 (Table S2), and within them, only the unique sequences were further considered. They were classified into three main polymer categories as shown in Figure 2. The highest number of sequences was in the polyester group (16,528 sequences) followed by C−C backbone plastics and heteroatomic backbone plastics groups (respectively 7069 and 5340 sequences, respectively). This is in line with the highest number of input-degrading genes related to the polyester group and also their general higher biodegradability.
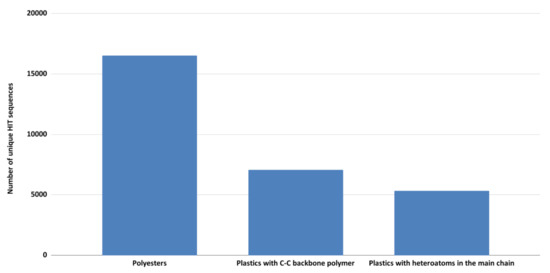
Figure 2.
Total number of unique HIT sequences, deriving from the comparison of 124-input degrading-genes against Rhodococcus genomes, were classified into three main polymer categories: polyesters, C−C backbone plastics, and heteroatomic backbone plastics.
Considering each polymer, the highest number of Rhodococcus DNA sequences encodes proteins putatively related to the degradation of PE, PBAT, PHB, PET, and PU; a few genes cover PHBV, PHV, PHPV, and PPL degradation (Figure 3).
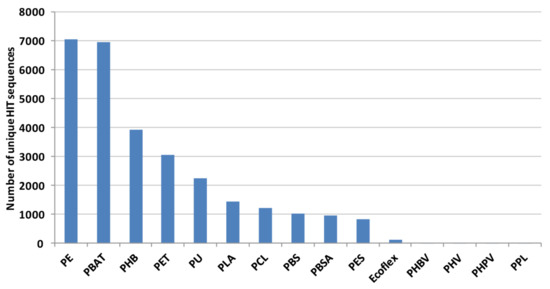
Figure 3.
Total number of unique HIT sequences identified in Rhodococcus genomes for each polymer material.
Overall, the results show the biodegradative potential of the Rhodococcus genus towards both synthetic and biodegradable plastics. Furthermore, these results evidenced a relation between gene products and the different species of Rhodococcus genus (Table S1).
Among Rhodococcus species, the highest number of genes putatively involved in plastic degradation belongs to R. equi, R. erythropolis, R. opacus, R. qingshengii, R. fascians, and R. rhodochrous (with 2643, 2464, 1786, 1777, 1640, and 1215 predicted genetic determinants, respectively). The lowest numbers of sequences predicted for plastic degradation are related to the following species: R. baikonurensis, R. artemisiae, R. nanhaiensis, R. sovatensis, R. canchipurensis, R. jialingiae, and R. percolatus (Figure 4). Nevertheless, these data should be evaluated considering the different number of genomes of species of the Rhodococcus genus.
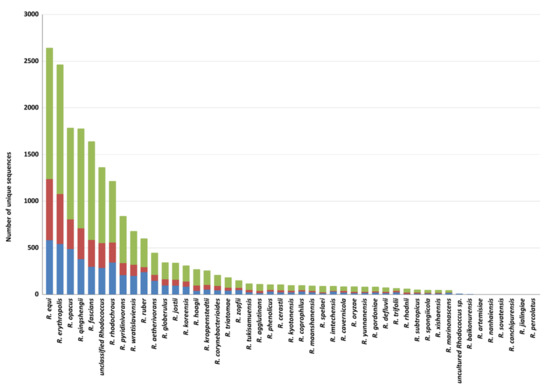
Figure 4.
Total number of unique HIT sequences identified in Rhodococcus species genomes for each polymer category: polyesters, heteroatomic backbone plastics, and C−C backbone plastics.
Moreover, these species present gene products mainly predicted for C−C backbone plastic degradation (Figure 5). Likewise, the category “uncultured Rhodococcus sp.” showed a few numbers of sequences predicted for plastic degradation and primarily against C-C backbone plastics. Among all the species, the average trend of unique HIT sequence number shows more than 50% of polyester degrading gene products, except for the species possessing the lowest number. The remaining 50% is mostly covered by PE-degrading gene products (Figure 5).
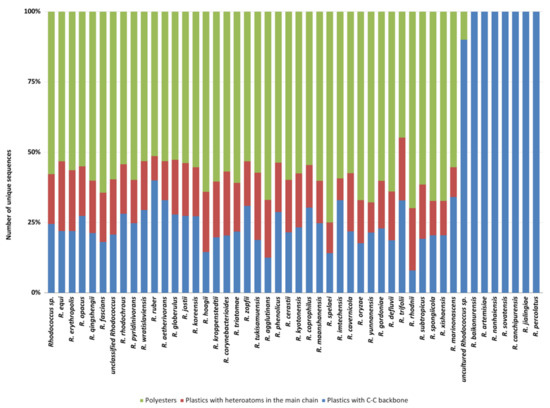
Figure 5.
Normalized number of unique HIT sequences identified in Rhodococcus species genomes for each polymer category: polyesters, heteroatomic backbone plastics, and C−C backbone plastics.
3.2. Rhodococcus Genus Degradative Potential towards C-C Backbone Polymer
The degradative potential of the Rhodococcus genus towards C-C backbone polymers was evaluated including only PE-degrading genes, since other polymers, such as polypropylene (PP), were correlated to similar genes associated with PE-degradation [44], or polystyrene (PS) and polyvinyl chloride (PVC), whose biodegradation is supported by few studies that did not focus on the molecular aspects and genetic determinants for these metabolisms [21]. The considered input genes encode alkane 1-monooxygenases, multicopper oxidases, cytochrome P450 hydroxylases, and transporters (possible membrane protein, integral membrane protein, and possible conserved integral membrane protein). The results of the bioinformatic analysis are shown in Figure 6 where the number of unique HIT sequences identified in Rhodococcus genomes is reported for different enzyme categories putatively related to C−C backbone plastic degradation. Figure 6 shows that the highest number of unique HIT sequences identified in Rhodococcus species belongs to the cytochrome P450 hydroxylase group (4864 sequences).
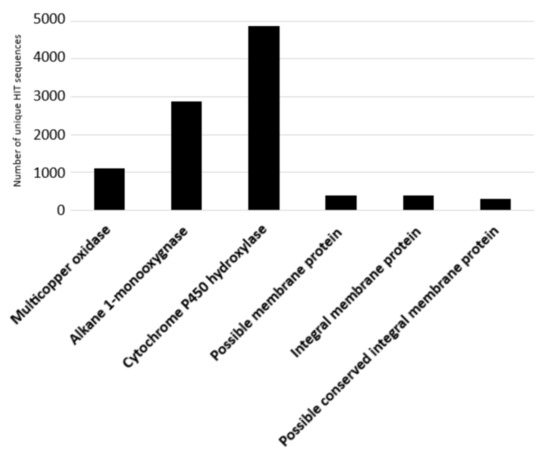
Figure 6.
Number of unique HIT sequences identified in Rhodococcus genomes for different enzyme categories putatively related to C−C backbone plastic degradation.
The clusterization analysis of all identified HIT sequences was performed by comparing multicopper oxidases, alkane monooxygenases, or cytochrome P450 hydroxylases, which were the most abundant sequences for their potential degradative role. They were clusterized into three different trees including only the unique HIT sequences of Rhodococcus that were 816, 940, and 1719 for the multicopper oxidase, alkane monooxygenase, and cytochrome P450 hydroxylase clusterization tree, respectively (Figures S1–S3). For each tree, the HIT sequences with the highest similarity with respect to the reference sequences were considered for the subtree generation. The considered reference sequences for the multicopper oxidase category were AII08809, AII11185, and AII11221, for alkane monooxygenase category it was AII08632, and for cytochrome P450 hydroxylase category it was AII08421. The first subtree showed mostly copper and multicopper oxidases (MmcO) belonging to the following species: Rhodococcus sp., R. opacus, R. wratislaviensis, R. marinonascens, R. imtechensis, R. jostii, and R. koreensis (Figure 7). The reference sequences clusterized into three different clades: AII08809 (multicopper oxidase I), grouped with a multicopper oxidase of R. wratislaviensis (WP_037231000) and a FtsP/CotA-like multicopper oxidase with cupredoxin domain of R. wratislaviensis (WP_112302511); AII11185 (multicopper oxidase II), clusterized with the clade of AII11221 reference; and AII11221 (multicopper oxidase III), grouped with two multicopper oxidases of Rhodococcus sp. (WP_095862380 and WP_133987587).

Figure 7.
Subtree clusterization of HIT sequences identified by oxidase input genes putatively involved in C-C backbone plastic degradation. Marked boxes evidence the reference input genes.
Considering the alkane monooxygenase subtree, eight clades can be distinguished comprising only alkane 1-monooxygenase (AII08632), belonging to the same species observed in the previous subtree (Figure 7) with the addition of R. rhodochrous and R. aetherivorans species (Figure 8). The closest sequences with respect to the reference input AII08632 belong to the R. wratislaviensis and R. jostii species.

Figure 8.
Subtree clusterization of HIT sequences identified by alkane monooxygenase input genes putatively involved in C-C backbone plastic degradation. Marked box evidences the reference input gene.
Cytochrome P450 hydroxylase subtree showed sequences belonging to Rhodococcus sp., R. opacus, R. wratislaviensis, R. imtechensis, R. jostii, and R. koreensis, which clusterized into eight clades (Figure 9). The reference sequence AII08421 principally clusterized with cytochrome P450 of R. wratislaviensis (WP_112300243) and the closest clade include cytochrome P450 of all the species mentioned in the previous subtrees except R. jostii (WP_012691738, WP_009475556, WP_144285016, WP_124393366, WP_095862123, WP_072937090, WP_185950062, WP_007299636, WP_087560732, WP_218267768, and WP_206009113).
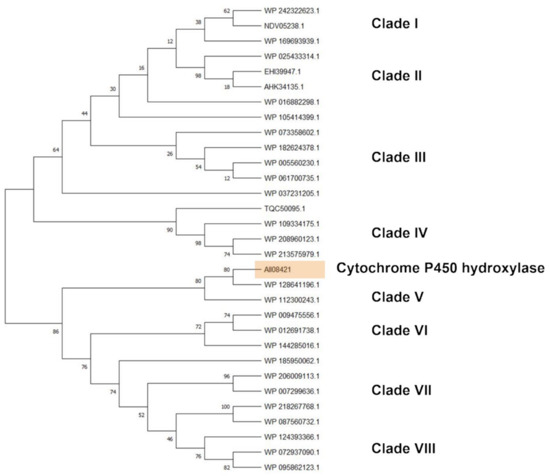
Figure 9.
Subtree clusterization of HIT sequences identified by cytochrome P450 hydroxylase input gene putatively involved in C-C backbone plastic degradation. Marked box evidences the reference input gene.
3.3. Degradative Potential of the Rhodococcus Genus towards the Degradation of Plastics with Heteroatoms in the Main Chain
Rhodococcus degrading potential for plastics with heteroatoms in the main chain was evaluated by selecting PET and PU degrading genes as references. The considered input genes encode cutinase, PU esterase, PET-hydrolase, triacylglycerol-lipase, polyester-hydrolase, polyurethanase, serine-hydrolase, and para-nitrobenzylesterase. The results of the comparison of Rhodococcus genomes against these reference sequences are shown in Figure 10, where the number of unique HIT sequences identified in Rhodococcus genomes is reported for different enzyme categories putatively related to heteroatomic backbone plastic degradation. The highest number of unique HIT sequences identified in Rhodococcus species belongs to PU esterase and para-nitrobenzylesterase groups (3205 and 2561 unique sequences, respectively).
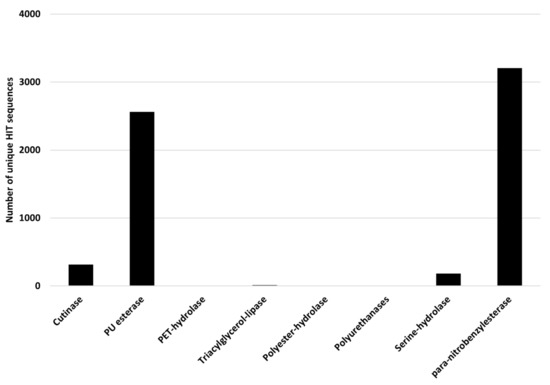
Figure 10.
Number of unique HIT sequences identified in Rhodococcus genomes for different enzyme categories putatively related to heteroatomic backbone plastic degradation.
In order to evaluate the potential of the diverse enzymes for PET and PU degradation, clusterization analyses of respectively 2938 and 2118 unique HIT sequences identified in Rhodococcus genomes for the two different polymers revealed two clusterization trees against the reference gene inputs (Figure S4 and Figure S5). Considering PET-degrading enzymes used as input sequences, a subtree clusterization of 110 HIT sequences was performed (Figure S6). Gene products classified as cutinase, PET-hydrolase, triacylglycerol-lipase, polyester-hydrolase, and serine-hydrolase putatively involved in PET degradation were included. para-Nitrobenzylesterases were excluded for the high number of HIT sequences and because they were clusterized on their own.
For PU-degrading genes, a PU subtree was generated. The most similar HIT sequences with respect to the reference sequences were considered for subtree generation (Figure 11). The figure shows the main clusters obtained. Sequences clustering closer to BAA76305 (PU esterase) reference protein were mostly deriving from Rhodococcus sp., R. opacus, R. wratislaviensis, R. jostii, R. aetherivorans, R. hoagii, R. erythropolis, and R. qingshengii. The reference sequence WP_088276085 (Polyester hydrolase) principally clusterized with Rhodococcus sp., R. rhodochrous, R. zopfii, R. triatomae, and R. fascians.
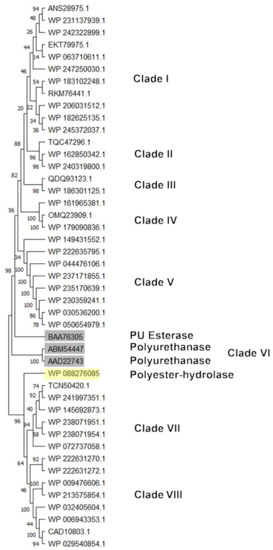
Figure 11.
Subtree clusterization of HIT sequences identified by PU esterase and polyurethanase input gene putatively involved in heteroatomic backbone plastic degradation. Marked boxes evidence the reference input genes.
3.4. Rhodococcus Genus Degradative Potential towards Polyesters
Polyesters are often considered biodegradable polymers [27]; thus, Rhodococcus degrading potential has been assessed for all genes related to PBAT, Ecoflex, PHB, PHA, PLA, PCL, PPL, PBS, PBSA, PES, PHBV, PHPV, and PHV degradation. The considered input genes encode PBS depolymerase, PHB-depolymerase, cutinase, esterase, PET-hydrolase, PLA-depolymerase, lipase, MCL-PHA-Depolymerase, polyesterase, hydrolase, and carboxylesterase. Figure 12 shows the results of the bioinformatic analysis where the number of unique HIT sequences identified in Rhodococcus genomes is reported for different enzyme categories putatively related to polyester degradation. The three enzymatic classes with the highest number of unique HIT sequences of Rhodococcus species belong to carboxylesterase (7007 sequences), PLA-depolymerase (5302 sequences), and PHB-depolymerase (4328 sequences) groups.
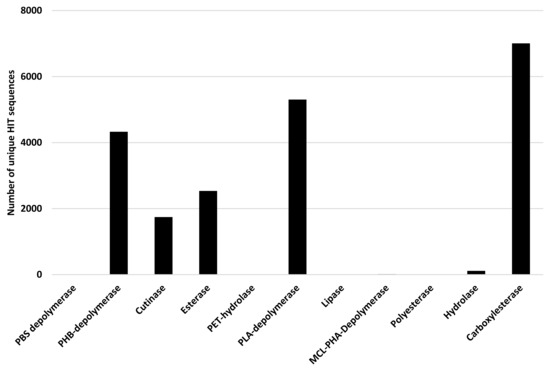
Figure 12.
Number of unique HIT sequences identified in Rhodococcus genomes for different enzyme categories putatively related to polyester degradation.
In this case, a clusterization tree was also generated, showing that all Rhodococcus putative polyester-degrading sequences were clusterized into a single tree against the reference gene inputs (Figure S7).
3.5. Dominant Rhodococcus Species and Main Enzyme Classes for Plastic Polymer Degradation
In order to predict the role in plastic degradation of different Rhodococcus species, the species presenting the most abundant number of unique sequences with a putative biodegradative potential (Figure 4) were used for score calculations. They were mostly isolated from diverse environmental niches, such as soils, contaminated environments, and water systems, some of them are pathogenic bacteria, and others are often detected in human infections. Since seventeen Rhodococcus species showed the highest number of sequences (although the different number of genomes for each Rhodococcus species), they were used for the following analyses, and they were: R. hoagii, R. ruber, R. pyridinivorans, R. qingshengii, R. opacus, R. erythropolis, R. aetherivorans, R. triatomae, R. fascians, R. jostii, R. rhodochrous, R. koreensis, R. equi, R. corynebacterioides, R. kroppenstedtii, R. wratislaviensis, and R. globerulus. The score was based on Rhodococcus sequence similarity with respect to the diverse gene targets chosen for the degradation of three groups of polymers (C−C backbone plastics, heteroatomic backbone plastics, and polyesters) and the sum of the scores resulted in Figure 13. It is noteworthy that among the 45 analysed Rhodococcus species, three out of four species often associated with pathogenic strains (R. hoagii, R. equi, and R. fascians) showed a high number of degrading genes and additionally high score levels. The highest scores are related to C−C backbone plastic-degrading sequences; in particular, R. hoagii showed the highest number of these enzymes despite a higher number of HIT sequences retrieved in R. equi.
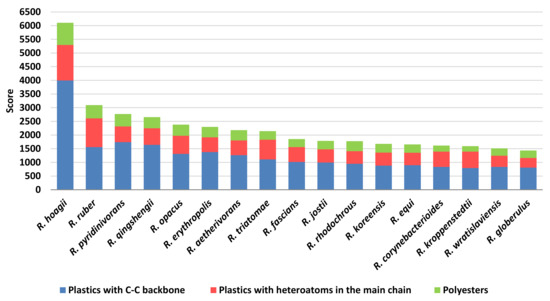
Figure 13.
Score values for the most prominent Rhodococcus species for each class of diverse polymer materials.
Among human infectious-associated species, R. triatomae presented the highest scores, especially with respect to C−C backbone plastic gene products.
Considering the environmental-associated isolated species, they presented average similar score levels, with some exceptions: R. ruber seemed to present the highest score value for PET and PU gene products despite the low number of identified HIT sequences with respect to other environmental-associated species; R. pyridinivorans and R. qingshengii revealed the highest scores towards PE degrading genes, and R. wratislaviensis showed the lowest scores towards polyester degrading genes. In this respect, the lowest score levels were detected for all the species against polyesters which were the highest polymer category in terms of the number of identified sequences.
Analyzing the score of the different enzymes identified for the biodegradation of synthetic and biodegradable plastics, multicopper oxidases, alkane 1-monooxygenases, and cytochrome P450 hydroxylases were the enzymes of major interest for C−C backbone plastic group (Figure 14). Alkane 1-monooxygenase class possesses the highest score value for R. erythropolis and R. qingshengii; multicopper oxidase class is prominent in R. erythropolis, R. qingshengii, R. opacus, and R. equi; cytochrome P450 hydroxylase category presented the highest score values for the R. opacus and R. erythropolis species. Worthy of being specified is the apparent contradiction for R. hoagii score values for C−C backbone plastic-degrading genes. Probably, the considerable score levels are primarily due to other enzymes considered, such as transporters and membrane proteins.
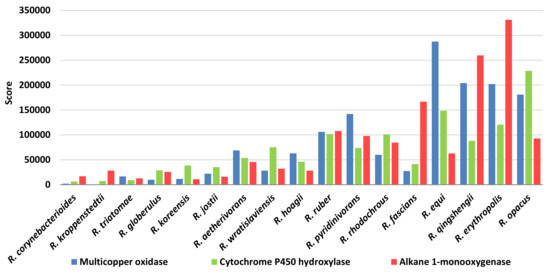
Figure 14.
Score values for the most prominent Rhodococcus species for the main enzymatic categories involved in C-C backbone plastic degradation.
The scores of the other two plastic-degrading enzyme categories related to heteroatomic backbone plastics and polyesters were simultaneously analyzed because of the overlapping capabilities of these enzymes (Figure 15). Above all, carboxylesterase showed the highest scores and, in particular, R. erythropolis, R. qingshengii, R. opacus, and R. equi were the most prominent species. A similar score-species pattern was observed for para-nitrobenzylesterase, PU esterases, and esterases, with the exclusion of R. qingshengii. PLA-depolymerase presented the highest score values for R. fascians species. The scores for PHB-depolymerase were mostly the highest in R. erythropolis, R. qingshengii, R. opacus, R. rhodochrous, and R. pyridinivorans. On the other hand, cutinases presented average low score values; the highest was shown for R. erythropolis, R. qingshengii, and R. fascians species.
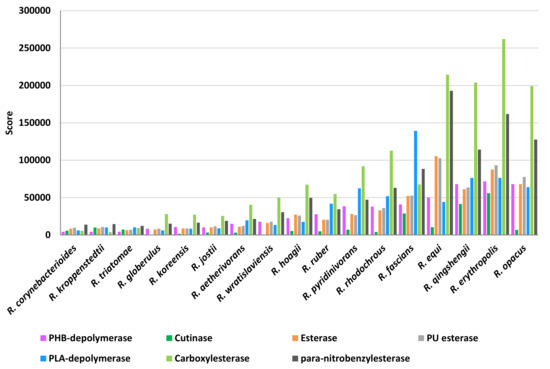
Figure 15.
Score values for the most prominent Rhodococcus species for the main enzymatic categories involved in PET, PU, and polyesters degradation.
4. Discussion
Increasing plastic production and consumption raised the consequent issue of its disposal management, which is becoming a compelling forefront [23,45]. For this reason, biodegradation of synthetic plastic has been increasingly studied, including the feasibility of bacteria/enzymes biodegradative processes [21,26,27,46,47]. In this context, among bacterial genera with extraordinary biodegradative capabilities, Rhodococcus genus was selected for its remarkable metabolic capabilities and strong persistence under stress conditions [14]. For the first time, in this work, Rhodococcus genomes were analyzed to predict genetic determinants with degrading capabilities towards synthetic and biodegradable plastics categorized into three classes: (i) C−C backbone plastics, (ii) heteroatomic backbone plastics, and (iii) polyesters. Comparative bioinformatics analysis was based on the selected gene products from several microorganisms related to plastic degradation. Their comparison against 669 Rhodococcus genomes showed that around 57% of unique HIT sequences were covered by polyester-like degrading enzymes (mostly PBAT and PHB), while 24% and 18% included C−C backbone plastics (PE) and heteroatomic backbone plastics groups (PET and PU), respectively. Preliminary analyses revealed amino acid sequences possessing peptide signals for extracellular secretion. This suggests their putative key role in the plastic degradative processes [17]. Remarkably, these results reflect the Rhodococcus genus biodegradative potential towards biodegradable plastics, especially towards PBAT and PHB. These outcomes are also evidenced by analyzing the single Rhodococcus species with the exclusion of the ones with the lowest number of HIT sequences.
On the other hand, score values highlighted PE-degrading sequences as the most probably involved especially for R. pyridinivorans, R. qingshengii, and R. hoagii genera [48,49], which have the highest PE-degrading potential. With respect to C−C backbone plastic gene products, R. triatomae presented the highest score if we consider only human-associated species, as mostly reported in the literature [50,51]. This high similarity for PE-degrading enzymes is probably due to the Rhodococcus strain origin of the majority of input sequences related to this class of polymers [17,31]. The association of the diverse species of Rhodococcus genus to diverse niches could be a useful hint to predict the most promising ecological niches, where rhodococci could be prominent in plastic degradation.
The subtree clustering analyses for PE-plastic gene products showed that the majority of homologous enzymes with respect to multicopper oxidases, alkane monooxygenases, or cytochrome P450 hydroxylases of diverse Rhodococcus species belong to the following species: R. opacus, R. wratislaviensis, R. marinonascens, R. imtechensis, R. jostii, R. koreensis, R. rhodochrous, and R. aetherivorans. In this regard, PE is considered one of the most used and wasted plastic due to its properties of non-degradability and durability [52], and only a few bacterial strains possess the ability to degrade, at least partially, diverse types of PE [17,29,30,31]. For this reason, these indications could be valuable for further investigations of the most interesting species; PE biodegradation could be performed by oxidation mediated by multicopper oxidases, probably through extracellular oxidation due to the preliminarily identified peptide signal sequences for extracellular secretion, and then smaller PE fragments can be internalized by transporters and other oxidative systems, including alkane monooxygenases or cytochrome P450 hydroxylases, could participate in further steps of intracellular PE-smaller fragment oxidation [18].
Rhodococcus degrading potential for plastics with heteroatoms in the main chain showed that the highest number of identified unique HIT sequences belong to PU esterase and para-nitrobenzylesterase and with a lower extent to cutinase. These enzymatic classes could putatively cover PET and PU biodegradation. Indeed, enzymatic hydrolysis is the route for the degradation of both these aromatic polyesters [21,34,53].
Although sequence clustering showed that PU degrading enzymes belonging to R. opacus, R. wratislaviensis, R. jostii, R. aetherivorans, R. hoagii, R. erythropolis, R. qingshengii, R. rhodochrous, R. zopfii, R. triatomae, and R. fascians were the closest to the reference, R. ruber presented the highest score value for both PET and PU gene products (despite the low number of identified HIT sequences with respect to other environmental-associated species).
Some categories of polyesters are considered biodegradable [27], comprising PBAT and PHB, which represent the polymers associated with the highest number of unique HIT sequences. In literature, the enzymes reported for the ability to degrade polyester-based plastics include polyester depolymerases, esterases, carboxylesterase, cutinase-like enzymes, and lipases [26,27]. In line with these results, the three main enzymatic classes predicted for their involvement in polyester biodegradation of Rhodococcus species belong to carboxylesterase (42% of total polyester predicted degrading enzymes), PLA-depolymerase (32%), and PHB-depolymerase (26%). Consistently, carboxylesterase, PLA-depolymerase, and PHB-depolymerase showed the highest scores for certain species. On the other hand, cutinases are present in low abundance among unique HIT sequences of Rhodococcus genomes, showing an average low score value.
Overall sequence comparison and score calculation were useful tools to weigh different Rhodococcus species and different genes putatively involved in the metabolism of diverse plastic polymers. In addition, tree clusterization allowed us to comprehend and distinguish the putative role of the new genes identified for the diverse Rhodococcus species. Notably, the combination of the similarity scores of enzymes from diverse microorganisms against Rhodococcus genomes with the clustering analysis poses the basis for evolutional relationships studies, and in some cases, can suggest events of horizontal gene transfer of Rhodococcus identified sequences.
The indications deriving from this study could be useful for future biotechnological applications at two levels: Rhodococcus strains can be exploited in plastic biodegradative processes and even some enzymatic functions could be further studied for catalytic reactions from a biotechnological point of view.
5. Conclusions
Since the current prominent plastic waste management problem at the global level, this research study aimed to shed light on Rhodococcus genus traits for its well-known remarkable biodegradative capacities. This work contributes to gain novel knowledge on the biodegradative potential of Rhodococcus species towards both synthetic and biodegradable polymers mediated by specific enzymatic classes. New insights on primary enzymatic classes involved in most produced and utilized plastic materials were shown. The sequence predictions derived from this study can be promising as an explorative survey to base the further sequence utilization as a reference for plastic contamination biomonitoring and the consequent plastic removal with biotechnological applications.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10091846/s1, Figure S1. Clusterization tree of HIT sequences identified by oxidase input genes from different microorganisms putatively involved in C-C backbone plastic degradation. Figure S2. Clusterization tree of HIT sequences identified by alkane monooxygenase input genes from different microorganisms putatively involved in C-C backbone plastic degradation. Figure S3. Clusterization tree of HIT sequences identified by cytochrome P450 hydroxylase input genes from different microorganisms putatively involved in C-C backbone plastic degradation. Figure S4. Clusterization tree of HIT sequences identified by input genes from different microorganisms putatively involved in PET degradation. Figure S5. Clusterization tree of HIT sequences identified by input genes from different microorganisms putatively involved in PU degradation. Figure S6. Subtree clusterization of HIT sequences identified by cutinase, PET-hydrolase, triacylglycerol-lipase, polyester-hydrolase, and serine-hydrolase input genes from different microorganisms putatively involved in PET degradation. Figure S7. Clusterization tree of HIT sequences identified by input genes from different microorganisms putatively involved in polyester degradation. Table S1. Total list of 124 genes used as input for the diverse polymer degradation capabilities deriving from different microorganisms. Table S2. Total list of HIT sequences identified by alignment against Rhodococcus genome sequences.
Author Contributions
Conceptualization, J.Z. and P.D.G.; Data curation, D.V.; Formal analysis, A.O.; Investigation, J.Z. and A.O.; Methodology, A.O.; Resources, P.D.G.; Software, D.V. and A.O.; Supervision, P.D.G.; Validation, J.Z. and A.O.; Visualization, J.Z. and A.O.; Writing—original draft, J.Z.; Writing—review and editing, J.Z. and P.D.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by 2021-ATE-0460 from the University of Milano-Bicocca.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated during this study are included in this article.
Conflicts of Interest
All other authors declare no conflict of interest.
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Saez, J.M.; Costa, J.S.D.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Tyumina, E.A.; Bazhutin, G.A.; Vikhareva, E.V.; Selyaninov, A.A.; Ivshina, I.B. Diclofenac as a factor in the change of Rhodococcus metabolism. IOP Conf. Ser. Mater. Sci. Eng. 2019, 487, 012027. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V.; Tyumina, E.A. Responses to Ecopollutants and pathogenization risks of saprotrophic Rhodococcus species. Pathogens 2021, 10, 974. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Di Canito, A.; Zampolli, J.; Orro, A.; D’Ursi, P.; Milanesi, L.; Sello, G.; Steinbüchel, A.; Di Gennaro, P. Genome-based analysis for the identification of genes involved in o-xylene degradation in Rhodococcus opacus R7. BMC Genom. 2018, 19, 587. [Google Scholar] [CrossRef]
- Zampolli, J.; Zeaiter, Z.; Di Canito, A.; Di Gennaro, P. Genome analysis and-omics approaches provide new insights into the biodegradation potential of Rhodococcus. Appl. Microbiol. Biotechnol. 2019, 103, 1069–1080. [Google Scholar] [CrossRef]
- Alvarez, H.M. Biology of Rhodococcus, 2nd ed.; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Zampolli, J.; Di Canito, A.; Manconi, A.; Milanesi, L.; Di Gennaro, P.; Orro, A. Transcriptomic analysis of Rhodococcus opacus R7 grown on o-xylene by RNA-Seq. Front. Microbiol. 2020, 11, 1808. [Google Scholar] [CrossRef]
- De Carvalho, C.C.; Costa, S.S.; Fernandes, P.; Couto, I.; Viveiros, M. Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front. Physiol. 2014, 5, 133. [Google Scholar] [CrossRef]
- Laczi, K.; Kis, Á.; Horváth, B.; Maróti, G.; Hegedüs, B.; Perei, K.; Rákhely, G. Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons. Appl. Microbiol. Biotechnol. 2015, 99, 9745–9759. [Google Scholar] [CrossRef] [Green Version]
- Orro, A.; Cappelletti, M.; D’Ursi, P.; Milanesi, L.; Di Canito, A.; Zampolli, J.; Collina, E.; Decorosi, F.; Viti, C.; Fedi, S.; et al. Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: Genetic determinants and metabolic abilities with environmental relevance. PLoS ONE 2015, 10, e0139467. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, M.; Fedi, S.; Zampolli, J.; Di Cainta, A.; D’Ursi, P.; Orro, A.; Viti, C.; Milanesi, L.; Zannoni, D.; Di Gennaro, P. Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Res. Microbiol. 2016, 167, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, M.; Zampolli, J.; Di Gennaro, P.; Zannoni, D. Genomics of Rhodococcus. In Biology of Rhodococcus, 2nd ed.; Microbiology monographs; Alvarez, H.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 16, pp. 23–60. [Google Scholar]
- Bell, K.S.; Philp, J.C.; Aw, D.W.J.; Christofi, N. The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.J.; Kulakov, L.A.; Allen, C.C. Biodegradation by members of the genus Rhodococcus: Biochemistry, physiology, and genetic adaptation. Adv. Appl. Microbiol. 2006, 59, 1–29. [Google Scholar] [PubMed]
- Zampolli, J.; Orro, A.; Manconi, A.; Ami, D.; Natalello, A.; Di Gennaro, P. Transcriptomic analysis of Rhodococcus opacus R7 grown on polyethylene by RNA-seq. Sci. Rep. 2021, 11, 21311. [Google Scholar] [CrossRef]
- Gravouil, K.; Ferru-Clément, R.; Colas, S.; Helye, R.; Kadri, L.; Bourdeau, L.; Bouziane, M.; Mercier, A.; Ferreira, T. Transcriptomics and lipidomics of the environmental strain Rhodococcus ruber point out consumption pathways and potential metabolic bottlenecks for polyethylene degradation. Environ. Sci. Technol. 2017, 51, 5172–5181. [Google Scholar] [CrossRef]
- Basik, A.A.; Sanglier, J.J.; Yeo, C.T.; Sudesh, K. Microbial degradation of rubber: Actinobacteria. Polymers 2021, 13, 1989. [Google Scholar] [CrossRef]
- Plastics Europe. Plastics-the Facts, 2019: An Analysis of European Plastics Production, Demand and waste Data Plastics Europe; Plastics Europe: Brussels, Belgium, 2019. [Google Scholar]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef]
- Yeung, C.W.; Teo, J.Y.; Loh, X.J.; Lim, J.Y. Polyolefins and polystyrene as chemical resources for a sustainable future: Challenges, advances, and prospects. ACS Mater. Lett. 2021, 3, 1660–1676. [Google Scholar] [CrossRef]
- Matjašič, T.; Simčič, T.; Medvešček, N.; Bajt, O.; Dreo, T.; Mori, N. Critical evaluation of biodegradation studies on synthetic plastics through a systematic literature review. Sci. Total. Environ. 2021, 752, 141959. [Google Scholar] [CrossRef]
- Lear, G.; Kingsbury, J.M.; Franchini, S.; Gambarini, V.; Maday, S.D.M.; Wallbank, J.A.; Pantos, O. Plastics and the microbiome: Impacts and solutions. Environ. Microbiome 2021, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total. Environ. 2021, 759, 143536. [Google Scholar] [CrossRef] [PubMed]
- Satti, S.M.; Shah, A.A. Polyester-based biodegradable plastics: An approach towards sustainable development. Lett. Appl. Microbiol. 2020, 70, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, A.K.; Mirończuk, A.M.; García-Martín, A.; Saborido, A.; de la Mata, I.; Arroyo, M. Biochemical properties and biotechnological applications of microbial enzymes involved in the degradation of polyester-type plastics. Biochim. Biophys. Acta. Proteins Proteom. 2020, 1868, 140315. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, D.; Wei, N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022, 40, 22–37. [Google Scholar] [CrossRef]
- Yoon, M.G.; Jeon, H.J.; Kim, M.N. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J. Bioremed. Biodegrad. 2012, 3, 1–8. [Google Scholar]
- Jeon, H.J.; Kim, M.N. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme–laccase–in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current status and application aspects. Acs. Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Howard, G.T.; Mackie, R.I.; Cann, I.K.O.; Ohene-Adjei, S.; Aboudehen, K.S.; Duos, B.G.; Childers, G.W. Effect of insertional mutations in the pueA and pueB genes encoding two polyurethanases in Pseudomonas chlororaphis contained within a gene cluster. J. Appl. Microbiol. 2007, 103, 2074–2083. [Google Scholar] [CrossRef]
- Bollinger, A.; Thies, S.; Knieps-Grünhagen, E.; Gertzen, C.; Kobus, S.; Höppner, A.; Jaeger, K.E. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri—Structural and functional insights. Front. Microbiol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [PubMed]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta. Crystallogr. D. Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Rana, A.K.; Thakur, M.K.; Saini, A.K.; Mokhta, S.K.; Moradi, O.; Rydzkowski, T.; Alsanie, W.F.; Wang, Q.; Grammatikos, S.; Thakur, V.K. Recent developments in microbial degradation of polypropylene: Integrated approaches towards a sustainable environment. Sci. Total Environ. 2022, 826, 154056. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Koutra, E.; Kornaros, M.; El-Sheekh, M.; Abdelkarim, E.A.; Zhu, D.; Sun, J. Degradation of conventional plastic wastes in the environment: A review on current status of knowledge and future perspectives of disposal. Sci. Total Environ. 2021, 771, 44719. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Elsamahy, T.; Al-Tohamy, R.; Zhu, D.; Mahmoud, Y.A.; Koutra, E.; Metwally, M.A.; Kornaros, M.; Sun, J. Plastic wastes biodegradation: Mechanisms, challenges and future prospects. Sci. Total Environ. 2021, 780, 146590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhaoa, Y.; Wang, D.; Yan, M.; Zhang, J.; Zhang, P.; Ding, T.; Chen, L.; Chen, C. Current technologies for plastic waste treatment: A review. J. Clean. Prod. 2021, 282, 124523. [Google Scholar] [CrossRef]
- Kämpfer, P.; Dott, W.; Martin, K.; Glaeser, S.P. Rhodococcus defluvii sp. nov., isolated from wastewater of a bioreactor and formal proposal to reclassify Corynebacterium hoagii and Rhodococcus equi as Rhodococcus hoagii comb. nov. Int. J. Syst. Bacteriol. 2014, 64, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Sangal, V.; Jones, A.L.; Sutcliffe, I.C. Charting stormy waters: A commentary on the nomenclature of the equine pathogen variously named Prescottella equi, Rhodococcus equi and Rhodococcus hoagii. Equine Vet. J. 2015, 47, 508–509. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A.F. Rhodococcus triatomae sp. nov., isolated from a blood-sucking bug. Int. J. Syst. Evol. Microbiol. 2005, 55, 1575–1579. [Google Scholar] [CrossRef]
- Majidzadeh, M.; Fatahi-Bafghi, M. Current taxonomy of Rhodococcus species and their role in infections. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2045–2062. [Google Scholar] [CrossRef]
- Scarascia-Mugnozza, G.; Sica, C.; Russo, G. Plastic materials in European agriculture: Actual use and perspectives. J. Agric. Eng. 2011, 3, 15–28. [Google Scholar] [CrossRef]
- Damayanti; Wu, H.-S. Strategic Possibility Routes of Recycled PET. Polymers 2021, 13, 1475. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).